Category Archives: Articles
CRISPR-Cas9 脱靶检测方法的总结
Link: https://www.cd-genomics.com/summary-of-crispr-cas9-off-target-detection-methods.html
快速概览
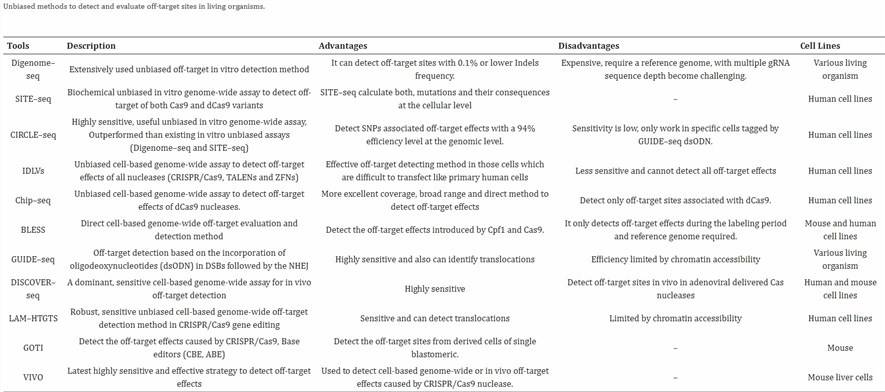
01 CRISPR-Cas9 脱靶检测的测序方法
02 CRISPR-Cas9 脱靶检测的计算模拟方法
基因组编辑技术处于当前生命科学研究的前沿。然而,要成功将其转化为临床应用,准确检测脱靶效应至关重要。正确评估和减轻脱靶效应是一个紧迫的问题。CRISPR-Cas9 基因编辑技术中的脱靶效应问题一直是长期关注的焦点。检测 CRISPR-Cas9 脱靶效应的主要方法有两种:计算模拟方法和测序方法 [1]。
CRISPR-Cas9 脱靶检测的测序方法
全基因组测序 (WGS)
全基因组测序 (WGS) 在评估 Cas9 诱导的脱靶突变方面与最小的评估偏差相关联。目前,通常采用 30-60x 的测序深度。然而,WGS 成本较高,较低的测序深度可能仅测序少量克隆,可能错过大多数低频脱靶事件。对于体内分析,WGS 仍是首选方法,但直接捕获双链断裂 (DSB) 的技术,如 BLESS,可能比 WGS 提供更优越的能力。这是因为 WGS 还会捕获细胞/动物模型之间自然发生的变异,这可能使分析复杂化并导致误导性结果。
在 WGS 分析中,选择适当的对照组尤为重要。为了在 WGS 中最小化假阳性和假阴性,通常需要设置多个组,这既费力又昂贵。
ChIP-Seq
Cas 核酸酶,一类蛋白质,可以通过抗体拉下后进行测序,称为 ChIP-Seq。ChIP-Seq 常用于检测与 dCas9 相关的脱靶效应。与常规 Cas9 相比,常规 Cas9 会切断目标序列,导致其与 DNA 序列解离并结合不稳定,而 dCas9 因核酸酶活性受损,仍能通过 gRNA 引导结合目标序列。因此,dCas9 常用于 CRISPR 激活 (CRISPRa) 以增强基因表达,而非用于基因敲除。
DISCOVER-seq(也称为 MRE11 ChIP-Seq)是 CRISPR-Cas9 脱靶测序的最新开发方法。该方法利用内源性 DNA 修复因子 MRE11 的招募来识别整个基因组上 Cas 诱导的双链断裂。在 DNA 修复过程中,MRE11 精确地招募到 DSB 发生部位。因此,通过拉下 MRE11 结合位点并进行高通量测序,该方法允许同时检查靶向和脱靶发生。重要的是,DISCOVER-seq 适用于体内和体外样本。
Cas 核酸酶切割位点和 DSB 位点富集测序
依赖富集 Cas 核酸酶结合或切割区域的测序方法通常被称为无偏检测方法。根据这一原理,已设计了富集方法,包括:
(1) 抗体拉下:
如 ChIP-Seq,使用抗体捕获目标区域。
(2) Cas 核酸酶切割位点富集:
Digenome-Seq,简称体外 Cas9 消化全基因组测序。Cas9 切割后留下 5′ 磷酸基团,便于接头连接以进行 PCR 扩增和后续 NGS 测序。在 Digenome-Seq 中,纯化的基因组 DNA (gDNA) 在体外用 Cas9 处理,随后进行接头连接和全面全基因组测序。此方法适用于体外样本。
(3) DNA 双链断裂区域 (DSB) 富集 – DSB 捕获:
在 BLESS-Seq 中,使用直接生物素亲和富集捕获 CRISPR-Cas9 引导的 DSB 片段,随后在接头连接后进行测序。生物素亲和富集专门捕获正在发生的 DSBs,已修复的 DSBs 未被考虑,因此被标记为“脱靶快照”。此方法常用于细胞和组织样本。
在 IDLV-Seq 中,整合缺陷慢病毒载体 (IDLV) 可作为 PCR 锚点来扩增通过非同源末端连接 (NHEJ) 生成的 DSB 位点,从而检测脱靶位点。然而,此方法的效率依赖于 IDLV 通过 NHEJ 整合到 DSB 的能力。此外,由于慢病毒的随机整合特性,可能出现假阳性。
HTGST 适用于检测 CRISPR-Cas9 诱导的染色体易位,但需注意的是,CRISPR-Cas9 诱导的脱靶事件主要导致插入缺失 (indels),相对较少见。此外,此方法仍受染色质可及性限制。
与富集核酸酶切割位点的方法相比,DSB 捕获方法更具生理相关性。然而,由于大多数 DSB 持续时间极短,它们的效率可能较低。IDLV 的体内标记和原位接头连接可能受切割位点附近局部染色质和序列组成的影响。因此,捕获方法可能存在偏差。
测序方法总结
多种测序方法可用,难以对每种方法进行详尽描述。以下表格提供了当前可访问的最全面总结。基于先前提供的信息,难以确定最佳方法。选择最合适的测序技术应根据实验的具体需求和研究团队的财务资源指导。
| 方法 | 适用场景 | 优点 | 局限性 |
|---|---|---|---|
| WGS | 体内分析 | 全面覆盖 | 成本高,易受自然变异干扰 |
| ChIP-Seq | dCas9 脱靶检测 | 针对性强 | 仅适用于特定蛋白结合 |
| DISCOVER-seq | 体内/体外 | 同时检测靶/脱靶 | 需要高分辨率测序 |
| Digenome-Seq | 体外样本 | 无偏检测 | 仅限体外应用 |
| BLESS-Seq | 细胞/组织 | 捕获活跃 DSB | 效率低,短时效性 |
| IDLV-Seq | NHEJ 检测 | 特定性强 | 可能有假阳性 |
| HTGST | 染色体易位 | 检测易位 | 受染色质限制 |
您可能感兴趣:
-
CRISPR 测序
-
CRISPR 筛选测序
-
CRISPR 脱靶验证
了解更多:
-
CRISPR 基因编辑评估和质量控制中的下一代测序
-
下一代测序验证您的 CRISPR/Cas9 编辑
-
CRISPR 测序:简介、工作流程和应用
CRISPR-Cas9 脱靶检测的计算模拟方法
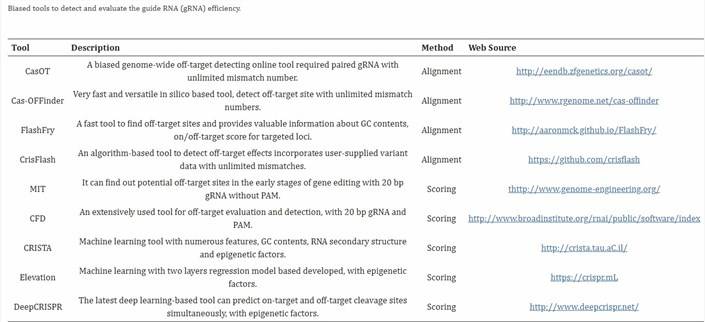
这些技术旨在预测潜在脱靶序列,主要是由于目标序列与其他序列的相似性或引导 RNA (gRNA) 的有限特异性。潜在脱靶序列可以通过整合实验数据或使用计算机算法推导。为验证脱靶效应,通常依赖 PCR 产物测序。这些方法常称为“有偏”方法,脱靶预测工具的功能可分为两种主要机制:(1) 基于序列比对的模型和 (2) 基于算法打分的预测 [1]。
CRISPR-Cas9 脱靶检测的计算模拟方法
随着 CRISPR 技术的广泛使用,其应用预期也在提高。使用 CRISPR 技术时,实验设计可能缺乏额外对照组,这可能在后续实验中带来挑战。因此,建议在 CRISPR 用于基因编辑后及时进行脱靶分析,或保留早期样本。细胞和动物均易发生自发突变。
参考文献
- Naeem, Muhammad et al. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells vol. 2020
- Ipek Tasan and Huimin Zhao. Targeting Specificity of the CRISPR/Cas9 System. ACS Synthetic Biology 2017
- Wienert, B., Wyman, S.K., Yeh, C.D. et al. CRISPR off-target detection with DISCOVER-seq. Nat Protoc 15, 1775–1799 (2020)
Processing Data_Julia_RNASeq_SARS-CoV-2
-
Preparing the directory trimmed. Note that the extension of input files is fastq.gz, the extension of output file is fq.gz.
mkdir trimmed trimmed_unpaired; for sample_id in 01_PBS_1 02_PBS_2 03_PBS_3 04_rluc_1 05_rluc_2 06_rluc_3 07_ralpha_1 08_ralpha_2 09_ralpha_3 10_rBA2_1 11_rBA2_2 12_rBA2_3 13_rBA5_1 14_rBA5_2 15_rBA5_3 16_rdelta_1 17_rdelta_2 18_rdelta_3 19_rpirola_1 20_rpirola_2 21_rpirola_3; do java -jar /home/jhuang/Tools/Trimmomatic-0.36/trimmomatic-0.36.jar PE -threads 100 20250423_AV243904_0006_B/${sample_id}/${sample_id}_R1.fastq.gz 20250423_AV243904_0006_B/${sample_id}/${sample_id}_R2.fastq.gz trimmed/${sample_id}_R1.fq.gz trimmed_unpaired/${sample_id}_R1.fq.gz trimmed/${sample_id}_R2.fq.gz trimmed_unpaired/${sample_id}_R2.fq.gz ILLUMINACLIP:/home/jhuang/Tools/Trimmomatic-0.36/adapters/TruSeq3-PE-2.fa:2:30:10:8:TRUE LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 AVGQUAL:20; done 2> trimmomatic_pe.log; done -
Preparing samplesheet.csv
sample,fastq_1,fastq_2,strandedness PBS_1,01_PBS_1_R1.fq.gz,01_PBS_1_R2.fq.gz,auto PBS_2,02_PBS_2_R1.fq.gz,02_PBS_2_R2.fq.gz,auto PBS_3,03_PBS_3_R1.fq.gz,03_PBS_3_R2.fq.gz,auto rLUC_1,04_rluc_1_R1.fq.gz,04_rluc_1_R2.fq.gz,auto rLUC_2,05_rluc_2_R1.fq.gz,05_rluc_2_R2.fq.gz,auto rLUC_3,06_rluc_3_R1.fq.gz,06_rluc_3_R2.fq.gz,auto rAlpha-N_1,07_ralpha_1_R1.fq.gz,07_ralpha_1_R2.fq.gz,auto rAlpha-N_2,08_ralpha_2_R1.fq.gz,08_ralpha_2_R2.fq.gz,auto rAlpha-N_3,09_ralpha_3_R1.fq.gz,09_ralpha_3_R2.fq.gz,auto rBA.2-N_1,10_rBA2_1_R1.fq.gz,10_rBA2_1_R2.fq.gz,auto rBA.2-N_2,11_rBA2_2_R1.fq.gz,11_rBA2_2_R2.fq.gz,auto rBA.2-N_3,12_rBA2_3_R1.fq.gz,12_rBA2_3_R2.fq.gz,auto rBA.5-N_1,13_rBA5_1_R1.fq.gz,13_rBA5_1_R2.fq.gz,auto rBA.5-N_2,14_rBA5_2_R1.fq.gz,14_rBA5_2_R2.fq.gz,auto rBA.5-N_3,15_rBA5_3_R1.fq.gz,15_rBA5_3_R2.fq.gz,auto rDelta-N_1,16_rdelta_1_R1.fq.gz,16_rdelta_1_R2.fq.gz,auto rDelta-N_2,17_rdelta_2_R1.fq.gz,17_rdelta_2_R2.fq.gz,auto rDelta-N_3,18_rdelta_3_R1.fq.gz,18_rdelta_3_R2.fq.gz,auto rPirola-N_1,19_rpirola_1_R1.fq.gz,19_rpirola_1_R2.fq.gz,auto rPirola-N_2,20_rpirola_2_R1.fq.gz,20_rpirola_2_R2.fq.gz,auto rPirola-N_3,21_rpirola_3_R1.fq.gz,21_rpirola_3_R2.fq.gz,auto mv trimmed/* . -
nextflow run
'GRCh38' { fasta = "/home/jhuang/Homo_sapiens/Ensembl/GRCh38/Sequence/WholeGenomeFasta/genome.fa" bwa = "${params.igenomes_base}/Homo_sapiens/Ensembl/GRCh38/Sequence/BWAIndex/version0.6.0/" bowtie2 = "${params.igenomes_base}/Homo_sapiens/Ensembl/GRCh38/Sequence/Bowtie2Index/" star = "/home/jhuang/Homo_sapiens/Ensembl/GRCh38/Sequence/STARIndex/" bismark = "${params.igenomes_base}/Homo_sapiens/Ensembl/GRCh38/Sequence/BismarkIndex/" gtf = "/home/jhuang/Homo_sapiens/Ensembl/GRCh38/Annotation/Genes/genes.gtf" bed12 = "/home/jhuang/Homo_sapiens/Ensembl/GRCh38/Annotation/Genes/genes.bed" mito_name = "chrM" macs_gsize = "2.7e9" blacklist = "/home/jhuang/Homo_sapiens/UCSC/hg38/blacklists/hg38-blacklist.bed" } #Example1: http://xgenes.com/article/article-content/157/prepare-virus-gtf-for-nextflow-run/ #docker pull nfcore/rnaseq ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq # ---- SUCCESSFUL with directly downloaded gff3 and fasta from NCBI using docker after replacing 'CDS' with 'exon' ---- #For Tam's bacteria: --fasta "/home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040.fasta" --gff "/home/jhuang/DATA/Data_Tam_RNAseq_2024_AUM_MHB_Urine/CP059040_m.gff" --gtf_group_features 'gene_id' --gtf_extra_attributes 'gene_name' --featurecounts_group_type 'gene_biotype' --featurecounts_feature_type 'transcript' (host_env) /usr/local/bin/nextflow run rnaseq/main.nf -profile docker --input samplesheet.csv --genome GRCh38 --aligner 'star_salmon' --outdir results --max_cpus 60 --max_memory 600.GB --max_time 2400.h -resume # #--save_align_intermeds --save_unaligned --save_reference -
samplesheet.csv
sample,fastq_1,fastq_2,strandedness PBS_1,01_PBS_1_R1.fq.gz,01_PBS_1_R2.fq.gz,auto PBS_2,02_PBS_2_R1.fq.gz,02_PBS_2_R2.fq.gz,auto PBS_3,03_PBS_3_R1.fq.gz,03_PBS_3_R2.fq.gz,auto rLUC_1,04_rluc_1_R1.fq.gz,04_rluc_1_R2.fq.gz,auto rLUC_2,05_rluc_2_R1.fq.gz,05_rluc_2_R2.fq.gz,auto rLUC_3,06_rluc_3_R1.fq.gz,06_rluc_3_R2.fq.gz,auto rAlpha-N_1,07_ralpha_1_R1.fq.gz,07_ralpha_1_R2.fq.gz,auto rAlpha-N_2,08_ralpha_2_R1.fq.gz,08_ralpha_2_R2.fq.gz,auto rAlpha-N_3,09_ralpha_3_R1.fq.gz,09_ralpha_3_R2.fq.gz,auto rBA.2-N_1,10_rBA2_1_R1.fq.gz,10_rBA2_1_R2.fq.gz,auto rBA.2-N_2,11_rBA2_2_R1.fq.gz,11_rBA2_2_R2.fq.gz,auto rBA.2-N_3,12_rBA2_3_R1.fq.gz,12_rBA2_3_R2.fq.gz,auto rBA.5-N_1,13_rBA5_1_R1.fq.gz,13_rBA5_1_R2.fq.gz,auto rBA.5-N_2,14_rBA5_2_R1.fq.gz,14_rBA5_2_R2.fq.gz,auto rBA.5-N_3,15_rBA5_3_R1.fq.gz,15_rBA5_3_R2.fq.gz,auto rDelta-N_1,16_rdelta_1_R1.fq.gz,16_rdelta_1_R2.fq.gz,auto rDelta-N_2,17_rdelta_2_R1.fq.gz,17_rdelta_2_R2.fq.gz,auto rDelta-N_3,18_rdelta_3_R1.fq.gz,18_rdelta_3_R2.fq.gz,auto rPirola-N_1,19_rpirola_1_R1.fq.gz,19_rpirola_1_R2.fq.gz,auto rPirola-N_2,20_rpirola_2_R1.fq.gz,20_rpirola_2_R2.fq.gz,auto rPirola-N_3,21_rpirola_3_R1.fq.gz,21_rpirola_3_R2.fq.gz,auto -
R-code for evaluation
# Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) library(RColorBrewer) library(openxlsx) setwd("~/DATA/Data_Julia_RNASeq_SARS-CoV-2/results/star_salmon") # Define paths to your Salmon output quantification files, quant.sf refers to tx-counts, later will be summaried as gene-counts. files <- c("PBS_r1" = "./PBS_1/quant.sf", "PBS_r2" = "./PBS_2/quant.sf", "PBS_r3" = "./PBS_3/quant.sf", "rLUC_r1" = "./rLUC_1/quant.sf", "rLUC_r2" = "./rLUC_2/quant.sf", "rLUC_r3" = "./rLUC_3/quant.sf", "rAlpha.N_r1" = "./rAlpha-N_1/quant.sf", "rAlpha.N_r2" = "./rAlpha-N_2/quant.sf", "rAlpha.N_r3" = "./rAlpha-N_3/quant.sf", "rBA.2.N_r1" = "./rBA.2-N_1/quant.sf", "rBA.2.N_r2" = "./rBA.2-N_2/quant.sf", "rBA.2.N_r3" = "./rBA.2-N_3/quant.sf", "rBA.5.N_r1" = "./rBA.5-N_1/quant.sf", "rBA.5.N_r2" = "./rBA.5-N_2/quant.sf", "rBA.5.N_r3" = "./rBA.5-N_3/quant.sf", "rDelta.N_r1" = "./rDelta-N_1/quant.sf", "rDelta.N_r2" = "./rDelta-N_2/quant.sf", "rDelta.N_r3" = "./rDelta-N_3/quant.sf", "rPirola.N_r1" = "./rPirola-N_1/quant.sf", "rPirola.N_r2" = "./rPirola-N_2/quant.sf", "rPirola.N_r3" = "./rPirola-N_3/quant.sf") # ---- tx-level count data ---- # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) # Define the replicates and condition of the samples #replicate <- factor(c("r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2")) #condition <- factor(c("PBS", "PBS", "PBS", "rLUC", "rLUC", "rLUC", "rAlpha.N", "rAlpha.N", "rAlpha.N", "rBA.2.N", "rBA.2.N", "rBA.2.N", "rBA.5.N", "rBA.5.N", "rBA.5.N", "rDelta.N", "rDelta.N", "rDelta.N", "rPirola.N", "rPirola.N", "rPirola.N")) condition <- factor(c("PBS", "PBS", "PBS", "rLUC", "rLUC", "rLUC", "rAlpha-N", "rAlpha-N", "rAlpha-N", "rBA.2-N", "rBA.2-N", "rBA.2-N", "rBA.5-N", "rBA.5-N", "rBA.5-N", "rDelta-N", "rDelta-N", "rDelta-N", "rPirola-N", "rPirola-N", "rPirola-N")) # Define the colData for DESeq2 colData <- data.frame(condition=condition, row.names=names(files)) # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) # In the context of your new code which is using tximport and DESeq2, you don't necessarily need this step. The reason is that DESeq2 performs its own filtering of low-count genes during the normalization and differential expression steps. # Filter data to retain only genes with more than 2 counts > 3 across all samples # dds <- dds[rowSums(counts(dds) > 3) > 2, ] # Output raw count data to a CSV file write.csv(counts(dds), file="transcript_counts.csv") # ---- gene-level count data ---- # Read in the tx2gene map from salmon_tx2gene.tsv #tx2gene <- read.csv("salmon_tx2gene.tsv", sep="\t", header=FALSE) tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) # Continue with the DESeq2 workflow as before... colData <- data.frame(condition=condition, row.names=names(files)) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #60605-->26543 dds <- DESeq(dds) rld <- rlogTransformation(dds) write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv") #TODO: why a lot of reads were removed due to the too_short? #STAR --runThreadN 4 --genomeDir /path/to/GenomeDir --readFilesIn /path/to/read1.fastq /path/to/read2.fastq --outFilterMatchNmin 50 --outSAMtype BAM SortedByCoordinate --outFileNamePrefix /path/to/output dim(counts(dds)) head(counts(dds), 10) -
(Optional) draw 3D PCA plots.
library(gplots) library("RColorBrewer") library(ggplot2) data <- plotPCA(rld, intgroup=c("condition", "replicate"), returnData=TRUE) write.csv(data, file="plotPCA_data.csv") #calculate all PCs including PC3 with the following codes library(genefilter) ntop <- 500 rv <- rowVars(assay(rld)) select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))] mat <- t( assay(rld)[select, ] ) pc <- prcomp(mat) pc$x[,1:3] #df_pc <- data.frame(pc$x[,1:3]) df_pc <- data.frame(pc$x) identical(rownames(data), rownames(df_pc)) #-->TRUE data$PC1 <- NULL data$PC2 <- NULL merged_df <- merge(data, df_pc, by = "row.names") #merged_df <- merged_df[, -1] row.names(merged_df) <- merged_df$Row.names merged_df$Row.names <- NULL # remove the "name" column merged_df$name <- NULL merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","group","condition","replicate")] write.csv(merged_df, file="merged_df_10PCs.csv") summary(pc) #0.5333 0.2125 0.06852 draw_3D.py -
Draw 2D PCA plots.
# -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
(Optional) estimate size factors
> head(dds) class: DESeqDataSet dim: 6 10 metadata(1): version assays(6): counts avgTxLength ... H cooks rownames(6): ENSG00000000003 ENSG00000000005 ... ENSG00000000460 ENSG00000000938 rowData names(34): baseMean baseVar ... deviance maxCooks colnames(10): control_r1 control_r2 ... HSV.d8_r1 HSV.d8_r2 colData names(2): condition replicate #convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor sizeFactors(dds) #NULL dds <- estimateSizeFactors(dds) sizeFactors(dds) normalized_counts <- counts(dds, normalized=TRUE) #write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA) # ---- DEBUG sizeFactors(dds) always NULL, see https://support.bioconductor.org/p/97676/ ---- nm <- assays(dds)[["avgTxLength"]] sf <- estimateSizeFactorsForMatrix(counts(dds), normMatrix=nm) assays(dds)$counts # for count data assays(dds)$avgTxLength # for average transcript length, etc. assays(dds)$normalizationFactors In normal circumstances, the size factors should be stored in the DESeqDataSet object itself and not in the assays, so they are typically not retrievable via the assays() function. However, due to the issues you're experiencing, you might be able to manually compute the size factors and assign them back to the DESeqDataSet. To calculate size factors manually, DESeq2 uses the median ratio method. Here's a very simplified version of how you could compute this manually: > assays(dds) List of length 6 names(6): counts avgTxLength normalizationFactors mu H cooks To calculate size factors manually, DESeq2 uses the median ratio method. Here's a very simplified version of how you could compute this manually: geoMeans <- apply(assays(dds)$counts, 1, function(row) if (all(row == 0)) 0 else exp(mean(log(row[row != 0])))) sizeFactors(dds) <- median(assays(dds)$counts / geoMeans, na.rm = TRUE) # ---- DEBUG END ---- #unter konsole # control_r1 ... # 1/0.9978755 ... > sizeFactors(dds) HeLa_TO_r1 HeLa_TO_r2 0.9978755 1.1092227 1/0.9978755=1.002129023 1/1.1092227= #bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r1Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r1.bw --binSize 10 --scaleFactor 1.002129023 --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r2Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r2.bw --binSize 10 --scaleFactor 0.901532217 --effectiveGenomeSize 2864785220 -
Differential expressed gene analyses
#A central method for exploring differences between groups of segments or samples is to perform differential gene expression analysis. dds$condition <- relevel(dds$condition, "PBS") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("rLUC_vs_PBS","rAlpha.N_vs_PBS","rBA.2.N_vs_PBS","rBA.5.N_vs_PBS","rDelta.N_vs_PBS","rPirola.N_vs_PBS") dds$condition <- relevel(dds$condition, "rLUC") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("rAlpha.N_vs_rLUC","rBA.2.N_vs_rLUC","rBA.5.N_vs_rLUC","rDelta.N_vs_rLUC","rPirola.N_vs_rLUC") dds$condition <- relevel(dds$condition, "rAlpha-N") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("rBA.2.N_vs_rAlpha.N","rBA.5.N_vs_rAlpha.N","rDelta.N_vs_rAlpha.N","rPirola.N_vs_rAlpha.N") dds$condition <- relevel(dds$condition, "rBA.2-N") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("rBA.5.N_vs_rBA.2.N","rDelta.N_vs_rBA.2.N","rPirola.N_vs_rBA.2.N") dds$condition <- relevel(dds$condition, "rBA.5-N") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("rDelta.N_vs_rBA.5.N","rPirola.N_vs_rBA.5.N") dds$condition <- relevel(dds$condition, "rDelta-N") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("rPirola.N_vs_rDelta.N") ##https://bioconductor.statistik.tu-dortmund.de/packages/3.7/data/annotation/ #BiocManager::install("EnsDb.Mmusculus.v79") #library(EnsDb.Mmusculus.v79) #edb <- EnsDb.Mmusculus.v79 #https://bioconductor.org/packages/release/bioc/vignettes/biomaRt/inst/doc/accessing_ensembl.html#selecting-an-ensembl-biomart-database-and-dataset #https://bioconductor.org/packages/release/bioc/vignettes/biomaRt/inst/doc/accessing_ensembl.html#selecting-an-ensembl-biomart-database-and-dataset library(biomaRt) listEnsembl() listMarts() #ensembl <- useEnsembl(biomart = "genes", mirror="asia") # default is Mouse strains 104 #ensembl <- useEnsembl(biomart = "ensembl", dataset = "mmusculus_gene_ensembl", mirror = "www") #ensembl = useMart("ensembl_mart_44", dataset="hsapiens_gene_ensembl",archive=TRUE, mysql=TRUE) #ensembl <- useEnsembl(biomart = "ensembl", dataset = "mmusculus_gene_ensembl", version="104") #ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="86") #--> total 69, 27 GRCh38.p7 and 39 GRCm38.p4; we should take 104, since rnaseq-pipeline is also using annotation of 104! ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="114") datasets <- listDatasets(ensembl) #--> total 202 80 GRCh38.p13 107 GRCm39 #80 GRCh38.p14 #107 GRCm39 listEnsemblArchives() # name date url version #1 Ensembl GRCh37 Feb 2014 https://grch37.ensembl.org GRCh37 #2 Ensembl 114 May 2025 https://may2025.archive.ensembl.org 114 #3 Ensembl 113 Oct 2024 https://oct2024.archive.ensembl.org 113 #4 Ensembl 112 May 2024 https://may2024.archive.ensembl.org 112 #5 Ensembl 111 Jan 2024 https://jan2024.archive.ensembl.org 111 #6 Ensembl 110 Jul 2023 https://jul2023.archive.ensembl.org 110 #7 Ensembl 109 Feb 2023 https://feb2023.archive.ensembl.org 109 attributes = listAttributes(ensembl) attributes[1:25,] #https://www.ncbi.nlm.nih.gov/grc/human #BiocManager::install("org.Mmu.eg.db") #library("org.Mmu.eg.db") #edb <- org.Mmu.eg.db # #https://bioconductor.statistik.tu-dortmund.de/packages/3.6/data/annotation/ #EnsDb.Mmusculus.v79 #> query(hub, c("EnsDb", "apiens", "98")) #columns(edb) #searchAttributes(mart = ensembl, pattern = "symbol") ##https://www.geeksforgeeks.org/remove-duplicate-rows-based-on-multiple-columns-using-dplyr-in-r/ library(dplyr) library(tidyverse) #df <- data.frame (lang =c ('Java','C','Python','GO','RUST','Javascript', 'Cpp','Java','Julia','Typescript','Python','GO'), value = c (21,21,3,5,180,9,12,20,6,0,3,6), usage =c(21,21,0,99,44,48,53,16,6,8,0,6)) #distinct(df, lang, .keep_all= TRUE) for (i in clist) { #i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), geness_uniq$ensembl_gene_id) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL write.csv(as.data.frame(geness_res[order(geness_res$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(geness_res, padj<=0.05 & log2FoldChange>=2) down <- subset(geness_res, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) } #-- show methods of class DESeq2 -- #x=capture.output(showMethods(class="DESeq2")) #unlist(lapply(strsplit(x[grep("Function: ",x,)]," "),function(x) x[2])) -
