-
nextflow
#under hamm
ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq
#Debug the following error: added "--minAssignedFrags 0 \\" to modules/nf-core/salmon/quant/main.nf option "salmon quant" and added "--min_mapped_reads 0" in the nextflow command
(rnaseq_2021) jhuang@hamm:~/DATA/Data_Denise_sT_RNASeq$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_JN707599 --fasta JN707599.fasta --gtf JN707599.gtf -profile test_full -resume --max_memory 124.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0
(rnaseq_2021) jhuang@hamm:~/DATA/Data_Denise_sT_RNASeq$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_LT --fasta JN707599.fasta --gtf LT.gtf -profile test_full -resume --max_memory 124.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0
(rnaseq_2021) jhuang@hamm:~/DATA/Data_Denise_sT_RNASeq$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_sT --fasta JN707599.fasta --gtf sT.gtf -profile test_full -resume --max_memory 124.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0
#DEBUG
#V_8_4_1_p602_d8_DonorI,V_8_4_2_p602_d8_DonorI.fastq.gz,,auto
#V_8_5_1_p602and604_d3_DonorI,V_8_5_1_602and604_d3_DonorI.fastq.gz,,auto
#V_8_5_2_p602and604_d3_DonorII,V_8_5_2_602and604_d3_DonorII.fastq.gz,,auto
#mamba install -c bioconda rsem
#mamba install fq
-
construct DESeqDataSet including host+viral gene expressions
# Import the required libraries
library("AnnotationDbi")
library("clusterProfiler")
library("ReactomePA")
library(gplots)
library(tximport)
library(DESeq2)
library("org.Hs.eg.db")
library(dplyr)
library(tidyverse)
setwd("~/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq/results_JN707599/star_salmon")
# Define paths to your Salmon output quantification files
files <- c("V_8_0_mock_DonorI" = "./V_8_0_mock_DonorI/quant.sf",
"V_8_0_mock_DonorII" = "./V_8_0_mock_DonorII/quant.sf",
"V_8_1_5_p601_d3_DonorII" = "./V_8_1_5_p601_d3_DonorII/quant.sf",
"V_8_1_5_p604_d3_DonorII" = "./V_8_1_5_p604_d3_DonorII/quant.sf",
"V_8_1_5_p601_d8_DonorII" = "./V_8_1_5_p601_d8_DonorII/quant.sf",
"V_8_1_5_p604_d8_DonorII" = "./V_8_1_5_p604_d8_DonorII/quant.sf",
"V_8_1_6_p601_d3_DonorI" = "./V_8_1_6_p601_d3_DonorI/quant.sf",
"V_8_1_6_p604_d3_DonorI" = "./V_8_1_6_p604_d3_DonorI/quant.sf",
"V_8_1_6_p601_d8_DonorI" = "./V_8_1_6_p601_d8_DonorI/quant.sf",
"V_8_1_6_p604_d8_DonorI" = "./V_8_1_6_p604_d8_DonorI/quant.sf",
"V_8_2_3_p600_d3_DonorII" = "./V_8_2_3_p600_d3_DonorII/quant.sf",
"V_8_2_3_p605_d3_DonorII" = "./V_8_2_3_p605_d3_DonorII/quant.sf",
"V_8_2_3_p600_d8_DonorII" = "./V_8_2_3_p600_d8_DonorII/quant.sf",
"V_8_2_3_p605_d8_DonorII" = "./V_8_2_3_p605_d8_DonorII/quant.sf",
"V_8_2_4_p600_d3_DonorI" = "./V_8_2_4_p600_d3_DonorI/quant.sf",
"V_8_2_4_p605_d3_DonorI" = "./V_8_2_4_p605_d3_DonorI/quant.sf",
"V_8_2_4_p600_d8_DonorI" = "./V_8_2_4_p600_d8_DonorI/quant.sf",
"V_8_2_4_p605_d8_DonorI" = "./V_8_2_4_p605_d8_DonorI/quant.sf",
"V_8_4_1_p602_d8_DonorII" = "./V_8_4_1_p602_d8_DonorII/quant.sf",
"V_8_4_1_p602_d8_DonorI" = "./V_8_4_1_p602_d8_DonorI/quant.sf",
"V_8_3_1_p600and601_d12_DonorI" = "./V_8_3_1_p600and601_d12_DonorI/quant.sf",
"V_8_3_1_p604and605_d12_DonorI" = "./V_8_3_1_p604and605_d12_DonorI/quant.sf",
"V_8_3_2_p600and601_d9_DonorII" = "./V_8_3_2_p600and601_d9_DonorII/quant.sf",
"V_8_3_2_p604and605_d9_DonorII" = "./V_8_3_2_p604and605_d9_DonorII/quant.sf",
"V_8_4_2_p602_d3_DonorI" = "./V_8_4_2_p602_d3_DonorI/quant.sf",
"V_8_4_2_p602_d3_DonorII" = "./V_8_4_2_p602_d3_DonorII/quant.sf",
"V_8_5_1_p602and604_d3_DonorI" = "./V_8_5_1_sTplusLT_d3_Donor1/quant.sf",
"V_8_5_2_p602and604_d3_DonorII" = "./V_8_5_2_sTplusLT_d3_Donor2/quant.sf")
# Import the transcript abundance data with tximport
txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE)
column_names <- colnames(txi$counts)
output_string <- paste(column_names, collapse = ", ")
cat(output_string, "\n")
#"V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII"
col_order <- c("V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII")
#reordered.txi <- txi[,col_order]
identical(column_names,col_order)
