-
CPU
-Maschine1- Model name: Intel(R) Xeon(R) CPU E5-1660 v3 @ 3.00GHz CPU family: 6 Model: 63 Thread(s) per core: 2 Core(s) per socket: 8 Socket(s): 1 # in total 16 -Maschine2- Model name: Intel(R) Xeon(R) CPU E5-2680 v4 @ 2.40GHz CPU family: 6 Model: 79 Thread(s) per core: 2 Core(s) per socket: 14 Socket(s): 2 # in total 56 -Maschine3- Model name: Intel(R) Xeon(R) Gold 6338 CPU @ 2.00GHz CPU family: 6 Model: 106 Thread(s) per core: 2 Core(s) per socket: 32 Socket(s): 2 # in total 128 -
Memory
-Maschine1- 65(G),786,164 -Maschine2- 131(G),876,772 -Maschine3- 1056(G),431,808 -
Log
jhuang@hamburg:~$ lscpu Architecture: x86_64 CPU op-mode(s): 32-bit, 64-bit Byte Order: Little Endian Address sizes: 46 bits physical, 48 bits virtual CPU(s): 16 On-line CPU(s) list: 0-15 Thread(s) per core: 2 Core(s) per socket: 8 Socket(s): 1 NUMA node(s): 1 Vendor ID: GenuineIntel CPU family: 6 Model: 63 Model name: Intel(R) Xeon(R) CPU E5-1660 v3 @ 3.00GHz Stepping: 2 CPU MHz: 1199.590 CPU max MHz: 3500,0000 CPU min MHz: 1200,0000 BogoMIPS: 5997.89 Virtualization: VT-x L1d cache: 256 KiB L1i cache: 256 KiB L2 cache: 2 MiB L3 cache: 20 MiB NUMA node0 CPU(s): 0-15 Vulnerability Itlb multihit: KVM: Mitigation: Split huge pages Vulnerability L1tf: Mitigation; PTE Inversion; VMX conditional cache flushes, SMT vulnerable Vulnerability Mds: Vulnerable: Clear CPU buffers attempted, no microcode; SMT vulnerable Vulnerability Meltdown: Mitigation; PTI Vulnerability Spec store bypass: Vulnerable Vulnerability Spectre v1: Mitigation; usercopy/swapgs barriers and __user pointer sanitization Vulnerability Spectre v2: Mitigation; Full generic retpoline, STIBP disabled, RSB filling Vulnerability Srbds: Not affected Vulnerability Tsx async abort: Not affected Flags: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush dts acpi mmx fxsr sse sse2 ss ht tm pbe syscall nx pdpe1gb rdtscp lm constant_tsc arch_perfmon pebs bts rep_good nopl xtopology nonstop_tsc cpuid aperfmperf pni pclmulqdq dtes64 monitor ds_cpl vmx smx est tm2 ssse3 sdb g fma cx16 xtpr pdcm pcid dca sse4_1 sse4_2 x2apic movbe popcnt aes xsave avx f16c rdrand lahf_lm abm cpuid_fault epb invpcid_single pti intel_ppin t pr_shadow vnmi flexpriority ept vpid ept_ad fsgsbase tsc_adjust bmi1 avx2 smep bmi2 erms invpcid cqm xsaveopt cqm_llc cqm_occup_llc dtherm ida arat p ln pts jhuang@hamburg:~$ free total used free shared buff/cache available Mem: 65786164 2048076 492364 54600 63245724 62947768 Swap: 20937724 524 20937200 jhuang@hamm:~$ lscpu Architecture: x86_64 CPU op-mode(s): 32-bit, 64-bit Address sizes: 46 bits physical, 48 bits virtual Byte Order: Little Endian CPU(s): 56 On-line CPU(s) list: 0-55 Vendor ID: GenuineIntel Model name: Intel(R) Xeon(R) CPU E5-2680 v4 @ 2.40GHz CPU family: 6 Model: 79 Thread(s) per core: 2 Core(s) per socket: 14 Socket(s): 2 Stepping: 1 CPU(s) scaling MHz: 37% CPU max MHz: 3300.0000 CPU min MHz: 1200.0000 BogoMIPS: 4800.21 Flags: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush dts acpi mmx fxsr sse sse2 ss ht tm pbe syscall nx pd pe1gb rdtscp lm constant_tsc arch_perfmon pebs bts rep_good nopl xtopology nonstop_tsc cpuid aperfmperf pni pclmulqdq dtes64 monitor ds_cpl vmx smx est tm2 ssse3 sdbg fma cx16 xtpr pdcm pcid dca sse4_1 sse4_2 x2apic movbe popcnt tsc_deadline_timer aes xsave avx f16c rdrand lahf_lm abm 3dnowprefetch cpuid_fault epb cat_l3 cdp_l3 invpcid_single pti intel_ppin ssbd ibrs