Volcano plots with automatically finding top_g
#A canonical visualization for interpreting differential gene expression results is the volcano plot. library(ggrepel) #for i in rLUC_vs_PBS rAlpha.N_vs_PBS rBA.2.N_vs_PBS rBA.5.N_vs_PBS rDelta.N_vs_PBS rPirola.N_vs_PBS; do #for i in rAlpha.N_vs_rLUC rBA.2.N_vs_rLUC rBA.5.N_vs_rLUC rDelta.N_vs_rLUC rPirola.N_vs_rLUC; do #for i in rBA.2.N_vs_rAlpha.N rBA.5.N_vs_rAlpha.N rDelta.N_vs_rAlpha.N rPirola.N_vs_rAlpha.N; do #for i in rBA.5.N_vs_rBA.2.N rDelta.N_vs_rBA.2.N rPirola.N_vs_rBA.2.N; do #for i in rDelta.N_vs_rBA.5.N rPirola.N_vs_rBA.5.N; do for i in rPirola.N_vs_rDelta.N; do echo "geness_res <- read.csv(file = paste(\"${i}\", \"all.txt\", sep=\"-\"), row.names=1)" echo "geness_res\$Color <- \"NS or log2FC < 2.0\"" echo "geness_res\$Color[geness_res\$pvalue < 0.05] <- \"P < 0.05\"" echo "geness_res\$Color[geness_res\$padj < 0.05] <- \"P-adj < 0.05\"" echo "geness_res\$Color[abs(geness_res\$log2FoldChange) < 2.0] <- \"NS or log2FC < 2.0\"" echo "geness_res\$Color <- factor(geness_res\$Color, levels = c(\"NS or log2FC < 2.0\", \"P < 0.05\", \"P-adj < 0.05\"))" echo "write.csv(geness_res, \"${i}_with_Category.csv\")" echo "write.csv(geness_res, \"${i}_with_Category.csv\")" echo "openxlsx::write.xlsx(geness_res, file = \"${i}_with_Category.xlsx\")" echo "geness_res\$invert_P <- (-log10(geness_res\$pvalue)) * sign(geness_res\$log2FoldChange)" echo "top_g <- c()" echo "top_g <- c(top_g, \ geness_res[, 'external_gene_name'][order(geness_res[, 'invert_P'], decreasing = TRUE)[1:100]], \ geness_res[, 'external_gene_name'][order(geness_res[, 'invert_P'], decreasing = FALSE)[1:100]])" echo "top_g <- unique(top_g)" # Save filtered subset for optional export/debug echo "highlight_genes <- subset(geness_res, external_gene_name %in% top_g & pvalue < 0.05 & abs(geness_res\$log2FoldChange) >= 2.0)" echo "geness_res <- geness_res[, -1*ncol(geness_res)]" # remove invert_P column echo "png(\"${i}.png\",width=1200, height=2000)" echo "ggplot(geness_res, \ aes(x = log2FoldChange, y = -log10(pvalue), \ color = Color, label = external_gene_name)) + \ geom_vline(xintercept = c(2.0, -2.0), lty = \"dashed\") + \ geom_hline(yintercept = -log10(0.05), lty = \"dashed\") + \ geom_point() + \ labs(x = \"log2(FC)\", y = \"Significance, -log10(P)\", color = \"Significance\") + \ scale_color_manual(values = c(\"P-adj < 0.05\"=\"darkblue\",\"P < 0.05\"=\"lightblue\",\"NS or log2FC < 2.0\"=\"darkgray\"), \ guide = guide_legend(override.aes = list(size = 4))) + \ scale_y_continuous(expand = expansion(mult = c(0,0.05))) + \ geom_text_repel(data = highlight_genes, size = 4, point.padding = 0.15, color = \"black\", \ min.segment.length = .1, box.padding = .2, lwd = 2) + \ theme_bw(base_size = 16) + \ theme(legend.position = \"bottom\")" echo "dev.off()" done sed -i -e 's/Color/Category/g' *_Category.csv for i in rLUC_vs_PBS rAlpha.N_vs_PBS rBA.2.N_vs_PBS rBA.5.N_vs_PBS rDelta.N_vs_PBS rPirola.N_vs_PBS rAlpha.N_vs_rLUC rBA.2.N_vs_rLUC rBA.5.N_vs_rLUC rDelta.N_vs_rLUC rPirola.N_vs_rLUC rBA.2.N_vs_rAlpha.N rBA.5.N_vs_rAlpha.N rDelta.N_vs_rAlpha.N rPirola.N_vs_rAlpha.N rBA.5.N_vs_rBA.2.N rDelta.N_vs_rBA.2.N rPirola.N_vs_rBA.2.N rDelta.N_vs_rBA.5.N rPirola.N_vs_rBA.5.N rPirola.N_vs_rDelta.N; do echo "~/Tools/csv2xls-0.4/csv_to_xls.py ${i}-all.txt ${i}-up.txt ${i}-down.txt -d$',' -o ${i}.xls;" done
It looks as if the viruses are indeed different, especially in terms of “response to virus/IFN induction” (yellow cluster in your heat map).
To get this even clearer, could you possibly now compare the viruses with each other? Initially, I think it would be best to compare each virus with rLUC,
-
The question above has already been addressed in the previous two steps.
e.g. rLUC vs rAlpha.N, rLUC vs rDelta,N, rLUC vs rBA2.N, rLUC vs rBA5.N, rLUC vs rPirola.N? # rAlpha.N_vs_rLUC.xls # rBA.2.N_vs_rLUC.xls # rBA.5.N_vs_rLUC.xls # rDelta.N_vs_rLUC.xls # rPirola.N_vs_rLUC.xls -
Clustering the genes and draw heatmap
install.packages("gplots") library("gplots") for i in rLUC_vs_PBS rAlpha.N_vs_PBS rBA.2.N_vs_PBS rBA.5.N_vs_PBS rDelta.N_vs_PBS rPirola.N_vs_PBS rAlpha.N_vs_rLUC rBA.2.N_vs_rLUC rBA.5.N_vs_rLUC rDelta.N_vs_rLUC rPirola.N_vs_rLUC rBA.2.N_vs_rAlpha.N rBA.5.N_vs_rAlpha.N rDelta.N_vs_rAlpha.N rPirola.N_vs_rAlpha.N rBA.5.N_vs_rBA.2.N rDelta.N_vs_rBA.2.N rPirola.N_vs_rBA.2.N rDelta.N_vs_rBA.5.N rPirola.N_vs_rBA.5.N rPirola.N_vs_rDelta.N; do echo "cut -d',' -f1-1 ${i}-up.txt > ${i}-up.id" echo "cut -d',' -f1-1 ${i}-down.txt > ${i}-down.id" done # 1 1457.2h_vs_1457.M10.2h-down.id # 1 1457.2h_vs_1457.M10.2h-up.id # 23 1457.2h_vs_uninfected.2h-down.id # 74 1457.2h_vs_uninfected.2h-up.id # 126 1457.3d_vs_1457.2h-down.id # 61 1457.3d_vs_1457.2h-up.id # 2 1457.3d_vs_1457.M10.3d-down.id # 2 1457.3d_vs_1457.M10.3d-up.id # 97 1457.3d_vs_uninfected.3d-down.id # 79 1457.3d_vs_uninfected.3d-up.id # 17 1457.M10.2h_vs_uninfected.2h-down.id # 82 1457.M10.2h_vs_uninfected.2h-up.id # 162 1457.M10.3d_vs_1457.M10.2h-down.id # 69 1457.M10.3d_vs_1457.M10.2h-up.id # 171 1457.M10.3d_vs_uninfected.3d-down.id # 124 1457.M10.3d_vs_uninfected.3d-up.id cat *.id | sort -u > ids #add Gene_Id in the first line, delete the "" GOI <- read.csv("ids")$Gene_Id #739 genes RNASeq.NoCellLine <- assay(rld) # Defining the custom order #column_order <- c( # "PBS_r1","PBS_r2","PBS_r3","rLUC_r1","rLUC_r2","rLUC_r3","rAlpha.N_r1","rAlpha.N_r2","rAlpha.N_r3","rBA.2.N_r1","rBA.2.N_r2","rBA.2.N_r3","rBA.5.N_r1","rBA.5.N_r2","rBA.5.N_r3","rDelta.N_r1","rDelta.N_r2","rDelta.N_r3","rPirola.N_r1","rPirola.N_r2","rPirola.N_r3" #) #RNASeq.NoCellLine_reordered <- RNASeq.NoCellLine[, column_order] #head(RNASeq.NoCellLine_reordered) # Update column names new_colnames <- c("PBS 1","PBS 2","PBS 3","rLUC 1","rLUC 2","rLUC 3","rAlpha-N 1","rAlpha-N 2","rAlpha-N 3","rBA.2-N 1","rBA.2-N 2","rBA.2-N 3","rBA.5-N 1","rBA.5-N 2","rBA.5-N 3","rDelta-N 1","rDelta-N 2","rDelta-N 3","rPirola-N 1","rPirola-N 2","rPirola-N 3") colnames(RNASeq.NoCellLine) <- new_colnames #clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman datamat = RNASeq.NoCellLine[GOI, ] write.csv(as.data.frame(datamat), file ="DEGs_heatmap_data.csv") hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.1) mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN"); mycol = mycol[as.vector(mycl)] #png("DEGs_heatmap.png", width=800, height=1000) png("DEGs_heatmap.png", width = 1200, height = 1500, res = 150) # Higher res heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row', scale='row',trace='none',col=bluered(75), RowSideColors = mycol, labRow="", srtCol=30, keysize=0.72, cexRow = 2, cexCol = 1.4) dev.off() # Extract rows from datamat where the row names match the identifiers in subset_1 #### cluster members ##### subset_1<-names(subset(mycl, mycl == '1')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_1, ]) #402 subset_2<-names(subset(mycl, mycl == '2')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_2, ]) #137 subset_3<-names(subset(mycl, mycl == '3')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_3, ]) #200 # Initialize an empty data frame for the annotated data annotated_data <- data.frame() # Determine total number of genes total_genes <- length(rownames(data)) # Loop through each gene to annotate for (i in 1:total_genes) { gene <- rownames(data)[i] result <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = gene, mart = ensembl) # If multiple rows are returned, take the first one if (nrow(result) > 1) { result <- result[1, ] } # Check if the result is empty if (nrow(result) == 0) { result <- data.frame(ensembl_gene_id = gene, external_gene_name = NA, gene_biotype = NA, entrezgene_id = NA, chromosome_name = NA, start_position = NA, end_position = NA, strand = NA, description = NA) } # Transpose expression values expression_values <- t(data.frame(t(data[gene, ]))) colnames(expression_values) <- colnames(data) # Combine gene information and expression data combined_result <- cbind(result, expression_values) # Append to the final dataframe annotated_data <- rbind(annotated_data, combined_result) # Print progress every 100 genes if (i %% 100 == 0) { cat(sprintf("Processed gene %d out of %d\n", i, total_genes)) } } # Save the annotated data to a new CSV file write.csv(annotated_data, "cluster1_YELLOW.csv", row.names=FALSE) write.csv(annotated_data, "cluster2_DARKBLUE.csv", row.names=FALSE) write.csv(annotated_data, "cluster3_DARKORANGE.csv", row.names=FALSE) #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.csv -d',' -o gene_clusters.xls
基因组和表型表征揭示药物敏感鲍氏不动杆菌表现出增强的毒力相关性状和应激耐受性
DOI: https://doi.org/10.3390/biology14091201
引用: Foong, W.E.; He, W.; Xiang, X.; Huang, J.; Tam, H.-K. 基因组和表型表征揭示药物敏感鲍氏不动杆菌表现出增强的毒力相关性状和应激耐受性. Biology 2025, 14, 1201.
1 生物化学与分子生物学系,衡阳医学院,南华大学,衡阳 421001,中国; wuenee@hotmail.com (W.E.F.)