condition = as.factor(c("untreated","untreated", "p601_d3","p604_d3", "p601_d8","p604_d8", "p601_d3","p604_d3","p601_d8","p604_d8", "p600_d3","p605_d3","p600_d8", "p605_d8", "p600_d3","p605_d3","p600_d8","p605_d8", "p602_d8","p602_d8", "p600and601_d9d12", "p604and605_d9d12","p600and601_d9d12","p604and605_d9d12", "p602_d3","p602_d3", "p602and604_d3","p602and604_d3"))
batch = as.factor(c("200420", "200420", "190927", "190927", "190927", "190927", "190228", "190228", "190228", "190228", "191008", "191008", "191008", "191008", "190228", "190228", "190228", "190228", "200817", "200817", "200420", "200420", "200817", "200817", "210302", "210302", "210504","210504"))
ids = as.factor(c("untreated_DonorI","untreated_DonorII", "p601_d3_DonorII","p604_d3_DonorII", "p601_d8_DonorII","p604_d8_DonorII", "p601_d3_DonorI","p604_d3_DonorI","p601_d8_DonorI","p604_d8_DonorI", "p600_d3_DonorII","p605_d3_DonorII","p600_d8_DonorII", "p605_d8_DonorII", "p600_d3_DonorI","p605_d3_DonorI","p600_d8_DonorI","p605_d8_DonorI", "p602_d8_DonorII","p602_d8_DonorI", "p600and601_d12_DonorI", "p604and605_d12_DonorI","p600and601_d9_DonorII","p604and605_d9_DonorII", "p602_d3_DonorI","p602_d3_DonorII", "p602and604_d3_DonorI","p602and604_d3_DonorII"))
donor = as.factor(c("DonorI","DonorII", "DonorII","DonorII", "DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorII","DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorI", "DonorI", "DonorI","DonorII","DonorII", "DonorI","DonorII", "DonorI","DonorII"))
# Define the colData for DESeq2
#colData = data.frame(row.names=column_names, condition=condition, donor=donor, batch=batch, ids=ids)
colData <- data.frame(condition=condition, donor=donor, row.names=names(files))
# -- transcript-level count data (for virus) --
# Create DESeqDataSet object
dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition+donor)
write.csv(counts(dds), file="transcript_counts.csv")
# -- gene-level count data (for virus) --
# Read in the tx2gene map from salmon_tx2gene.tsv
tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE)
# Set the column names
colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name")
# Remove the gene_name column if not needed
tx2gene <- tx2gene[,1:2]
# Import and summarize the Salmon data with tximport
txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE)
dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition+donor)
#dds <- dds[rowSums(counts(dds) > 3) > 2, ] #60605-->26543
write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv")
# -- merge the raw counts of human and microbe --
setwd("~/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq/results/featureCounts")
d.raw<- read.delim2("merged_gene_counts_2_f3_f5.txt",sep="\t", header=TRUE, row.names=1)
colnames(d.raw)<- c("V_8_0_mock_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_5_p604_d3_DonorII","V_8_1_6_p604_d3_DonorI","V_8_2_3_p605_d8_DonorII","V_8_0_mock_DonorII","V_8_1_5_p601_d8_DonorII","V_8_2_3_p605_d3_DonorII","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d8_DonorII","V_8_2_4_p600_d8_DonorI","V_8_2_4_p600_d3_DonorI","V_8_1_5_p604_d8_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_6_p601_d3_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_4_p605_d3_DonorI","V_8_4_1_p602_d8_DonorI","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_2_4_p605_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_4_1_p602_d8_DonorII","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII", "V_8_5_1_p602and604_d3_DonorI", "V_8_5_2_p602and604_d3_DonorII") #26364
col_order <- c("V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII", "V_8_1_5_p604_d3_DonorII", "V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII", "V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI", "V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII", "V_8_2_3_p605_d8_DonorII", "V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI", "V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI", "V_8_3_1_p600and601_d12_DonorI", "V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII", "V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII", "V_8_5_1_p602and604_d3_DonorI", "V_8_5_2_p602and604_d3_DonorII")
reordered.raw <- d.raw[,col_order]
write.csv(reordered.raw, file="merged_gene_counts_2_f3_f5_reordered.txt")