ibpb stibp tpr_shadow vnmi fl expriority ept vpid ept_ad fsgsbase tsc_adjust bmi1 hle avx2 smep bmi2 erms invpcid rtm cqm rdt_a rdseed adx smap intel_pt xsaveopt c qm_llc cqm_occup_llc cqm_mbm_total cqm_mbm_local dtherm ida arat pln pts md_clear flush_l1d Virtualization features: Virtualization: VT-x Caches (sum of all): L1d: 896 KiB (28 instances) L1i: 896 KiB (28 instances) L2: 7 MiB (28 instances) L3: 70 MiB (2 instances) NUMA: NUMA node(s): 2 NUMA node0 CPU(s): 0-13,28-41 NUMA node1 CPU(s): 14-27,42-55 Vulnerabilities: Itlb multihit: KVM: Mitigation: Split huge pages L1tf: Mitigation; PTE Inversion; VMX conditional cache flushes, SMT vulnerable Mds: Mitigation; Clear CPU buffers; SMT vulnerable Meltdown: Mitigation; PTI Mmio stale data: Mitigation; Clear CPU buffers; SMT vulnerable Retbleed: Not affected Spec store bypass: Mitigation; Speculative Store Bypass disabled via prctl and seccomp Spectre v1: Mitigation; usercopy/swapgs barriers and __user pointer sanitization Spectre v2: Mitigation; Retpolines, IBPB conditional, IBRS_FW, STIBP conditional, RSB filling, PBRSB-eIBRS Not affected Srbds: Not affected Tsx async abort: Mitigation; Clear CPU buffers; SMT vulnerable jhuang@hamm:~$ free total used free shared buff/cache available Mem: 131876772 1433640 834304 85032 129608828 129289928 Swap: 5194740 317252 4877488 jhuang@WS-2290C:~$ lscpu Architecture: x86_64 CPU op-mode(s): 32-bit, 64-bit Address sizes: 46 bits physical, 57 bits virtual Byte Order: Little Endian CPU(s): 128 On-line CPU(s) list: 0-127 Vendor ID: GenuineIntel Model name: Intel(R) Xeon(R) Gold 6338 CPU @ 2.00GHz CPU family: 6 Model: 106 Thread(s) per core: 2 Core(s) per socket: 32 Socket(s): 2 Stepping: 6 CPU max MHz: 3200,0000 CPU min MHz: 800,0000 BogoMIPS: 4000.00 Flags: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush dts acpi mmx fxsr sse sse2 ss ht tm pbe syscall nx pd pe1gb rdtscp lm constant_tsc art arch_perfmon pebs bts rep_good nopl xtopology nonstop_tsc cpuid aperfmperf pni pclmulqdq dtes64 moni tor ds_cpl vmx smx est tm2 ssse3 sdbg fma cx16 xtpr pdcm pcid dca sse4_1 sse4_2 x2apic movbe popcnt tsc_deadline_timer aes xsave avx f16c rdrand lahf_lm abm 3dnowprefetch cpuid_fault epb cat_l3 invpcid_single ssbd mba ibrs ibpb stibp ibrs_enhanced tpr_shadow flexpri ority ept vpid ept_ad fsgsbase tsc_adjust bmi1 avx2 smep bmi2 erms invpcid cqm rdt_a avx512f avx512dq rdseed adx smap avx512ifma clfl ushopt clwb intel_pt avx512cd sha_ni avx512bw avx512vl xsaveopt xsavec xgetbv1 xsaves cqm_llc cqm_occup_llc cqm_mbm_total cqm_mbm_loc al split_lock_detect wbnoinvd dtherm ida arat pln pts vnmi avx512vbmi umip pku ospke avx512_vbmi2 gfni vaes vpclmulqdq avx512_vnni av x512_bitalg tme avx512_vpopcntdq la57 rdpid fsrm md_clear pconfig flush_l1d arch_capabilities Virtualization features: Virtualization: VT-x Caches (sum of all): L1d: 3 MiB (64 instances) L1i: 2 MiB (64 instances) L2: 80 MiB (64 instances) L3: 96 MiB (2 instances) NUMA: NUMA node(s): 2 NUMA node0 CPU(s): 0-31,64-95 NUMA node1 CPU(s): 32-63,96-127 Vulnerabilities: Gather data sampling: Mitigation; Microcode Itlb multihit: Not affected L1tf: Not affected Mds: Not affected Meltdown: Not affected Mmio stale data: Mitigation; Clear CPU buffers; SMT vulnerable Retbleed: Not affected Spec rstack overflow: Not affected Spec store bypass: Mitigation; Speculative Store Bypass disabled via prctl Spectre v1: Mitigation; usercopy/swapgs barriers and __user pointer sanitization Spectre v2: Mitigation; Enhanced / Automatic IBRS, IBPB conditional, RSB filling, PBRSB-eIBRS SW sequence Srbds: Not affected Tsx async abort: Not affected jhuang@WS-2290C:~$ free total used free shared buff/cache available Mem: 1056431808 6613676 867797608 67436 182020524 1043552932 Swap: 2097148 0 2097148