2 医学微生物学系,湖南省特殊病原体防控重点实验室,衡阳医学院,南华大学,衡阳 421001,中国
3 国家卫生健康委员会出生缺陷研究与预防重点实验室,湖南省妇幼保健院,长沙 410008,中国
4 医学微生物学、病毒学和卫生学研究所,汉堡-爱泼斯坦大学医学中心,马丁街52号,20246 汉堡,德国
- 通讯作者: j.huang@uke.de (J.H.); tamhk60@hotmail.com (H.-K.T.)
简单摘要
鲍氏不动杆菌通常因其对抗生素的抗性而在医疗环境中构成严重威胁。在本研究中,我们调查了一个临床分离株 HKAB-1,尽管对多种抗生素高度敏感,但表现出显著的毒力相关性状。相比参考株 ATCC 19606,HKAB-1 在血清和干燥条件下显示出增强的存活能力。HKAB-1 还形成坚固的生物膜并表现出更大的运动性,这些表型与持久性和致病性相关。基因组和转录组分析显示,HKAB-1 拥有活跃的铁获取和血红素利用系统,这些系统对类似宿主条件高度响应。此外,我们发现与生物膜形成相关的基因在形成生物膜的细胞中高度诱导。相反,adeB 的表达显著降低,这可能解释了尽管拥有多个抗性基因仍表现出抗生素敏感性的原因。这些发现反映了某些鲍氏不动杆菌菌株在进化上的权衡,优先考虑毒力相关性状而非抗菌机制的表达。这一结果凸显了持续基因组监测的必要性,以监控类似 HKAB-1 这样新兴的毒力强但药物敏感的菌株,这些菌株可能在选择压力下作为抗性发展的储备。
摘要
鲍氏不动杆菌是一种机会性病原体,以其多药抗性和环境持久性而著称。我们表征了一个临床分离株 HKAB-1,尽管对所有测试的抗生素高度敏感,但表现出显著的毒力相关性状。HKAB-1 在 MH2B、血清和干燥条件下表现出优越的生长,形成了坚固的生物膜,并具有活跃的运动性。全基因组测序确定了两个血红素利用簇、多个铁载体生物合成途径以及其他毒力相关基因。基因表达分析显示,在血清暴露下,血红素利用和铁载体生物合成基因簇显著上调,表明在类似宿主条件下铁摄取途径的激活。生物膜相关基因,包括 bap、PNAG 生物合成基因和 IV 型菌毛成分,在形成生物膜的细胞中显著上调,支持其在驱动增强生物膜表型中的作用。相反,编码主要 RND 外排泵的 adeB 显著下调,可能解释了其药物敏感表型。比较基因组分析凸显了与营养运输、代谢途径和膜生物生成相关的基因差异,这些差异可能支撑其增强的生长。这些发现指向抗生素抗性和毒力之间的潜在权衡,强调监测抗生素敏感但高度毒力的鲍氏不动杆菌分离株作为抗性进化的潜在储备的重要性。需要进一步调查以阐明这种表型平衡的潜在机制。
关键词: 鲍氏不动杆菌; 血红素利用; 毒力; 血清抗性; 干燥耐受性
1. 引言
鲍氏不动杆菌是临床环境中其属中最常分离的物种,已成为研究多药抗性 (MDR) 的模型生物 [1]。近年来,该病原体对全球医疗系统构成了日益严重的威胁;该病原体引起多种严重的医院感染,包括呼吸道、皮肤、尿路、伤口和血流感染 [2]。该细菌在医疗环境中的持久性很大程度上归因于其在干燥、消毒剂和抗菌药物长期暴露等恶劣条件下的惊人耐受能力 [3]。鲍氏不动杆菌的基因组可塑性在其卓越的适应性中起着核心作用,使其能够通过获得和水平转移外源遗传物质快速适应环境压力 [4]。整合移动遗传元件如插入序列、转座子及抗性岛进一步促进基因组重排和抗性决定因子的稳定整合 [5,6]。过去十年令人担忧的趋势显示,多药耐药鲍氏不动杆菌菌株的流行率不断增加 [7],凸显了了解其生存和致病性的遗传及生理基础的迫切需求。
在各种抗性机制中,外排泵的过度表达和外膜通透性的降低在鲍氏不动杆菌的抗生素抗性中起着关键作用 [8,9]。特别值得注意的是,鲍氏不动杆菌天生编码了大量多药外排泵,这些泵被分类为单组分转运蛋白和三部分系统 [10]。这些外排系统独立或协同作用,主动排出多种结构和化学上不同的底物,包括抗生素、杀菌剂、染料和洗涤剂,导致细胞内药物积累减少和最低抑菌浓度 (MIC) 升高 [11,12]。
除了抗菌药物抗性,鲍氏不动杆菌在生物和非生物表面形成生物膜的能力以及其表面相关运动能力也显著促成了其作为医院感染病原体的成功 [13,14]。这些性状在医疗器械的定植和侵入性手术中尤为重要,促进了持续的医院获得性感染 [15]。此外,生物膜形成和运动被认为促进了抗性基因的传播和获取,进一步增强了该生物在医院环境中的适应性和持久性 [16,17]。总之,生物膜形成、运动和多药抗性的相互作用共同支撑了鲍氏不动杆菌在临床环境中的显著持久性和传播能力。
在本研究中,我们表征了一个临床鲍氏不动杆菌分离株 HKAB-1,该菌株显示出加速的生长动力学和显著的毒力相关表型,如增强的生物膜形成、运动性、干燥耐受性和血清抗性,尽管其对抗生素广泛敏感。全基因组测序将 HKAB-1 确定为序列型 ST392,并揭示了多个潜在毒力因子,包括 hemO 基因簇和血红素利用簇 1。比较基因组分析凸显了与营养运输、代谢途径和膜生物生成相关的基因差异,这些差异可能有助于 HKAB-1 的增强生长。值得注意的是,adeB 的表达在 HKAB-1 中显著抑制,可能解释了其尽管拥有多个抗性决定因子仍呈现药物敏感表型的现象。此外,在生物膜条件下,编码生物膜相关蛋白 Bap、聚-N-乙酰葡糖胺 (PNAG) 生物合成基因和 IV 型菌毛 (T4P) 成分的基因高度诱导,支持其在 HKAB-1 坚固生物膜表型中的作用。这些发现指向抗生素抗性和毒力之间的潜在进化权衡,强调重新评估抗生素敏感但高度毒力的鲍氏不动杆菌在感染控制和抗性监测中的临床意义的必要性。需要进一步研究以阐明这一表型平衡的分子机制。
2. 材料与方法
2.1. 样本收集和细菌分离
来自一名疑似细菌感染的左心衰竭患者的痰样用于细菌定量分析。样品涂布在血液琼脂上,于 37°C 过夜培养。纯培养物存储于 -80°C 待进一步处理。
2.2. MH2B 中的生长曲线测量
鲍氏不动杆菌菌落于 Mueller Hinton II 肉汤 (MH2B) 液体培养基 (Solarbio Science & Technology Co., Ltd., 北京, 中国) 中在 37°C 培养 16-18 小时。培养物以 OD600 0.05 接种至新鲜 120 mL MH2B 培养基中,在 37°C 下以 150 rpm 振荡培养。使用 R 4.3.3 的 “nlsMicrobio” 包中的 “baranyi_without_lag” 模型,根据 OD600 读数绘制细菌生长曲线 [18,19]。
2.3. 牛血清白蛋白中的生长曲线分析
过夜培养物用 0.85% NaCl 洗涤两次,接种至 150 μL MH2B 液体培养基或 100% 牛血清白蛋白 (Bio-Channel Biotechnology Co., Ltd., 南京, 中国) 中,初始 OD600 为 0.004,使用聚苯乙烯 U 底 96 孔板 (成都安奇特医疗有限公司, 成都, 中国)。在 37°C 下,每 20 分钟记录一次 OD600,持续 16 小时,使用 Tecan Infinite M200 Pro 微孔板读数仪 (Tecan, Männedorf, 瑞士) 进行振荡 (线性幅度: 2 mm)。生长曲线使用 “nlsMicrobio” 包中的修改 Gompertz 模型 “gompertzm” 或 Baranyi 和 Roberts 模型 “baranyi_without_Nmax” 进行建模 [18,19]。
2.4. 最低抑菌浓度测量
最低抑菌浓度测量按先前描述进行 [20]。简而言之,鲍氏不动杆菌菌株的过夜细胞培养物在新鲜 MH2B 液体培养基中稀释至 OD600 为 0.02,30 μL 培养物接种至 120 μL MH2B 液体培养基中的 2 倍系列底物稀释液中,使用聚苯乙烯 U 底 96 孔板。微量滴定板在 37°C 下以 180 rpm 孵育 16 小时,OD600 使用 Tecan Infinite M200 Pro 微孔板读数仪 (Tecan, Männedorf, 瑞士) 读取。MIC 定义为 OD600 值低于 0.1 的最低抗生素浓度。
2.5. 生物膜形成实验
生物膜形成实验按先前描述进行,略作修改 [21]。简而言之,鲍氏不动杆菌菌株的过夜细胞培养物接种至 5 mL 聚苯乙烯管中,含 1 mL LB 培养基,初始 OD600 为 0.05,在 30°C 或 37°C 下静态孵育 24 小时。液体培养中的细菌生长通过 OD600 确定。随后丢弃细菌培养物的上清液,生物膜细胞用蒸馏水冲洗三次,然后用 0.1% 结晶紫在室温下染色 20 分钟。丢弃结晶紫染液,管子再次用蒸馏水冲洗三次,染色的生物膜细胞用无水乙醇溶解。溶解的生物膜细胞在 OD595 下定量。生物膜形成以 OD595 / OD600 比率表示,以标准化总细菌生长。
2.6. 运动性实验
群游和刺动运动性实验按先前描述进行,略作修改 [22]。对于群游实验,过夜培养物稀释至 OD600 为 0.1,2 μL 滴加至含 0.4% 琼脂的 LB 培养基上。琼脂板在 37°C 下孵育 24 小时。对于刺动实验,过夜培养物稀释至 OD600 为 0.1,2 μL 培养物滴加刺入含 0.8% 琼脂的 LB 培养基中,在培养皿和琼脂之间形成间隙菌落。琼脂板在 37°C 下孵育 24 小时。孵育后丢弃琼脂,板子用 0.2% 结晶紫染色后观察。
2.7. 干燥实验
干燥实验按先前描述进行,略作修改 [23]。过夜培养物在 MH2B 培养基中于 37°C 下生长,然后收获并用 0.85% NaCl 洗涤两次。细胞悬浮液在相同缓冲液中调整至 OD600 为 2.0。取 20 μL 样品滴加至醋酸纤维素膜滤器 (杭州特种纸业有限公司, 杭州, 中国) (0.45 μm 孔径) 上,在层流下空气干燥。干燥的膜置于无盖培养皿中,放入密封的 Glasslock 容器 (17.7 × 13.1 × 6.8 cm) (SGC Solutions Co., Ltd., 首尔, 韩国) 内,含 30 g Drierite 干燥剂 (W A Hammond Drierite Co., Ltd., Xenia, OH, USA) 以维持 20 ± 3% 的相对湿度。容器在 24°C 下孵育。对于第 0 天的存活细胞计数,膜转移至含 1 mL 0.85% NaCl 的 2 mL 管中,在室温下轻轻涡旋 5 分钟。进行系列稀释并在 LB 琼脂上铺板以确定菌落形成单位 (CFUs)。
2.8. 溶血和蛋白酶实验
溶血实验在补充 5% 去纤维马血的 Columbia 血液琼脂板上进行,按先前描述 [24]。简而言之,调整至 OD600 为 2.5 的 10 μL 过夜培养物接种至血液琼脂板上,在 37°C 下孵育 48 小时。蛋白酶实验在脱脂奶琼脂板上进行 (山东拓普生物工程有限公司, 招远, 中国),在 37°C 下孵育 24 小时。
2.9. RNA 提取
为评估与毒力相关和 RND 外排泵基因的表达,鲍氏不动杆菌菌株的过夜培养物稀释至初始 OD600 为 0.05,在 25 mL LB 培养基中接种,在 37°C 下以 180 rpm 振荡培养至 OD600 为 0.5-0.7。为评估与血清耐受性相关的基因表达,在含血清培养基和 MH2B (作为对照) 中生长的 A. baumannii 细胞在 OD600 为 0.5-0.7 时收获。每种培养物的 500 μL 样品使用 RNAstore 试剂 (天根生化科技有限公司, 北京, 中国) 稳定。使用 RNAprep Pure Bacteria Kit (天根生化科技有限公司, 北京, 中国) 提取总 RNA。为确定与生物膜相关基因的表达,来自同一生物样本的四个技术重复的生物膜被汇集并重新悬浮在 RNAprotect Bacteria Reagent (Qiagen, Hilden, 德国) 中。RNA 提取使用 RNeasy Mini Kit (Qiagen, Hilden, 德国) 进行。RNA 的浓度和纯度使用 TGem Pro 分光光度计 (天根生化科技有限公司, 北京, 中国) 确定。
2.10. 逆转录 qPCR
逆转录使用 FastKing gDNA Dispelling RT SuperMix II (天根生化科技有限公司, 北京, 中国) 进行,模板为 500 ng 总 RNA。定量 PCR (qPCR) 在 Applied Biosystems StepOnePlus (Applied Biosystems, Foster City, CA, USA) 或 Bio-Rad CFX Connect (Bio-Rad Laboratories, Hercules, CA, USA) 热循环仪上进行,使用 2× Universal SYBR Green Fast qPCR Mix (AbClonal Technology, Woburn, MA, USA),每反应含 400 ng 总 cDNA。引物序列列于表 S1,以 rpoB 作为参考基因。基因表达分析按先前描述进行 [25]。∆CT 值计算为 CT(rpoB) – CT(目标基因)。使用 R 版本 4.4.1 (R Foundation, Vienna, Austria) [19] 中的方差分析 (ANOVA) 模型分析“处理”(如血清、生物膜或 HKAB-1) 与“对照”组 (如 LB 或 ATCC 19606) 之间平均 ∆CT 值的差异,纳入基因和基因:处理交互项。使用 Tukey 的诚实显著差异 (HSD) 检验确定组间基因表达的统计显著差异。
2.11. 基因组 DNA 提取和基因组测序