# Confirm the identification of the headers of two files before merging!
#kate ./results/featureCounts/merged_gene_counts_2_f3_f5_reordered.txt
#"","V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII"
#results_JN707599/star_salmon/gene_counts.csv
#"","V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII"
#cat ../Data_Denise_RNASeq/results/featureCounts/merged_gene_counts_2_f3_f5_reordered.txt ../Data_Denise_RNASeq/results_JN707599/star_salmon/gene_counts.csv > merged_gene_counts.csv
#~/Tools/csv2xls-0.4/csv_to_xls.py merged_gene_counts.csv -d',' -o raw_gene_counts.xls;
setwd("~/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq_Merged/")
d.raw <- read.csv("merged_gene_counts.csv", header=TRUE, row.names=1)
dds <- DESeqDataSetFromMatrix(countData=d.raw, colData=colData, design=~condition+donor)
dim(counts(dds))
head(counts(dds), 10)
rld <- rlogTransformation(dds)
# draw simple pca and heatmap
library(gplots)
library("RColorBrewer")
#mat <- assay(rld)
#mm <- model.matrix(~condition, colData(rld))
#mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm)
#assay(rld) <- mat
# -- pca --
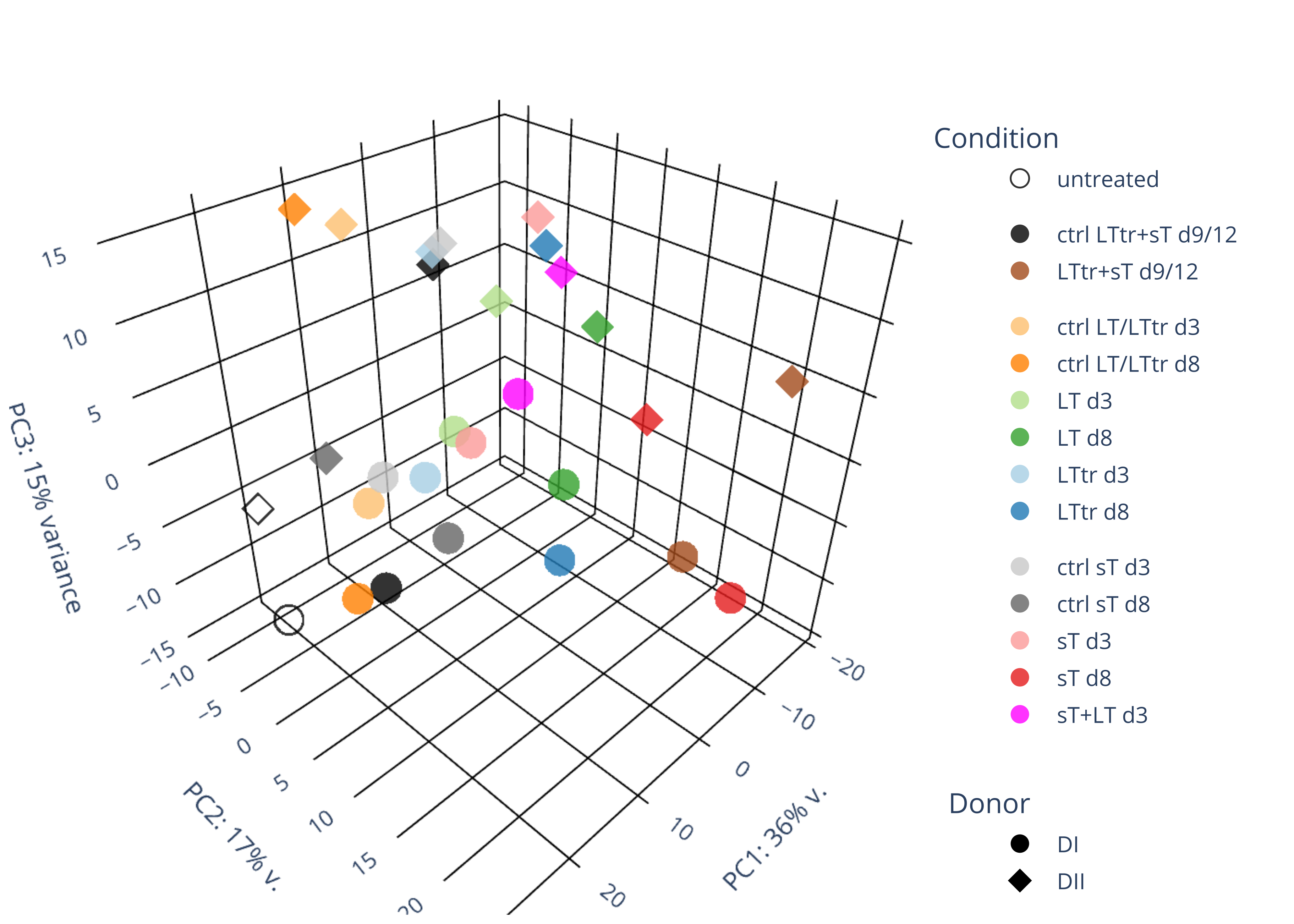
png("pca.png", 1200, 800)
plotPCA(rld, intgroup=c("condition"))
dev.off()
# -- heatmap --
png("heatmap.png", 1200, 800)
distsRL <- dist(t(assay(rld)))
mat <- as.matrix(distsRL)
hc <- hclust(distsRL)
hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100)
heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13))
dev.off()
-
(corrected of last step) construct DESeqDataSet including host+viral gene expressions
column_names <- colnames(d.raw)
col_order <- c("V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII")
identical(column_names,col_order)
condition = as.factor(c("untreated","untreated", "p601_d3","p604_d3", "p601_d8","p604_d8", "p601_d3","p604_d3","p601_d8","p604_d8", "p600_d3","p605_d3","p600_d8", "p605_d8", "p600_d3","p605_d3","p600_d8","p605_d8", "p602_d8","p602_d8", "p600and601_d9d12", "p604and605_d9d12","p600and601_d9d12","p604and605_d9d12", "p602_d3","p602_d3", "p602and604_d3","p602and604_d3"))