Author Archives: gene_x
GO terms for S. epidermidis
-
download go terms from Gene Ontology https://geneontology.org/docs/download-ontology/
import obonet # Load the OBO file url = 'http://purl.obolibrary.org/obo/go/go-basic.obo' graph = obonet.read_obo(url) # Open a file to write to with open('go_terms.csv', 'w') as f: f.write("id,name,namespace\n") # Write header for node_id, data in graph.nodes(data=True): name = data.get('name', '') namespace = data.get('namespace', '') f.write(f"{node_id}\t{name}\t{namespace}\n") # Write data -
prepare the blastp output
python3 filter_uniref_by_taxid.py uniref100.fasta uniref100_taxid1282.fasta makeblastdb -in uniref90_taxid1282.fasta -dbtype prot makeblastdb -in uniref50_taxid1282.fasta -dbtype prot; makeblastdb -in uniref100_taxid1282.fasta -dbtype prot; # -strand both -max_target_seqs 1 -taxidlist taxid_list.txt blastp -query 1585_CDS.fasta -db /home/jhuang/REFs/uniref100_taxid1282.fasta -out 1585_CDS_on_uniref100.blastp -num_threads 100 -outfmt 6 -evalue 1.0e-30 grep -v "UniRef100_UPI" 1585_CDS_on_uniref100.blastp > 1585_CDS_on_uniref100_.blastp python3 add_RefSeq-ID.py 1585_genes_annotated_with_GO.csv 1585.csv 1585_genes_annotated_with_GO_RefSeq-ID.csv python3 add_UniProt-ID.py #Input are 1585_CDS_on_uniref100_.blastp and 1585_genes_annotated_with_GO_RefSeq-ID.csv; Output is 1585_genes_annotated_with_GO_RefSeq-ID_UniProt-ID.csv. python3 add_Translation.py 1585_genes_annotated_with_GO_RefSeq-ID_UniProt-ID.csv 1585.csv 1585_genes_annotated_with_GO_RefSeq-ID_UniProt-ID_Translation.csv # check if they are running correctly cut -f1 -d',' 1585_genes_annotated_with_GO_NCBI-ID.csv > temp1 cut -f1 -d',' 1585_genes_annotated_with_GO_NCBI-ID_UniProt-ID_Translation.csv > temp2 diff temp1 temp2 -
code of filter_uniref_by_taxid.py
“) sys.exit(1) # Get input and output file paths from command-line arguments input_file = sys.argv[1] output_file = sys.argv[2] # Filter UniRef50 database based on TaxID=1282 filter_uniref50(input_file, output_file) print(“Filtered UniRef50 database saved to:”, output_file)import sys def filter_uniref50(input_file, output_file): with open(input_file, 'r') as in_handle: with open(output_file, 'w') as out_handle: record_started = False for line in in_handle: if line.startswith(">"): if "TaxID=1282 " in line: record_started = True out_handle.write(line) else: record_started = False elif record_started: out_handle.write(line) if __name__ == "__main__": # Check command-line arguments if len(sys.argv) != 3: print("Usage: python3 filter_uniref_by_taxid.py -
code of add_RefSeq-ID.py
“) #python3 add_NCBI-ID.py 1585_genes_annotated_with_GO.csv 1585.csv 1585_genes_annotated_with_GO_NCBI-ID.csv else: input_file1 = sys.argv[1] input_file2 = sys.argv[2] output_file = sys.argv[3] main(input_file1, input_file2, output_file)import pandas as pd import sys def main(input_file1, input_file2, output_file): # Load the first table table1 = pd.read_csv(input_file1) # Load the second table table2 = pd.read_csv(input_file2) # Merge the tables on SeqName from table1 and Locus Tag from table2 merged_table = pd.merge(table1, table2[['Locus Tag', 'inference']], left_on='SeqName', right_on='Locus Tag', how='left') # Drop the extra 'Locus Tag' column that is redundant after merge merged_table.drop(columns=['Locus Tag'], inplace=True) # Save the merged table to a new CSV file merged_table.to_csv(output_file, index=False) if __name__ == "__main__": if len(sys.argv) != 4: print("Usage: python script.py -