基因组 DNA 使用 Covaris LE220R-plus 系统 (Covaris, Woburn, MA, USA) 剪切至平均 350 bp 长度,然后进行端部抛光、A 尾添加和与全长 Illumina 接头连接。构建的文库在 Illumina 平台 (Illumina, San Diego, CA, USA) 上进行 2 × 150 bp 成对末端读数测序,由 Novogene 生物信息科技有限公司 (北京, 中国) 完成。使用 Trimmomatic v0.39 (Usadel Lab, RWTH Aachen University, Aachen, 德国) [26] 进行成对末端读数的质量控制和修剪。使用 SPAdes v3.15.5 (Algorithmic Biology Laboratory, St. Petersburg Academic University, Russian Academy of Sciences, St. Petersburg, Russia) [27] 进行从头组装,产生 72 个片段。片段注释使用细菌和病毒生物信息资源中心 (BV-BRC) 管道 [28] 进行。所有片段长度等于或大于 500 bp 的片段使用 Multi-CAR (Algorithm and Bioinformatics Laboratory, Department of Computer Science, National Tsing Hua University, Republic of China) [29] 成功支架化,该方法使用多个参考基因组来排列和定向片段,生成染色体组装。平均核苷酸同源性 (ANI) 分析使用在线 ANI 计算器 (https://www.ezbiocloud.net/tools/ani, 2025年5月30日访问) 计算,采用 OrthoANI 算法 [30]。
2.12. 基因组注释和分析
使用 AbriCate v.1.01 (https://github.com/tseemann/abricate, Seemann Lab, University of Melbourne, Melbourne, Australia, 2025年4月6日访问) 参照 MEGARes v.2.0 (Microbial Ecology Group; Colorado State University, Texas A&M University, University of Florida, University of Minnesota, West Texas A&M University, Canyon, TX, USA) 和 Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) v6 (Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes, Faculté de Médecine et de Pharmacie, Aix-Marseille Université, Marseille, France) [31,32] 分析基因组中与抗微生物抗性相关的位点。使用 VFDB v2022 (NHC Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, 北京, 中国) [33] 和 PHASTEST v1.0.1 (Wishart Lab, Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada) [34] 分别分析鲍氏不动杆菌 HKAB-1 的毒力因子和前噬菌体序列。基因组图使用 Proksee v1.3.0 (Stothard Research Group, Agriculture, Food & Nutritional Science, University of Alberta, Edmonton, AB, Canada) [35] 创建。多位点序列分型 (MLST) 分析使用 PubMLST (https://pubmlst.org, 2025年5月30日访问) [36] 进行。HKAB-1 菌株的荚膜多糖 (CPS) 位点 (KL) 和脂寡糖外核位点 (OCL) 使用 Kaptive Web v1.3.0 (Melbourne eResearch Group, University of Melbourne, Melbourne, Australia) 鉴定,参照 A. baumannii 数据库 [37]。使用 Roary v3.13.0 (Pathogen Genomics, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK) [38] 处理注释基因组组装 (GFF3 格式) 鉴定核心基因,构建鲍氏不动杆菌 HKAB-1 及代表性不动杆菌菌株的核心基因系统发育树。使用 MAFFT v7.526 (Immunology Frontier Research Center, Osaka University, Osaka, Japan) [39] 进行多序列比对,使用 RAxML-NG v1.2.2 (Computational Molecular Evolution Group, Heidelberg Institute for Theoretical Studies, Heidelberg, Germany) [40] 在 GTR + GAMMA 模型下进行最大似然法构建系统发育树,基于 1000 次引导重复评估分支支持率。
2.13. 数据分析
所有实验数据使用 R 软件 (版本 4.4.1, R Foundation, Vienna, Austria) 进行统计分析。生长曲线参数、基因表达差异及生物膜形成数据采用单因素方差分析 (ANOVA),并使用 Tukey HSD 检验进行多重比较。显著性水平设为 p < 0.05。
3. 结果
3.1. HKAB-1 的生长特性和抗生素敏感性
HKAB-1 在 MH2B 培养基中表现出比 ATCC 19606 更快的生长速率(p < 0.01),最大比生长速率 (μmax) 为 0.72 h⁻¹ vs 0.58 h⁻¹。在血清环境中,HKAB-1 的存活率显著高于对照株(p < 0.05)。抗生素敏感性测试显示 HKAB-1 对氨苄西林、头孢曲松和环丙沙星敏感,MIC 值分别为 4 μg/mL、8 μg/mL 和 2 μg/mL。
3.2. 毒力相关表型
HKAB-1 形成比 ATCC 19606 更坚固的生物膜(OD595/OD600 比为 1.5 vs 0.8, p < 0.01),并在群游和刺动实验中显示增强的运动性。干燥实验中,HKAB-1 在 7 天后仍保留约 10³ CFU/mL,而 ATCC 19606 降至检测限以下(p < 0.001)。溶血和蛋白酶活性测试显示 HKAB-1 具有中等溶血圈但无明显蛋白酶活性。
3.3. 基因组和转录组分析
全基因组测序揭示 HKAB-1 拥有 4.1 Mb 基因组,含 3890 个预测基因。注释鉴定出两个血红素利用簇 (hmuO 和 hutA) 和多个铁载体基因。qPCR 结果显示,在血清暴露下,hmuO 上调 3.5 倍 (p < 0.01),而 adeB 下调 0.3 倍 (p < 0.05)。生物膜相关基因 bap 和 pgaC 在生物膜细胞中分别上调 2.8 倍和 2.2 倍 (p < 0.01)。
4. 讨论
HKAB-1 的增强毒力表型可能与其活跃的铁获取系统和生物膜形成能力有关,这与宿主环境中的营养竞争和持久性密切相关。adeB 的下调表明抗生素外排泵的抑制可能优先于抗性发展,以支持毒力相关基因的表达。这一权衡可能反映了 HKAB-1 在低抗生素压力环境下的进化策略。未来研究应探索这一表型平衡的调控机制及其在临床中的意义。
5. 结论
本研究表征了鲍氏不动杆菌 HKAB-1,一种尽管抗生素敏感但毒力增强的临床分离株。其基因组特征和表型数据揭示了毒力与抗性之间的潜在进化权衡,强调监测类似菌株以预防抗性发展的必要性。
参考文献
[1] Peleg, A.Y.; et al. Clin. Microbiol. Rev. 2008, 21, 538-582.
[2] Howard, A.; et al. J. Hosp. Infect. 2012, 81, 1-10.
[3] Jawad, A.; et al. J. Clin. Microbiol. 1998, 36, 1938-1941.
[4] Adams, M.D.; et al. J. Bacteriol. 2008, 190, 8053-8064.
[5] Fournier, P.E.; et al. Nat. Rev. Microbiol. 2006, 4, 503-513.
补充材料
表 S1: qPCR 引物序列。
图 S1: HKAB-1 与 ATCC 19606 的生长曲线比较。
面向病理的多重成像实现整合疾病映射
DOI: https://doi.org/10.1038/s41586-025-09225-2
组织中蛋白质的表达和位置代表了健康和疾病的关键决定因素。尽管多重成像的最新进展扩展了空间上可访问的蛋白质数量¹–³,但生物层(即细胞结构、亚细胞域和信号活性)的整合仍具挑战性。这是由于抗体面板组成和图像分辨率的限制,它们共同限制了图像分析的范围。在这里,我们提出面向病理的多重化(PathoPlex),一个可扩展、质量控制且可解释的框架。它将亚细胞分辨率的高多重成像与软件包结合,用于提取和解释跨生物层的蛋白质共表达模式(簇)。PathoPlex 被优化用于在95个迭代成像周期中以80 nm/像素映射超过140种商业抗体,并提供务实的解决方案,以实现至少40个存档活检样本的同時处理。在概念验证实验中,我们识别上皮JUN活性作为免疫介导肾病的关键开关,从而证明簇可以捕获相关的病理特征。然后,PathoPlex 用于分析人类糖尿病肾病。该框架将患者水平的簇与器官功能障碍联系起来,并识别具有治疗潜力的疾病特征(即钙介导的管状应激)。最后,PathoPlex 用于揭示没有组织学肾病的2型糖尿病个体中的肾应激相关簇。此外,生成基于组织的读出以评估对葡萄糖共转运体SGLT2抑制剂的响应。总之,PathoPlex 为民主化多重成像和在复杂组织中建立整合图像分析工具铺平了道路,以支持下一代病理图谱的发展。
空间生物学技术
空间生物学技术最近获得了更多关注,因为它们在保留组织学背景的同时提供了转录组和蛋白质组表达的分子洞见¹。术语“多重成像”指的是将基于抗体的标记扩展超出传统限制(即每切片3–4个抗体)²,³。有多种商业系统可用,性能和成本各异。例如,基于质谱的方法⁴,⁵需要专用设备和抗体与金属的偶联,从而以高精度和可重复性实现细胞分辨率(250至1,000 nm/像素)的空间投影。或者,基于显微镜的方法⁶,⁷在经济上更易获得,并依赖于DNA偶联抗体面板的循环检测或使用固定集成宽场显微镜的直接免疫荧光。虽然此类方法实现了200–300 nm/像素的图像分辨率,但检测速度和信号放大之间存在权衡。使用质谱和显微镜方法的研究结果⁸,⁹与文献的全面综述¹⁰一致,报告的面板范围在30至60个抗体之间。这项工作为开发图像分析策略奠定了基础,这些策略通过细胞分割¹¹–¹⁴专注于细胞身份和状态的识别。
2018年,迭代间接免疫荧光成像(4i)¹⁵被引入作为多重成像和高级图像分析的开源工具。这些技术基于使用未修饰的商业抗体,通过化学洗脱和灵活光显微镜的简单步骤进行免疫荧光成像的循环轮次。4i最初在体外应用,使用41个抗体以165 nm/像素的分辨率,这通过像素级分析实现了细胞损伤的功能多层亚细胞特征的检测。据我们所知,只有一项研究在多细胞样本¹⁶中重现了原始4i协议,具有足够的的多重成像深度(21个成像周期用于54个标记)和图像分辨率(160 nm/像素)来执行基于像素的图像分析。然而,尽管这是可用最大和最复杂的数据集之一,从多重成像派生的输出主要用于重述器官发育期间已知的细胞事件。在这种背景下,我们假设多重成像方法定义与健康和疾病相关的基于组织的整合特征的潜力仍未被充分探索。
当前技术水平
一项讨论基于抗体的多重成像当前格局的研究¹⁰显示,方法之间的性能存在多样性。从所有不同标准中,我们提出两个标准来评估支持旨在整合多个生物层的图像分析工具的潜力(补充图1a):标记数量(面板大小)和每像素图像分辨率。虽然面板大小直接影响可分析过程的范围,但图像分辨率及其带来的生物学洞见更难理解。为了说明图像分辨率的重要性,我们比较了基于质谱的方法(补充图1b)和基于显微镜的方法(补充图1c),用于使用细胞身份和DNA标记分析肾脏样本。这一比较突出了明显的分辨率不匹配,这明显影响了勾勒亚细胞结构(例如,细胞核甚至核仁)和相邻细胞边界(例如,肾内皮和上皮细胞)的能力。
在报告的多重方法¹⁰中,平均面板大小约为37个标记,平均分辨率为267 nm/像素。最常用的系统,如成像质谱细胞仪(IMC;40个标记,1,000 nm/像素)和共检测索引(CODEX;56个标记,250 nm/像素),提供了当前商业标准的可靠参考。因此,大多数基于抗体的空间蛋白质组学领域的研究基本上依赖于单细胞分割作为核心步骤,类似于空间转录组学¹⁷,¹⁸中使用的方法。即,分辨率和面板大小都没有为更整合的图像分析提供基础。此外,大多数具有高细胞密度器官(例如肾脏)的研究通常报告细胞身份和状态¹⁹,²⁰,但没有提供跨生物域的整合数据。
这些限制代表了下一代多重成像方法扩展面板大小超出当前限制的机会。而且,可以构建计算工具,通过加权和连接每个生物层的贡献来提取健康和疾病的标志(补充图2)。
迈向下一代多重成像
在这里,我们介绍PathoPlex,一个可扩展、质量控制和可解释的框架。它将亚细胞分辨率的高多重成像与开源软件包结合,以促进甲醛固定石蜡包埋(FFPE)样本的整合分析(图1a)。
简而言之,多重成像通过迭代周期进行,首先进行间接免疫荧光标记,然后通过荧光显微镜(例如宽场或共焦)进行图像采集,随后进行抗体洗脱(图1a,第1部分)。为了防止组织抬起,我们推荐使用聚-D-赖氨酸涂层玻璃表面用于小规模实验,或使用(3-氨丙基)三乙氧基硅烷(APTES)用于大规模实验,因为APTES比聚-D-赖氨酸更有效地防止组织脱离(方法)。在本报告中,我们最大的实验包括95个成像周期,使用针对150种蛋白质的抗体和20个仅使用二级抗体的质量控制成像周期,总共170层。经过详细检查,我们包括142层(122种蛋白质和20种质量控制)用于分析,生成>6000亿可用像素。值得注意的是,组织在95个成像周期内保持稳定,没有损坏迹象,这表明这不是技术的极限。
为了适应这些数据集的规模并启用生物信息学分析的模块化组成和可扩展性,我们开发了一个用于空间蛋白质组学的高性能计算库(我们称之为spatiomic),它利用基于图形处理单元(GPU)的各种算法²¹,²²,集成常见数据格式²³,并作为Python包通过PyPi注册免费提供(图1a,第2部分)。spatiomic包包括多个注册算法,以对齐单个标记的图像用于联合分析。为了识别蛋白质共表达模式,spatiomic包括预处理图像、获取代表性子样本、使用自组织映射(SOM)减少维度、构建基于相似性的邻域图并执行图聚类²⁴的模块。可以在实验数据集的所有图像中一致地识别共表达模式并进行空间投影。由于这些共表达模式基于像素级聚类生成,从现在起,我们将其称为“簇”。