batch = as.factor(c("200420", "200420", "190927", "190927", "190927", "190927", "190228", "190228", "190228", "190228", "191008", "191008", "191008", "191008", "190228", "190228", "190228", "190228", "200817", "200817", "200420", "200420", "200817", "200817", "210302", "210302", "210504","210504"))
ids = as.factor(c("untreated_DonorI","untreated_DonorII", "p601_d3_DonorII","p604_d3_DonorII", "p601_d8_DonorII","p604_d8_DonorII", "p601_d3_DonorI","p604_d3_DonorI","p601_d8_DonorI","p604_d8_DonorI", "p600_d3_DonorII","p605_d3_DonorII","p600_d8_DonorII", "p605_d8_DonorII", "p600_d3_DonorI","p605_d3_DonorI","p600_d8_DonorI","p605_d8_DonorI", "p602_d8_DonorII","p602_d8_DonorI", "p600and601_d12_DonorI", "p604and605_d12_DonorI","p600and601_d9_DonorII","p604and605_d9_DonorII", "p602_d3_DonorI","p602_d3_DonorII", "p602and604_d3_DonorI","p602and604_d3_DonorII"))
donor = as.factor(c("DonorI","DonorII", "DonorII","DonorII", "DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorII","DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorI", "DonorI", "DonorI","DonorII","DonorII", "DonorI","DonorII", "DonorI","DonorII"))
colData <- data.frame(condition=condition, donor=donor, row.names=column_names)
setwd("~/DATA/Data_Denise_sT_PUBLISHING/Data_Denise_RNASeq_Merged_Corrected/")
d.raw <- read.csv("merged_gene_counts_corrected.csv", header=TRUE, row.names=1)
dds <- DESeqDataSetFromMatrix(countData=d.raw, colData=colData, design=~condition+donor)
dim(counts(dds))
head(counts(dds), 10)
rld <- rlogTransformation(dds)
# draw simple pca and heatmap
library(gplots)
library("RColorBrewer")
#mat <- assay(rld)
#mm <- model.matrix(~condition, colData(rld))
#mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm)
#assay(rld) <- mat
# -- pca --
png("pca.png", 1200, 800)
plotPCA(rld, intgroup=c("condition"))
dev.off()
# -- heatmap --
png("heatmap.png", 1200, 800)
distsRL <- dist(t(assay(rld)))
mat <- as.matrix(distsRL)
hc <- hclust(distsRL)
hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100)
heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13))
dev.off()
-
select the differentially expressed genes
#untreated_DonorI,untreated
#untreated_DonorII,untreated
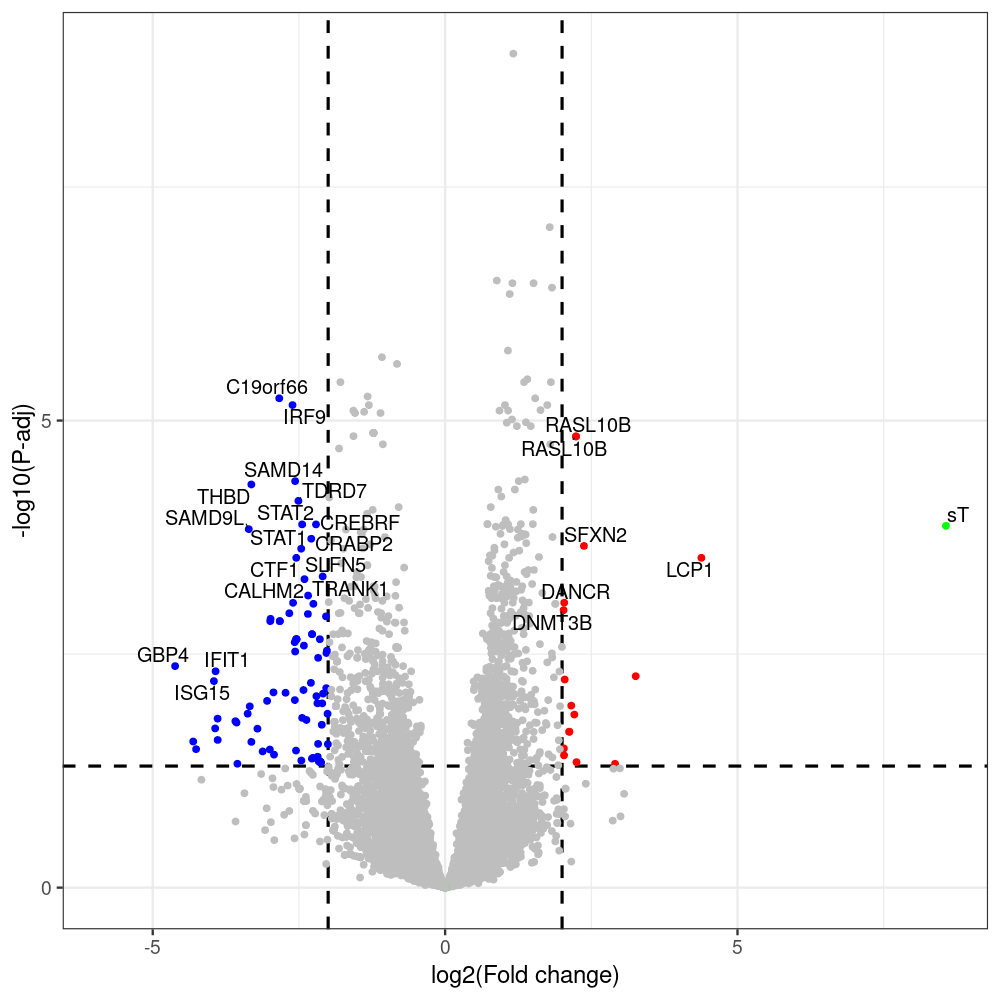
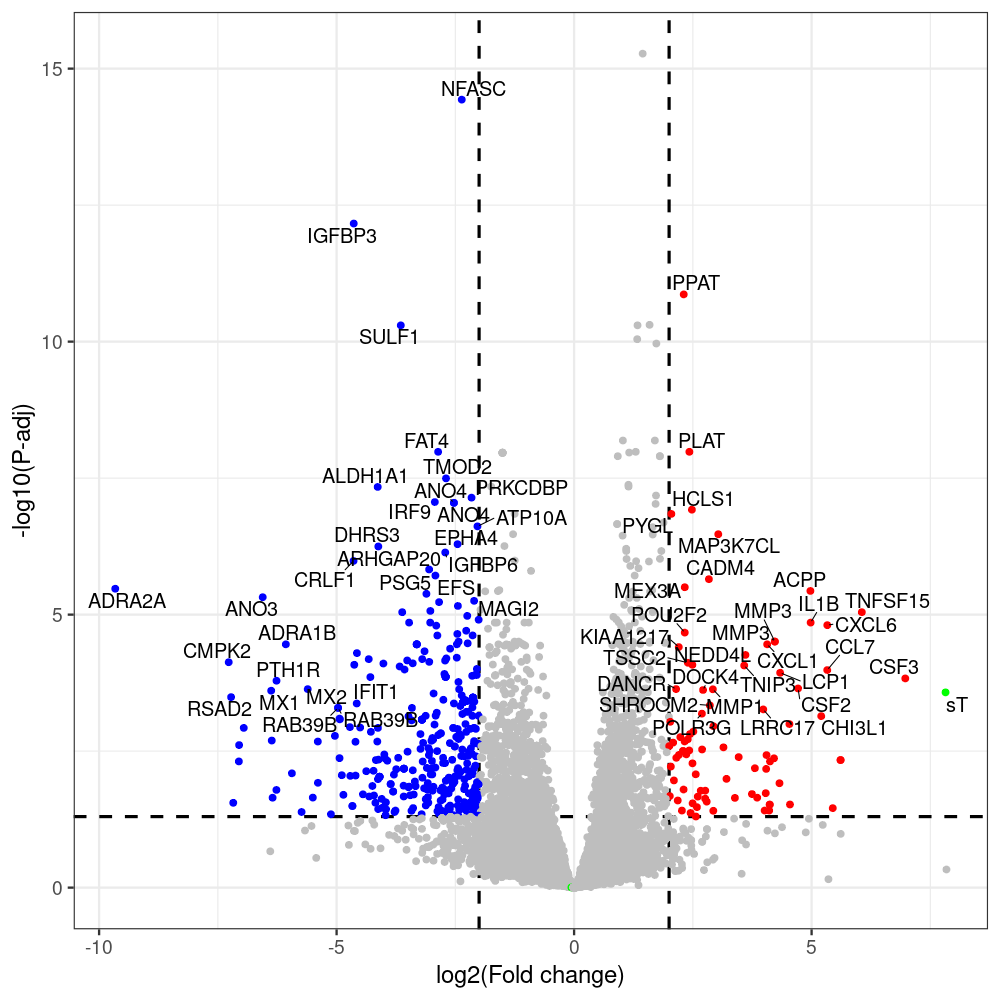
#p601_d3_DonorII,mCherry control
#p604_d3_DonorII,sT
#p601_d8_DonorII,mCherry control
#p604_d8_DonorII,sT
#p601_d3_DonorI,mCherry control
#p604_d3_DonorI,sT
#p601_d8_DonorI,mCherry control
#p604_d8_DonorI,sT
#
#p600_d3_DonorII,GFP control
#p605_d3_DonorII,LTtr
#p600_d8_DonorII,GFP control
#p605_d8_DonorII,LTtr
#p600_d3_DonorI,GFP control
#p605_d3_DonorI,LTtr
#p600_d8_DonorI,GFP control
#p605_d8_DonorI,LTtr
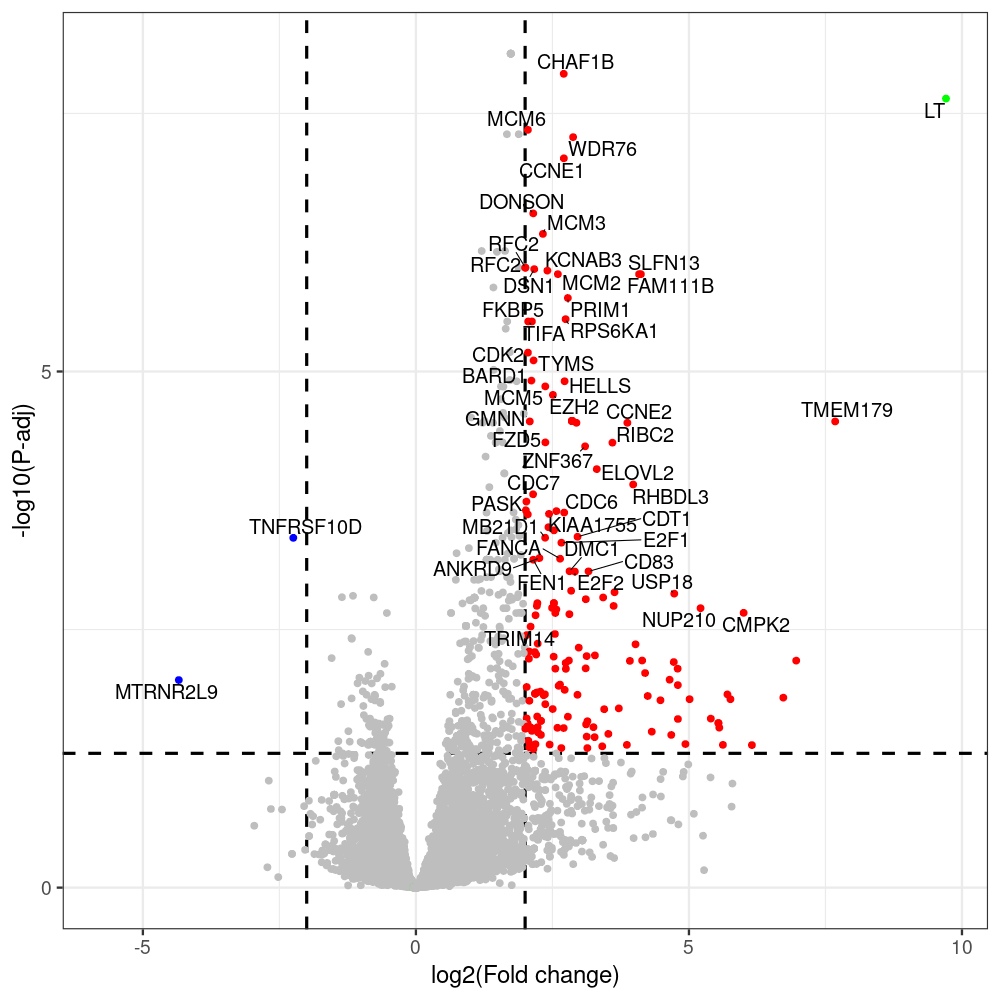
#p602_d3_DonorI,LT
#p602_d3_DonorII,LT
#p602_d8_DonorII,LT
#p602_d8_DonorI,LT
#
#p600and601_d12_DonorI,GFP+mCherry control
#p604and605_d12_DonorI,sT+LTtr
#p600and601_d9_DonorII,GFP+mCherry control
#p604and605_d9_DonorII,sT+LTtr
#
#p602and604_d3_DonorI,sT+LT
#p602and604_d3_DonorII,sT+LT
#CONSOLE: mkdir /home/jhuang/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq_Merged/degenes
#---- relevel to control ----
dds$condition <- relevel(dds$condition, "p601_d3")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("p604_d3_vs_p601_d3")
dds$condition <- relevel(dds$condition, "p600_d3")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("p605_d3_vs_p600_d3")
dds$condition <- relevel(dds$condition, "p600_d3")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("p602_d3_vs_p600_d3")
dds$condition <- relevel(dds$condition, "p601_d8")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("p604_d8_vs_p601_d8")
dds$condition <- relevel(dds$condition, "p600_d8")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("p605_d8_vs_p600_d8")
dds$condition <- relevel(dds$condition, "p600_d8")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("p602_d8_vs_p600_d8")
library(biomaRt)
listEnsembl()
listMarts()
#--> total 69, 27 GRCh38.p7 and 39 GRCm38.p4