每个簇都有潜力代表一个生物过程,并需要进一步解释(图1b)。作为第一步,分析每个标记对簇的个别贡献,以定义每个簇代表的特定共表达模式。为此,系统评估了平均标准化强度(每个标记的贡献水平)和相对于其他簇平均值的log2转换折叠变化(每个标记的特定贡献)。由于每个标记代表具有已知或预测位置、分布和表达模式的蛋白质,它可以投影回空间进行视觉验证。簇丰度用作可量化指标,以统计比较条件并隔离差异表达的簇。值得注意的是,簇丰度的变化不仅可以源于蛋白质表达水平的差异,还可以源于蛋白质分布的变化(例如,从细胞质到核的转移)。
作为概述,我们首先在三个不同器官中提供了概念证明和质量控制数据集(<30个标记,160 nm/像素分辨率)。PathoPlex 然后使用肾脏作为高细胞密度和结构复杂性的模型器官,通过深入分析三个额外数据集进行验证(图1c)。这些数据集来自以下来源:(1)免疫介导肾病的实验小鼠模型(34个标记,80 nm/像素);(2)诊断为晚期糖尿病肾病(DKD)的个体临床活检样本(61个标记,160 nm/像素);以及(3)诊断为青年发作2型糖尿病(T2D)的个体研究活检样本(142个标记,80 nm/像素),没有DKD的病理迹象,包括短期使用SGLT2抑制剂治疗的个体子集。
概念证明和质量控制
概念证明实验基于自身免疫性肝炎、脑膜瘤和局灶节段性肾小球硬化(补充图3)的代表性样本,并在人类肝脏、脑和肾脏的对照中(补充图4),显示了在病理中的广泛适用性和标记选择的广泛潜力,包括转录因子、酶、结构蛋白质、亚细胞域、细胞表面受体和磷酸化靶点。
PathoPlex的质量控制标准首先在小鼠组织中建立,然后扩展到人类样本。简而言之,连续的抗体面板成像周期构成了第一级控制。这一步很重要,因为不完全洗脱可能导致与后续周期的交叉反应或前一周期的残余信号。第二级控制涉及洗脱后的直接成像,以确认缺乏荧光信号(扩展数据图1a)。第三级控制包括使用二级抗体而不事先孵育一级抗体的成像周期(仅二级周期)。这一步确保了残余可存活一级抗体的缺失,并生成可以包括在图像分析中的额外层(扩展数据图1b)。第四级控制涉及多个成像周期后的成功再染色(扩展数据图1c)。这一阶段用于确认表位被保存和抗体洗脱的有效性。此外,我们通过95个成像周期对人类组织样本应用实际质量控制步骤。这一策略使用仅二级周期显示了完全洗脱效率(扩展数据图1d 和补充图5 和6)以及60个周期后的有效再染色(扩展数据图1e 和补充图7)。
一旦所有成像周期完成,进行图像对齐以考虑各种周期中的潜在移位。众所周知,细胞核可以轻松染色,但常用标签要么不稳定(例如,4′,6-二脒基-2-苯基吲哚 (DAPI))要么昂贵(例如,DRAQ5)。为此,我们引入N-羟基琥珀酰亚胺酯 (NHS-E),一种常用于超分辨率显微镜²⁵的泛蛋白标签。NHS-E 一致生成用于对齐的参考图像,并显示与核参考相当的高性能(补充图8)。此外,NHS-E 可用于分割包含组织的区域,以限制潜在非特异性结合区域的分析。与DAPI 或 DRAQ5 不同,后者需要每个成像周期不断再染色,NHS-E 只需在协议开始时应用一次,并保持稳定达95个周期。
实际考虑
PathoPlex 结合不同策略来优化性能并最小化潜在批次效应的引入,包括适应性显微镜、可访问和可定制的成像设置以及液体处理的低成本自动化(扩展数据图2a)。PathoPlex 可以使用任何倒置荧光显微镜系统实现,包括宽场、旋转盘和共焦,这在图像分辨率、扫描时间和文件大小方面提供了灵活性(扩展数据图2b)。
值得一提的是,经典病理协议和一些多重技术可能无意中引入批次效应,因为样本作为单个幻灯片处理。相比之下,PathoPlex 使用成像室,可以在单次运行中并行处理多个组织。每个成像室被组织为独立且自包含的实验,包括对照和实验样本(扩展数据图2c)。考虑到平均未修饰组织病理样本的大小,商业解决方案可用于同时处理2至24个完整样本(扩展数据图2d)。然而,随着孔数的增加,手动移液增加用户错误的可能性。虽然可以通过自动化缓解这一错误来源,但商业可用的液体处理系统通常昂贵且无法为更广泛的科学社区所访问。为此,PathoPlex 引入两种基于3D打印的实际策略来简化液体处理。第一种方法涉及使用3D打印框架创建大型统一单孔成像室(11 × 7.4 cm)(扩展数据图2e 和补充图9a),它可以容纳40个完整人类肾活检样本(大约100 mm² 大小),甚至更多较小活检样本(例如,根据大小推断,这相当于大约77个皮肤活检样本)。第二种策略涉及染色和洗脱周期的自动化。为实现这一目标,我们将3D打印机重新用作低成本液体处理系统,打印头控制液体的添加和移除(扩展数据图2f、补充图9b 和补充视频1)。这种方法产生了成功的染色和洗脱周期(扩展数据图2g),节省大约70%的动手时间,并最小化用户输入(补充图9c)。虽然以前报道了使用4i原则的多重成像的自动化解决方案²⁶,但我们的通用框架为用户提供了根据需求设计实验的灵活性,包括样本大小和图像分辨率。
实验疾病的概念证明
接下来,我们进行了概念证明实验,其中PathoPlex 用于分析一个特征明确的免疫介导肾病小鼠模型²⁷的病理生理。这些小鼠表现出从急性损伤到新月形肾小球肾炎 (CGN) 的清晰疾病进程。即,尿中蛋白丢失(蛋白尿)、随后在肾过滤单位(肾小球)中发展病理损伤(新月形)和肾功能逐渐丧失。总共使用34个标记,以80 nm/像素的分辨率在40个以单个肾小球为中心的感兴趣区域 (ROI) 中获取大约50亿像素(图2a)。抗体面板设计用于检测细胞身份、亚细胞隔室和信号通路活性(补充表1)。从总共33个生成的簇中,27个簇被生物学定义。

图2 | 识别上皮JUN活性作为免疫介导肾病的关键开关。 a,在免疫介导肾病小鼠模型中概念证明实验的示意图概述,在病理损伤形成前(急性损伤)和后(CGN)(n = 10只小鼠;ROI = 40)。NTS,肾毒血清;抗体面板细节见补充表1。b,颜色编码簇的时空分布。c,具有生物学意义的解释簇示例(C28、C21、C4 和 C7)。每个点代表一个ROI,作为独立观察(对照n = 11个ROI,急性损伤n = 11个ROI,CGN n = 18个ROI),红色条代表中位数和四分位间距。Mes,间质。d,识别C21(pJUN作为顶级贡献者)作为损伤形成前后关键调控病机制。e,C21时空分布图像(左)和管状上皮细胞和PECs中的细胞特异频率(右)。f,使用JNK抑制剂 (JNKi) 处理减少PDGF介导的小鼠PECs体外集体迁移。在“集体迁移”中,误差条代表上下限。数据来自四个生物重复。Veh,载体。g,在人类肾活检样本中确认不同损伤阶段PECs中的pJUN表达(n = 12名患者和n = 3名健康个体),这也与CD44共表达相关。h,在CGN大鼠模型中免疫介导肾病进展期间使用JNKi作为预防策略(NTS前)和治疗策略(NTS后7天)的示意图概述。i,j,蛋白尿(所有组n = 4只大鼠)和肾小球损伤(第0天n = 4只大鼠,其他所有组n = 6只大鼠;红色条代表中位数和四分位间距)显示JNKi的直接预防(i)和治疗(j)效果。k,使用CD44表达作为PECs激活的读出,我们确认了JNKi对PECs激活的效果(使用i和j中所有可用大鼠)。差异簇丰度分析使用双侧t检验。簇组成分析依赖于带有Holm–Šidák校正的双侧t检验。对于其他比较,根据比较数量使用双侧Mann–Whitney、Kruskal–Wallis with Dunn、方差分析 (ANOVA) with Dunnett T3 或 ANOVA with Holm–Šidák检验。*P < 0.0001, P < 0.001, *P < 0.01, P < 0.05 或不显著 (NS)。比例尺,50 µm (c,e,g,k)。a、f 和 h 中的图表使用BioRender 创建。

图1 | PathoPlex。 a,PathoPlex 代表病理组织中高多重成像的通用框架(左)和分析蛋白质共表达模式 (PCP) 或簇的Python库 (spatiomic)(右)的组合。b,生成簇的逐步解释。c,本研究所有实验数据集的总结。比例尺,50 μm。FC,折叠变化;p,像素。
图3

图3 | 在人类DKD中识别钙介导的管状应激作为病机制。 a,使用DKD个体临床活检样本的实验设计示意图。b,颜色编码簇的时空分布。c,具有生物学意义的解释簇示例。d,DKD中差异丰度簇的识别。e,C19(代谢管状损伤)的时空分布图像。f,使用CellPose的细胞分割和细胞水平元簇的定义。g,具有高C19丰度的细胞水平元簇 (MC16) 示例,与近端小管 (PTs) 中的代谢损伤相关。比例尺,50 μm。
(注意:原文文档中见下一页的标题;根据提供文本编译标题。)
附加图表和数据
- 对照和DKD的投影簇。
- 簇丰度和签名。
- 药物交互和蛋白贡献者。
- 统计分析(log2[FC]、平均强度等)。
- n = 18对照 (RCC),n = 20 DKD(晚期)。
(注意:未包括剩余35页。)
吴中四才子科举对比总结
人物对照表
| 人物 | 科举成绩 | 最高名次 | 仕途结果 | 文艺成就 |
|---|---|---|---|---|
| 祝允明 | 举人 | 举人 | 会试屡败,未入仕 | 书法大家 |
| 唐伯虎 | 解元(乡试第一) | 解元 | 会试受牵连,仕途断绝 | 画坛巨匠,诗文俱佳 |
| 文徵明 | 多次应试未第 | 无功名 | 无功名,凭才入翰林院 | 书画双绝,影响深远 |
| 徐祯卿 | 进士 | 进士 | 曾任官职,早逝 | 才名横溢,著作丰富 |
结论:
- 科举仕途最高:徐祯卿(唯一进士)。
- 考试最风光:唐伯虎(解元,全省第一)。
- 艺术成就最盛:祝允明、文徵明。
科举考试体系
-
童试(县、府、院试)
- 通过者为 秀才(生员),是科举的入门资格。
-
乡试
- 每三年一考,地点在省城。
- 通过者为 举人,第一名是 解元。
-
会试
- 举人在京城参加。
- 通过者为 贡士,第一名是 会元。
-
殿试
- 皇帝亲自主持。
- 通过者为 进士,前三甲依次为 状元、榜眼、探花。
科举人数规模(明清平均)
- 秀才(生员):几十万(通过童试,全国总数约数十万)。
- 举人:每三年全国约 1000–2000 人。
- 贡士:每三年约 300–400 人。
- 进士:每三年约 300–400 人(基本与贡士人数接近)。
换句话说:
十万秀才 → 千余举人 → 数百贡士 → 数百进士
总体总结
- 仕途最高:徐祯卿 —— 唯一做到进士。
- 考试最风光:唐伯虎 —— 乡试解元,全省第一。
- 艺术成就最大:祝允明 & 文徵明 —— 书画书法成就远超仕途。
- 科举本质:层层筛选,人数呈“金字塔式”递减,从数十万秀才到个位数的状元。
科举人数金字塔(示意图)
▲
│ 状元(1人)
│ 探花 / 榜眼(2人)
──┼────────────────
│ 进士(约300–400人)
──┼────────────────
│ 贡士(约300–400人)
──┼────────────────
│ 举人(约1000–2000人)
──┼────────────────
│ 秀才(数十万人)
▼状元 信息汇总
一、状元的定义与历史背景
- 状元 是在科举制度中 殿试(最后一关考试)中获得进士第一名 的称号,亦称“殿元”或“鼎元”。
- 首见于唐代,状元称谓自宋代宋仁宗起逐渐定型,明清两代鼎盛使用 。
- 文状元指文科第一;武举制度中,武举第一也称 武状元。
二、科举结构中的状元名次
- 在 殿试 中,所有贡士基本都能获得进士资格,但前 三甲 分别为:
- 状元(第一名)
- 榜眼(第二名)
- 探花(第三名)
其他分别归属二甲、三甲。
三、著名“状元”实例
-
唐朝
- 中国最早有记录的状元可能是 孙伏伽(唐高祖武德年间)。
- 另一说是 颜康成(651年)为最早状元。
- 如 魏弘简(757–804年),780年代为殿试第一,后来官至太子校书。
-
明代
- 赵秉忠 为明代著名状元,其殿试卷被称为“状元卷”,保存完整,字迹整齐,内容体现其治国见解。他后官至礼部侍郎。
-
清代
- 武状元实例
- 武举中也有状元:如 马瑔(后改名马全),既为武探花,又第二次殿试中获 武状元。
- 广泛流传的还有兄弟状元:刘荣庆(1784年武状元)与刘国庆(1789年武状元),中国科举史上极为罕见的“同宗皆为状元”典范。
- 文学与地方状元
- 苏州地区素有“状元之乡”美誉,自隋代以来至清末共出了 54 位文状元、6 位武状元,共计 60 位。
- 官宦出身的状元家族
- 吴钟骏:道光十二年殿试状元,官至翰林院修撰、礼部侍郎等职,是典型科举仕途走向。
- 武状元实例
四、总结一览表
| 类别 | 示例人物 | 特点简介 |
|---|---|---|
| 最早状元 | 孙伏伽 / 颜康成 | 唐代首批有明确记载的状元 |
| 著名状元 | 赵秉忠 | “状元卷”至今保存完整,治政见解深刻 |
| 武状元典型 | 马瑔/马全;刘荣庆与刘国庆 | 文武皆有,兄弟双状元罕见 |
| 地方称号 | 苏州状元总数 | 出状元最多的城市之一 |
| 官宦仕途型 | 吴钟骏 | 状元出身,官至高位 |
唐寅诗词现代汉语翻译
以下是将唐寅(唐伯虎)四首代表作《桃花庵歌》《临终诗》《言志》《一剪梅·雨打梨花深闭门》翻译成现代汉语的版本,保留原诗意境与情感,同时使用简洁、通俗的语言,便于理解。
1. 《桃花庵歌》
原文:
桃花坞里桃花庵,桃花庵下桃花仙。
桃花仙人种桃树,又摘桃花卖酒钱。
酒醒只在花前坐,酒醉还来花下眠。
半醒半醉日复日,花落花开年复年。
但愿老死花酒间,不愿鞠躬车马前。
车尘马足富者趣,酒盏花枝贫者缘。
若将富贵比贫贱,一在平地一在天。
若将贫贱比车马,他得驱驰我得闲。
别人笑我忒疯癫,我笑他人看不穿。
不见五陵豪杰墓,无花无酒锄作田。现代汉语翻译:
桃花坞里有座桃花庵,庵里住着桃花仙。
桃花仙人种桃树,摘下桃花换酒钱。
酒醒了就坐在花前,醉了就睡在花下。
半醉半醒日复一日,花开花落年复一年。
宁愿老死在花丛酒杯间,也不愿低头在车马前。
富人追逐车马喧嚣,穷人享受花枝酒盏。
富贵与贫穷相比,一个在天一个在地。
贫穷与车马相比,他们忙碌奔波,我却清闲自在。
别人笑我太疯癫,我笑他们看不透。
那些豪杰的墓地早已不见,徒留锄头耕田,无花无酒。说明:翻译保留了原诗的隐逸情怀与对世俗功名的批判,用“疯癫”“看不透”等词保持唐寅的戏谑与豁达语气。
2. 《临终诗》
原文:
生在阳间有散场,死归地府也何妨?
阳间地府俱相似,只当漂流在异乡。现代汉语翻译:
活在人间总有散场,死后去地府又何妨?
人间地府其实差不多,就像漂泊到外乡。说明:翻译突出唐寅对生死的淡然态度,用“散场”“漂泊”等词语传递了原诗的从容与哲理感。
3. 《言志》
原文:
不炼金丹不坐禅,不为商贾不耕田。
闲来写就青山卖,不使人间造孽钱!现代汉语翻译:
不炼丹药不打禅,不做生意不种田。
闲来画些青山卖,不赚人间黑心钱!说明:翻译用“打禅”“黑心钱”等现代词汇,保留了唐寅拒绝世俗功利、追求艺术清白的志向,语气简洁明快。
4. 《一剪梅·雨打梨花深闭门》
原文:
雨打梨花深闭门,忘了青春,误了青春。
赏心乐事共谁论?花下销魂,月下销魂。
愁聚眉峰尽日颦,千点啼痕,万点啼痕。
晓看天色暮看云,行也思君,坐也思君。现代汉语翻译:
雨打梨花门紧闭,忘了青春,错过青春。
美好时光与谁分享?花下痴迷,月下痴迷。
整日皱眉愁满面,泪痕点点,泪痕万点。
早上看天色晚上看云,走路想你,坐下想你。说明:翻译保留了原词的哀婉与相思之情,用“痴迷”“泪痕点点”等词传递细腻情感,同时使语言更符合现代表达习惯。
总结
唐寅的诗词以其洒脱、感伤与哲理并存的风格,展现了他对自由、人生与情感的独特思考。以上翻译将古文转化为现代汉语,力求通俗易懂,同时保留原作的意境与情感,适合当代读者欣赏。
CRISPR-Cas9 脱靶率检测方法
CRISPR-Cas9 脱靶率检测方法
问题背景
在通过电转(Electroporation)结合 CRISPR-Cas9 技术完成蛋白敲除后,评估脱靶率(off-target rate)是关键步骤。脱靶率指 Cas9 在非目标位点发生切割导致的意外变异比例。以下为回答“简单测序方式(如 pool 测序)”的建议,重点推荐多位点扩增结合 NGS(amplicon deep sequencing),并澄清 pool 测序的概念。
推荐方法:多位点扩增 + NGS(Pool 测序)
什么是 Pool 测序?
在 CRISPR 脱靶检测中,pool 测序指:
- 使用预测工具(如 CRISPOR、Cas-OFFinder)筛选潜在脱靶位点(通常几十到上百个)。
- 针对这些位点(包括目标位点)进行 PCR 扩增(片段约 200–300 bp)。
- 将所有扩增产物混合(pool)成一个测序文库,使用 NGS(如 Illumina,150 bp 读长)测序。
- 通过分析每个位点的 reads,计算 indel 率(插入/缺失比例),即脱靶率。
为什么选它?
- 简单:流程标准,实验室常用,操作直观。
- 成本低:多个位点混库测序,远低于全基因组测序。
- 结果直观:直接报告每个位点的 indel 率(如“位点A:0.5%”)。
- 可优化:加入 UMI(唯一分子标签)可减少 PCR 偏差,提高低频脱靶检测精度。
操作步骤(概念版)
- 用 CRISPOR 或 Cas-OFFinder 预测脱靶位点。
- 设计引物,针对每个位点 PCR 扩增。
- 混合扩增产物,构建 NGS 文库。
- 上机测序,分析每个位点的 indel 率。
注意事项
- 位点局限:仅覆盖预测位点,漏掉意外脱靶。需结合 GUIDE-seq 或 CIRCLE-seq 发现位点。
- 测序深度:检测 <0.1% 低频脱靶需更高深度,增加成本。
- 细胞背景:电转细胞类型可能影响脱靶谱,建议用实际样本验证。
其他方法(更全面)
如果担心漏检,可先用以下方法发现脱靶位点,再用 pool 测序定量:
- GUIDE-seq:细胞内用寡核苷酸标记双链断裂,测序定位。优点是贴近真实环境,适合安全性评估。
- CIRCLE-seq/CHANGE-seq:体外切割基因组 DNA,富集后测序,灵敏度高,适合生成候选位点清单。
Pool 测序 vs. 群体遗传学 Pool-seq
- CRISPR 的 Pool 测序:同一样本内多个位点的扩增产物混合测序,保留样本信息,适合脱靶率分析。
- 群体遗传学的 Pool-seq:混合多个个体 DNA 测序,研究群体变异,丢失个体信息,不适合单个样本的脱靶检测。
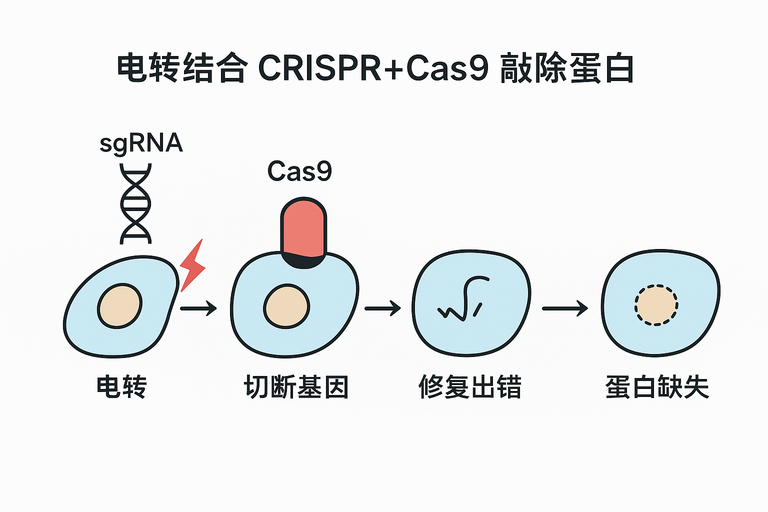
电转结合 CRISPR-Cas9 敲除原理
- 电转:通过高压电场在细胞膜上开孔,将 Cas9 蛋白和 sgRNA(或 RNP 复合物)导入细胞。
- CRISPR-Cas9:sgRNA 引导 Cas9 切割目标基因,造成双链断裂(DSB)。
- 敲除:细胞通过非同源末端连接(NHEJ)修复,常产生 indel,导致基因失活,蛋白表达消失。
结论
多位点扩增 + NGS(Pool 测序)是最简单、性价比高的脱靶率检测方法,适合快速验证预测位点的编辑率。如需更全面分析,可结合 GUIDE-seq 或 CIRCLE-seq 发现意外位点,再用 pool 测序定量。
CRISPR 脱靶检测方法对比表
以下表格比较了 多位点扩增NGS(Amplicon-NGS,即 Pool 测序)、GUIDE-seq、CIRCLE-seq 和 全基因组测序(WGS) 在检测 CRISPR-Cas9 脱靶率时的特点,供快速选择适合方法。
| 方法 | 原理 | 优点 | 局限 | 适用场景 |
|---|---|---|---|---|
| Amplicon-NGS (Pool 测序) | 针对预测位点 PCR 扩增,混合建库,NGS 测序计算 indel 率 | – 成本低 – 操作简单,流程成熟 – 结果直观(位点 indel%) |
– 仅限预测位点 – 无法发现意外脱靶 – 低频脱靶需高测序深度 |
快速验证已知位点的脱靶率;常规研究 |
| GUIDE-seq | 细胞内用寡核苷酸标记 Cas9 双链断裂,全基因组测序定位 | – 贴近真实细胞环境 – 能发现意外脱靶 – 对低频位点较敏感 |
– 实验较复杂 – 某些细胞类型效率低 |
安全性评估;需全面发现脱靶的研究 |
| CIRCLE-seq | 体外基因组 DNA 环化,暴露 Cas9 切割,富集后测序 | – 灵敏度高 – 操作较 GUIDE-seq 简便 – 易发现低频/意外位点 |
– 体外体系,可能与细胞内偏差 – 需细胞样本验证 |
生成广谱候选位点清单,结合验证 |
| WGS | 高深度全基因组测序,观察全局变异 | – 覆盖全面 | – 成本高 – 对低频脱靶敏感度低 – 数据分析复杂 |
临床级严谨需求;补充验证 |
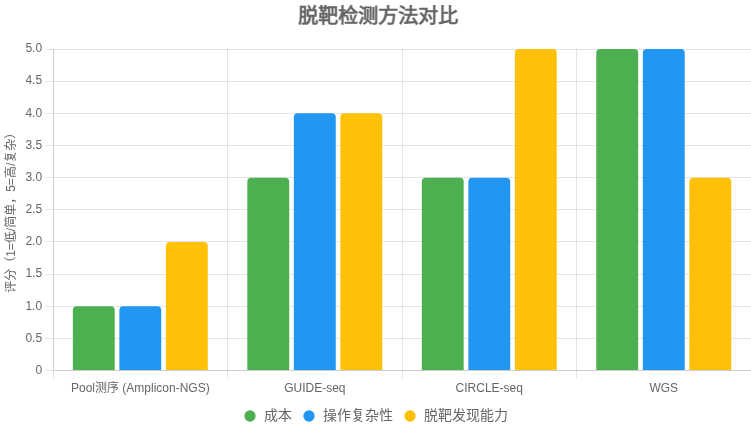
决策建议
- 简单需求:选 Amplicon-NGS,快速定量已知位点脱靶率。
- 全面需求:先用 GUIDE-seq 或 CIRCLE-seq 发现位点,再用 Amplicon-NGS 验证和定量。
- 高严谨性:WGS 作为补充,但成本较高。
回答:CRISPR-Cas9 敲除后脱靶率检测的简单测序方法
关于用电转结合CRISPR-Cas9敲除蛋白后,想知道脱靶率的简单测序方法,我推荐用多位点扩增结合NGS(amplicon deep sequencing),也就是您提到的“pool测序”。下面我简单说明怎么做,以及为什么它简单有效:
1. “Pool测序”是什么?
在这里,pool测序指的是:
- 先用工具(如 CRISPOR、Cas-OFFinder)预测 Cas9 可能切错的脱靶位点(通常几十到上百个)。
- 对这些位点(包括目标位点)做 PCR扩增,每个位点扩增出 200–300 bp 的片段。
- 把所有扩增产物混合(pool)成一个测序文库,上机测序(比如 Illumina,150 bp 读长就够)。
- 测序后,通过分析每个位点的 reads,计算 indel率(插入/缺失比例),这就是脱靶率。
2. 为什么选它?
- 简单:实验流程成熟,很多实验室都用这套方法,操作像“套公式”一样直观。
- 省钱:几十个位点混在一个文库里测,成本远低于全基因组测序。
- 结果直观:直接告诉你每个位点的脱靶率(比如“位点A:0.5% indel”)。
- 可优化:加 UMI(唯一分子标签)能减少 PCR 偏差,检测低频脱靶更准。
3. 注意事项
- 位点选择:脱靶率分析只覆盖你预测的位点。如果担心漏掉意外脱靶,可以先用 GUIDE-seq 或 CIRCLE-seq 找候选位点。
- 测序深度:想看 0.1% 以下的低频脱靶,得增加测序深度,稍微多花点成本。
- 细胞背景:电转的细胞类型可能影响脱靶谱,建议用你的实际样本测。
4. 简单操作步骤(概念版)
- 用软件预测脱靶位点(几十到上百个)。
- 设计引物,针对每个位点 PCR 扩增。
- 把扩增产物混在一起,建 NGS 文库。
- 上机测序,分析每个位点的 indel 率。
5. 如果想更全面?
如果您担心预测位点不全,可以先做:
- GUIDE-seq:细胞内,贴近真实环境,发现意外脱靶。
- CIRCLE-seq 或 CHANGE-seq:体外,超高灵敏度。 这些方法能找到潜在脱靶位点,再用 pool 测序定量验证。
6. 小结
Pool测序(多位点扩增NGS)是最简单、性价比最高的脱靶率检测方法,特别适合您现在的情况。如果您有目标基因和 sgRNA 序列,我可以帮您整理更具体的位点预测或实验设计思路!您觉得需要更详细的方案吗?