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="104")
#GRCh37.p13
ensembl <- useEnsembl("ensembl","hsapiens_gene_ensembl", host="https://grch37.ensembl.org")
datasets <- listDatasets(ensembl)
# -- 1. export res_df containing both human and virus genes --
#for (i in clist) {
i<-clist[1]
contrast = paste("condition", i, sep="_")
res = results(dds, name=contrast)
res <- res[!is.na(res$log2FoldChange),]
res_df <- as.data.frame(res)
write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-"))
up <- subset(res_df, padj<=0.05 & log2FoldChange>=2)
down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2)
write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-"))
write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-"))
#}
# -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database --
#> dim(geness) [1] 21938 9 using GRCh38.p7
#> dim(geness) [1] 23517 9 using GRCh37.p13
#for (i in clist) {
#i<-clist[1]
contrast = paste("condition", i, sep="_")
res = results(dds, name=contrast)
res <- res[!is.na(res$log2FoldChange),]
geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'external_gene_name',
values = rownames(res),
mart = ensembl)
geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE)
#merge by column by common colunmn name, in the case "GENEID"
res$geneSymbol = rownames(res)
#identical(rownames(res), rownames(geness_uniq))
res_df <- as.data.frame(res)
geness_res <- merge(geness_uniq, res_df, by.x="external_gene_name", by.y="geneSymbol")
dim(geness_res) #22044 15
rownames(geness_res) <- geness_res$ensembl_gene_id
geness_res$ensembl_gene_id <- NULL
#}
# -- 3. prepare annatete virus genes --
virus_genes <- c("LT", "sT", "VP1", "VP2", "VP3")
virus_rows <- res_df[rownames(res_df) %in% virus_genes, ]
virus_rows$external_gene_name <- rownames(virus_rows)
virus_rows$chromosome_name <- "JN707599"
# Define default values based on data type
default_values <- list(
character = NULL,
numeric = 0,
integer = 0L,
logical = FALSE
)
# Ensure that virus_rows has the same columns as geness_res
for (col in colnames(geness_res)) {