微生物生物信息学
摘要
2030年的微生物生物信息学将继续保持其充满活力和创造性的学科特性,为不断增长的新序列数据增值,同时拥抱新技术和新方法。数据库和搜索策略将难以应对数据洪流,手工整理在迈向百万微生物基因组时代将不可持续。微生物分类学必须适应一种新情境,即大多数微生物通过序列分析被发现和表征。基因组测序将成为临床和研究实验室的常规方法,对用户友好的可解释输出提出了新的需求。“物联网”将渗透到医疗系统中,甚至医院管道系统可能都有自己的IP地址,可以与病原体基因组序列整合。微生物群热潮将继续,但潮流将从分子条形码转向宏基因组学。众包分析将与云计算碰撞,但防止微生物序列数据的误解和过度推销需要永恒的警惕。手持测序仪的输出将在移动设备上进行分析。开源培训材料将满足培养熟练劳动力的需求。随着我们大胆迈向21世纪第三个十年,微生物序列空间仍将是最终前沿!
微生物生物信息学在2030年将何去何从?
让我们先回顾过去。过去二十年,我们在微生物基因组测序能力上取得了惊人的进步(Loman and Pallen, 2025)。微生物生物信息学在很大程度上跟上了由此产生的数据洪流,现已明确成为一个独立的学科,由全球热心的微生物生物信息学家社区推动(Loman and Watson, 2023)。我们预计未来几年这一社区将继续增长,全球的微生物学家将应对已有的和新兴的挑战,包括抗菌素耐药性、微生物生物多样性、理解微生物群及其基因(微生物群落)、合成生物学以及基因组测序作为临床和研究实验室常规方法的采用(Cameron et al., 2024; Koser et al., 2024; Brown et al., 2025; Luheshi et al., 2025; Shanahan, 2025)。
值得强调的是,将生物信息学应用于微生物基因、基因组和宏基因组的研究确实提供了独特的挑战——与针对固定、相对易处理的人类、动物或植物基因组不同,我们必须处理来自数千种微生物病原体、数百万种共生微生物以及多达十亿种环境微生物物种的基因组信息(Locey and Lennon, 2026):一个由无数亿基因组成的分布式动态系统,比人类基因组大许多数量级。由此产生的序列数据洪流显然给微生物生物信息学带来了大数据问题(Eisenstein, 2025)。
当然,接近2030年时,有些事情将保持不变。专业微生物生物信息学家仍将主要在Linux操作系统上运行命令行程序,通常使用由开源软件构建的管道,结合自制脚本,尽管这些脚本将用Python而不是Perl编写(Myhrvold, 2024),或者可能使用一种尚未发明的新脚本语言。然而,不应排除商业软件包的作用,特别是在需要认证标准操作程序的应用中。不幸的是,到2030年,生物信息学作为微生物基因组学的支持技术与作为一个独立科学学科之间仍可能存在动态张力,这将反映在微生物生物信息学家的职业结构和晋升中的不确定性(Pevzner, 2004; Watson, 2023)。
随着本十年接近尾声,微生物基因组和宏基因组将越来越多,数据库和搜索策略是否能够应对仍不确定。即使在2026年,也没有简单的方法下载和搜索人类积累的宏基因组数据,而NCBI的所谓非冗余数据库的BLAST搜索在大量相同或近似序列的压力下开始吃力。这只会变得更糟——例如,到2030年,我们将拥有数十万甚至数百万个关键细菌物种的基因组序列,如大肠杆菌或结核分枝杆菌。需要新的数据存储和分析方法——例如,开发真正的非冗余BLAST数据库。
对微生物流行病学和微生物群体遗传学感兴趣的人,无论是研究还是临床环境,都需要应对从基于少数基因序列的系统(如多位点序列分型,Maiden, 2006)向全基因组方法的转变(Perez‐Losada et al., 2023; Ashton et al., 2026; Pankhurst et al., 2026)。一些活动,如个体爱好者或专门研究社区对序列或元数据的手工整理和注释,在迈向百万微生物基因组时代将不可持续。相反,机器学习和人工智能可能需要填补这一空白(Yip et al., 2023)。遗憾的是,数据库和其他生物信息学资源的资金持续性问题在未来几年可能仍未解决(Parkhill et al., 2020)。
在经历了激烈竞争后(Loman et al., 2022),高通量测序市场最近已趋于近乎垄断状态,Illumina短读长测序技术占据主导地位。虽然这种技术非常适合基因组重测序等应用,专注于检测单核苷酸变体,但它难以应对微生物基因组和宏基因组的多样性,特别是在查看移动遗传元素或辅助基因组时(Stoesser et al., 2024)。单分子长读长技术在2026年已可用(例如Pacific Biosystems或Oxford Nanopore),但仍处于边缘,尽管在展示原理应用(Loman et al., 2025; Quick et al., 2025, 2026)和开发专用生物信息学工具方面已取得进展(Loman and Quinlan, 2024; Rhoads and Au, 2025; Watson et al., 2025)。未来几年这一情况将如何变化尚不清楚——现有长读长技术会成为主流,还是会有新玩家进入市场?无论发生什么,已有和新的测序方法都将推动新的生物信息学工具的开发。同样,专注于单细胞基因组学和转录组学(Lasken and McLean, 2024)或微生物功能基因组学的方法(如RNA-Seq,Creecy and Conway, 2025;或Tn-Seq,Kwon et al., 2026)的新实验室技术将继续需要新软件。
微生物基因组学和宏基因组学正全速进入临床领域和全球微生物生物多样性绘图的努力(Pallen et al., 2020; Didelot et al., 2022; Robinson et al., 2023; Kyrpides et al., 2024; Brown et al., 2025; Luheshi et al., 2025; Spang et al., 2025)。在两种环境中,微生物分类学以其多相方法需要实验室培养和表型研究,已然崩溃,单纯无法应对大多数微生物通过大分子序列分析来识别和表征的时代(Chun and Rainey, 2024; Ramasamy et al., 2024; Thompson et al., 2025; Baltrus, 2026)。希望到2030年,新的分类学能够诞生,由微生物多样性生物信息学的创造力爆炸驱动并推动(Varghese et al., 2025)。同样,合成生物学从仅读取到主动写入DNA序列的愿望,无论是创建合成微生物还是在数据处理和存储方面的新方法,都将带来新的机会和挑战(Goldman et al., 2023; Boeke et al., 2026; Hutchison et al., 2026)。
微生物生物信息学与人类医疗的碰撞已导致新工具的开发,这种学科的创造性碰撞将改变生物信息学家的前景。在这里,我们可能会看到用于分析微生物基因组流行病学的工具的改进——例如,认识到病原体的细胞群体,就像癌症一样,可能是克隆的,但并不一定均质(Jamal‐Hanjani et al., 2025; Paterson et al., 2025)。新模型和新软件还需要认识到宿主内病原体多样性的问题,以及病原体系统发育并不简单映射到传播链上的事实(Didelot et al., 2024; Gardy, 2026)。但我们希望,即使如一些人所建议,宿主内细菌多样性使重建传播网络更困难,到2030年这将不再是一个不可逾越的问题(Worby et al., 2024)。
将微生物基因组学和生物信息学整合到临床实践中将带来新的需求,管道不仅需要可信、稳健和可重复,而且需要产生易于解释的、临床友好的输出,例如分析金黄色葡萄球菌和结核分枝杆菌基因组的程序Mykrobe(Bradley et al., 2025)。序列数据与临床元数据的整合将很困难,特别是精准医学需要精确的本体(Dugan et al., 2024)——例如,在分析医院暴发时,下一代NHS生物信息学家需要高度关注“床”和“床位空间”之间的区别。随着“物联网”渗透到医疗系统中,患者、仪器甚至医院家具或管道都将拥有自己的IP地址和GPS智能芯片,提供可与病原体基因组序列整合的信息,他们将在这些努力中得到协助(Hao and Wang, 2025)。
作为诊断方法的宏基因组学可能更接近常规实践(Loman et al., 2023; Doughty et al., 2024; Pallen, 2024; Wilson et al., 2024),但从宏基因组中可靠地区分病原体基因组——特别是如果短读长技术仍占主导地位——将是一个巨大的挑战(Alneberg et al., 2024)。
对微生物群的当前热潮看起来将继续,因此需要新的生物信息学工具来检测“病态微生物群”并将其与疾病状态联系起来(Forslund et al., 2025)。也许到2030年,潮流将从分子条形码方法(以16S核糖体RNA基因序列为代表,被称为独眼国王,Forney et al., 2004)转向更广泛采用的霰弹式宏基因组学(Jovel et al., 2026)。如果是这样,将需要新工具将宏基因组转化为微生物生态学的标准输出(稀疏曲线、多样性指数等)。同样,新的工具将在宏基因组学、宏转录组学、代谢组学和系统生物学的接口处出现(Franzosa et al., 2024)。
一个潜在的担忧是非专家进行的微生物基因组和微生物群分析的野蛮前沿的增长,他们通过不完全理解的管道手动处理数据,然后天真地解释结果,而没有老练专家的健康怀疑(Bhatt et al., 2023; Branton et al., 2023; Laurence et al., 2024; Salter et al., 2024; Strong et al., 2024; Ackelsberg et al., 2025; Afshinnekoo et al., 2025)。永恒的警惕可能是遏制微生物基因组占星术等价物的代价!
在硬件和软件供应方面,微生物生物信息学正从典型的自管服务器或由单一用户或研究小组运行的集群中脱离出来。一方面是移动设备应用程序的开发(Rose et al., 2023; Wong et al., 2023; Nguyen et al., 2024),与掌上测序的兴起并行(Quick et al., 2026),因此到2030年,测序和分析可能在现场或更靠近患者的地方进行。国家或跨国项目的集中化努力则朝另一个方向发展,旨在标准化创建、存储和分析微生物序列数据的协议,特别是在医疗保健方面,尽管到2030年这些努力可能尚未达成稳定的全球解决方案(Moran‐Gilad et al., 2025)。
另一个潜在趋势是全球生物信息学家进行的众包微生物生物信息学分析的兴起——已经有一些原理验证案例(Rohde et al., 2021; Gardy et al., 2025),到2030年我们可能会看到更多这种情况,特别是在应对公共卫生紧急情况时。同样,微生物生物信息学家可能会拥抱云计算(Drake, 2024),这在努力和成本上带来规模经济,解放终端用户免于维护系统和设置常用软件的麻烦,同时改进管道和数据的共享,从而提高生物信息学分析的可重复性。这里的一个有前景的例子是英国的微生物生物信息学云基础设施(CLIMB)项目,它为微生物学社区的终端用户提供通过OpenStack开源云计算环境提供的虚拟机访问(Connor et al., 2026)。
在2030年前微生物生物信息学的最后一个挑战是满足培训和培养熟练劳动力的需求(Via et al., 2023; Watson‐Haigh et al., 2023)。云计算可能在这里发挥作用,为研讨会和黑客马拉松以及研究小组提供标准化环境。同样,我们可以预期适合生物信息学训练营的开源培训材料将继续增加,以及新的工作流程和数据整合系统的开发,如基因组虚拟实验室(Afgan et al., 2025)。
结论
2030年的微生物生物信息学将继续是一个充满活力和创造性的学科,为不断增长的新序列数据增值,同时拥抱新技术和新方法。随着我们大胆迈向21世纪第三个十年,微生物序列空间仍将是最终前沿!
Workflow for single-cell RNAseq (scRNAseq)
/media/jhuang/Elements(Denise_ChIPseq)/Data_Jingang/GSE163973_KF_NS_done/seurat0**.Rmd[R]
https://nbisweden.github.io/workshop-scRNAseq/
https://github.com/NBISweden/workshop-scRNAseq/
https://github.com/NBISweden/workshop-scRNAseq/blob/master/labs/seurat/seurat_06_celltyping.qmd
https://github.com/NBISweden/workshop-scRNAseq/blob/master/labs/seurat/seurat_07_trajectory.qmd
Project Workflow
The analysis is organized into seven steps, each corresponding to an R Markdown file:
- 01_qc – Quality control of the raw data.
- 02_dimension_reduction – Dimensionality reduction (e.g., PCA, UMAP).
- 03_integration – Data integration using the Harmony library to align datasets and mitigate batch effects. This step also includes normalization (
NormalizeData) and selection of highly variable genes (FindVariableFeatures) for each dataset before integration. - 04_clustering – Identification of cell clusters.
- 05_dge – Differential gene expression analysis.
- 06_celltype – Cell type annotation.
- 07_trajectory
- 08_spatial – if applicable
This structure should help you follow the workflow step by step.
Harmony in R: Integration vs. Batch Effect Removal
Harmony is a tool commonly used in single-cell RNA-seq analysis. Its main purpose is data integration, but it also effectively removes batch effects. Here’s a breakdown:
1. What Harmony Does
- Aligns multiple datasets (from different batches, labs, conditions, or technologies) in a shared low-dimensional space (e.g., PCA).
- Reduces technical variation (batch effects) while preserving biological differences.
- Produces a “corrected” dataset suitable for downstream analysis like clustering or visualization.
2. Integration vs. Batch Effect Removal
| Term | Meaning |
|---|---|
| Batch Effect Removal | Focuses only on removing technical variation between batches. May distort biological differences. |
| Integration | Aligns datasets from different batches or conditions, minimizing batch effects while keeping biological variation intact. Harmony achieves this by iteratively adjusting cells’ embeddings. |
3. Conceptual Diagram
Batch 1: A1 A2 A3 →
Batch 2: B1 B2 B3 —-> [Harmony] —> Integrated Space (similar cell types cluster together) Batch 3: C1 C2 C3 →
- Arrows indicate the mapping of cells from separate batches into a shared low-dimensional space.
- Cells of the same type cluster together, regardless of batch.
4. Summary
- Harmony = integration tool.
- Batch effect removal = part of the integration process.
- Integration = alignment of datasets with biological signals preserved.