if (!col %in% colnames(virus_rows)) {
data_type <- class(geness_res[[col]])[1]
default_value <- default_values[[data_type]]
virus_rows[[col]] <- rep(default_value, nrow(virus_rows))
}
}
missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows))
for (col in missing_cols) {
virus_rows[[col]] <- NA # Or another default value as appropriate
}
# Reorder columns in virus_rows to match the order in geness_res
virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE]
# -- 4. merge them together --
#for (i in clist) {
merged_df <- rbind(geness_res, virus_rows)
merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),])
write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-"))
up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2)
down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2)
viral <- merged_df_sorted[merged_df_sorted$external_gene_name %in% c("LT", "sT", "VP1", "VP2", "VP3"), ]
write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-"))
write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-"))
write.csv(as.data.frame(viral), file = paste(i, "viral_annotated.txt", sep="-"))
#}
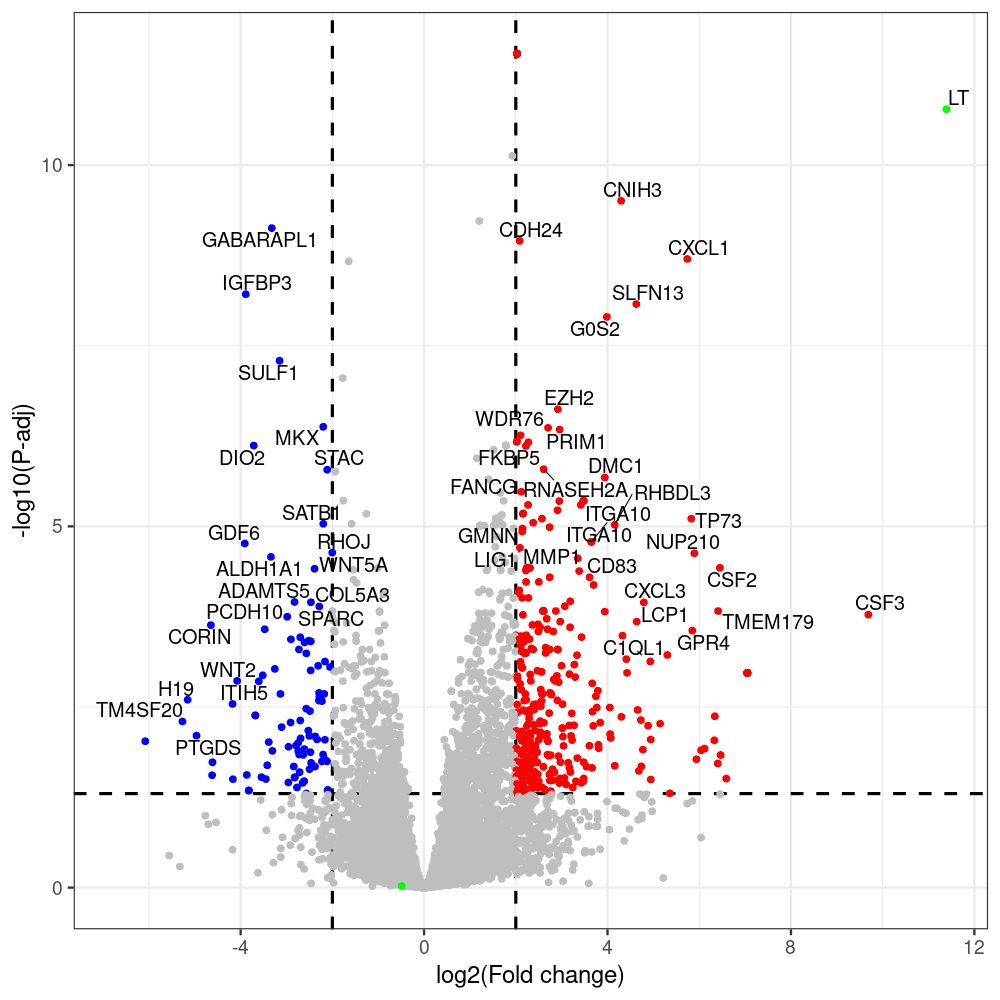
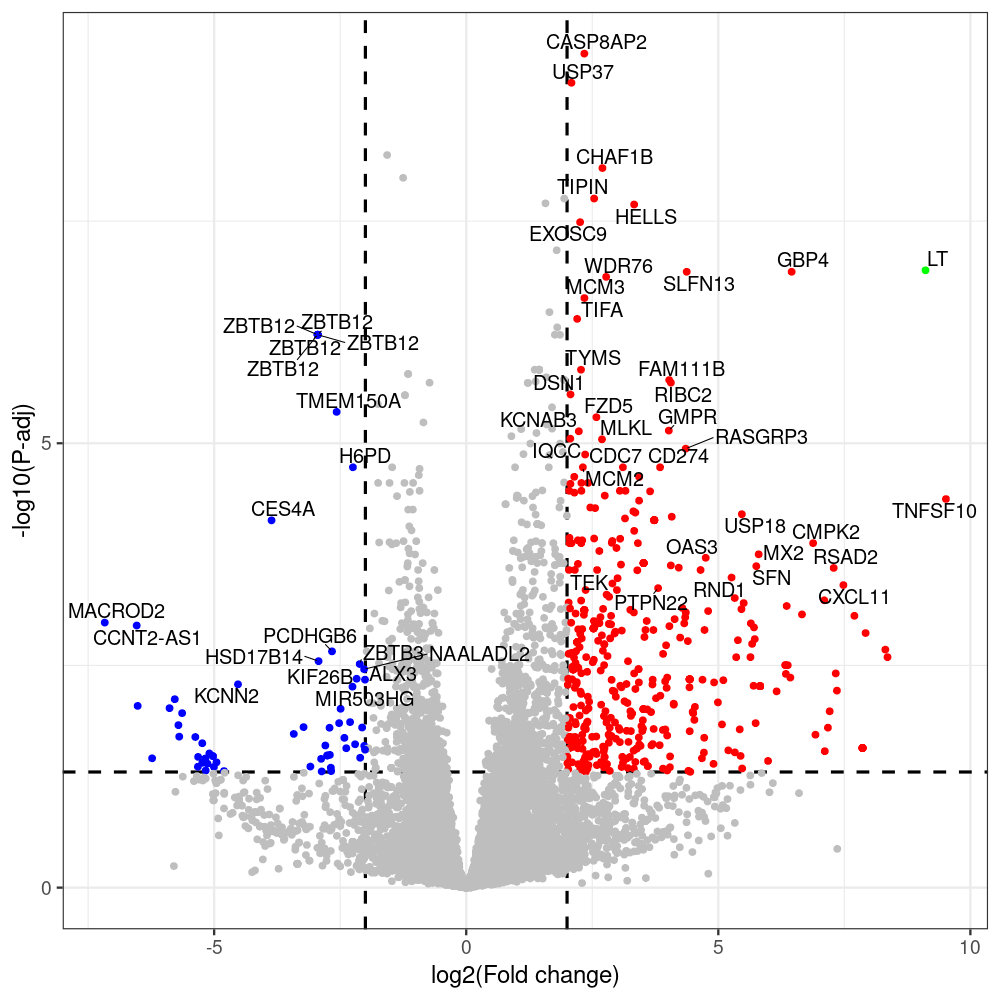
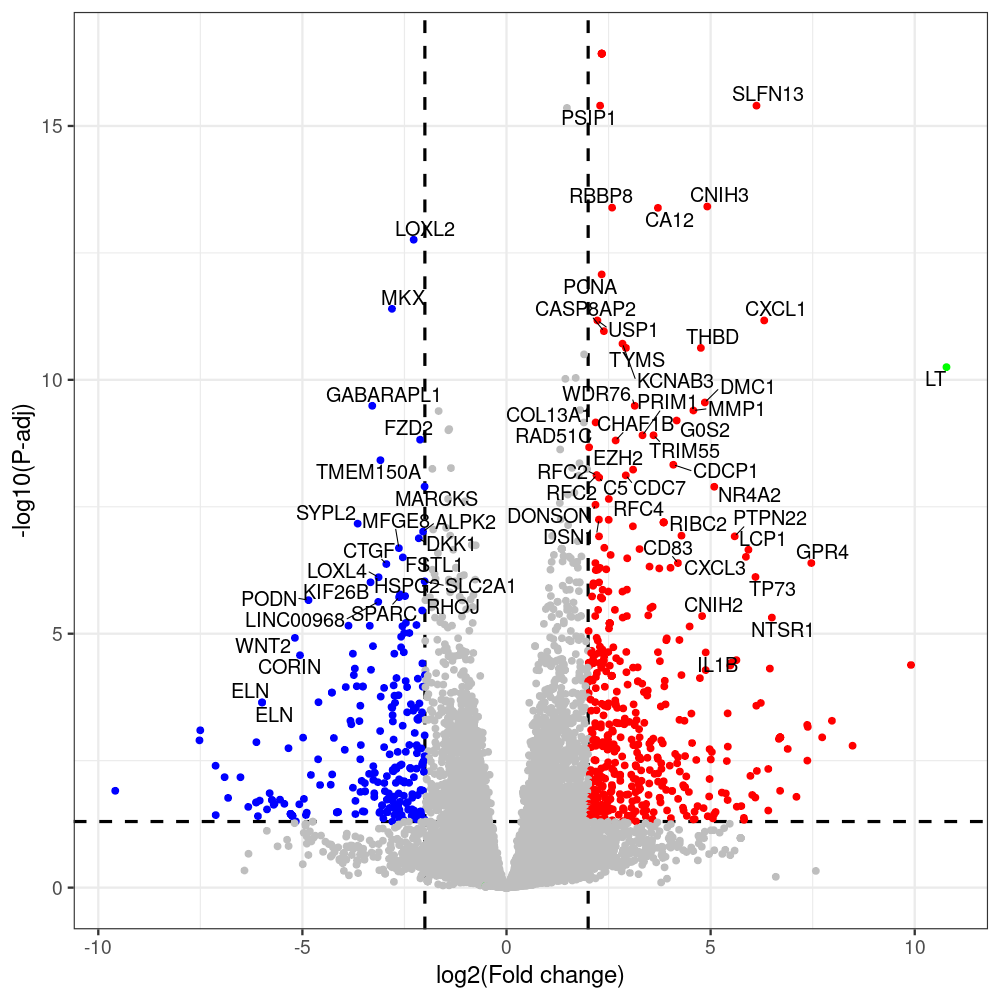
# -- 5. draw graphics --
library(ggrepel)
#geness_res <- read.csv(file = "p605_d8_vs_p600_d8-all_annotated.txt", sep=",", row.names=1)
#i<-"p605_d8_vs_p600_d8"
geness_res <- merged_df_sorted
# Color setting
geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray",
ifelse(geness_res$log2FoldChange > 0, "red", "blue"))
# Predefined genes colored in green
predefined_genes <- c("LT", "sT", "VP1", "VP2", "VP3")
geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green"
geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange)
top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:200],
geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:200]))
# Define the original and compressed ranges
original_range <- c(80, 115)
compressed_range <- c(80.0, 90.0)
# Calculate breaks for the y-axis
y_breaks_below <- seq(0, 80, by=5)
y_breaks_compressed <- c(80.0, 90.0)
y_breaks_above <- c()
y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above)
y_labels_below <- seq(0, 80, by=5)
y_labels_compressed <- c(80, 115)
y_labels_above <- c()
y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above)
# Adjust the p-values based on the ranges
geness_res$adjusted_pvalue <- with(geness_res,
ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2],
((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1],
ifelse(-log10(padj) > original_range[2],
-log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]),
-log10(padj))))
# Create the plot
png(paste(i, "png", sep="."), width=1000, height=1000)
ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) +
geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) +
geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) +
geom_point(size = 3) +
labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") +
scale_color_identity() +
geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)),
size = 7,
point.padding = 0.15,
color = "black",
min.segment.length = .1,
box.padding = .2,
lwd = 2) +
theme_bw(base_size = 24) +
theme(legend.position = "bottom") +
#annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") +
#annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") +
#annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) +
#annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) +
scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels))
dev.off()
#Under DIR degenes under KONSOLE and delete "start_position","end_position","strand" and so on.
#~/Tools/csv2xls-0.4/csv_to_xls.py viral_gene_counts.csv p604_d8_vs_p601_d8-viral_annotated.txt p605_d8_vs_p600_d8-viral_annotated.txt p602_d8_vs_p600_d8-viral_annotated.txt p604_d3_vs_p601_d3-viral_annotated.txt p605_d3_vs_p600_d3-viral_annotated.txt p602_d3_vs_p600_d3-viral_annotated.txt -d$',' -o viral_raw_counts_and_comparisons.xls;
#dpt: days post-transfection