code of add_UniProt-ID.py
import csv # Read blastp results and create a dictionary mapping CDS IDs to lists of UniRef100 IDs def read_blastp_results(blastp_file): cds_to_uniref100 = {} with open(blastp_file, 'r') as blastp_handle: blastp_reader = csv.reader(blastp_handle, delimiter='\t') for row in blastp_reader: cds_id, uniref100_id = row[0], row[1] if cds_id not in cds_to_uniref100: cds_to_uniref100[cds_id] = [] cds_to_uniref100[cds_id].append(uniref100_id) return cds_to_uniref100 # Add UniRef100 IDs as the last column in the input CSV file def add_uniref100_column(input_file, output_file, cds_to_uniref100): with open(input_file, 'r') as input_handle: with open(output_file, 'w', newline='') as output_handle: reader = csv.reader(input_handle) writer = csv.writer(output_handle) for row in reader: cds_id = row[0] if cds_id in cds_to_uniref100: uniref100_ids = '|'.join(cds_to_uniref100[cds_id]) row.append(uniref100_ids) else: row.append("") # If no UniRef100 IDs found, add empty string writer.writerow(row) if __name__ == "__main__": blastp_file = "1585_CDS_on_uniref100_.blastp" # Replace with your blastp results file input_file = "1585_genes_annotated_with_GO_NCBI-ID.csv" output_file = "1585_genes_annotated_with_GO_NCBI-ID_UniProt-ID.csv" # Read blastp results and create dictionary mapping CDS IDs to lists of UniRef100 IDs cds_to_uniref100 = read_blastp_results(blastp_file) # Add UniRef100 IDs as the last column in the input CSV file add_uniref100_column(input_file, output_file, cds_to_uniref100) print("UniRef100 IDs added to the input CSV file:", output_file) -
code of add_Translation.py
“) #python3 add_NCBI-ID_to_1585-table.py 1585_genes_annotated_with_GO.csv 1585.csv 1585_genes_annotated_with_GO_Translation.csv else: input_file1 = sys.argv[1] input_file2 = sys.argv[2] output_file = sys.argv[3] main(input_file1, input_file2, output_file)import pandas as pd import sys def main(input_file1, input_file2, output_file): # Load the first table table1 = pd.read_csv(input_file1) # Load the second table table2 = pd.read_csv(input_file2) # Merge the tables on SeqName from table1 and Locus Tag from table2 merged_table = pd.merge(table1, table2[['Locus Tag', 'Translation']], left_on='SeqName', right_on='Locus Tag', how='left') # Drop the extra 'Locus Tag' column that is redundant after merge merged_table.drop(columns=['Locus Tag'], inplace=True) # Save the merged table to a new CSV file merged_table.to_csv(output_file, index=False) if __name__ == "__main__": if len(sys.argv) != 4: print("Usage: python script.py
Processing transposon insertion-site deep sequencing (Tn-seq) data
-
run tpp https://transit.readthedocs.io/en/latest/transit_running.html
#-maxreads 10000 or not_given for take all! #-primer AGATGTGTATAAGAGACAG the default primer of Tn5 is TAAGAGACAG! #-primer-start-window 0,159 set 0,159 as default! python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R1_001.fastq -reads2 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R2_001.fastq -output 10_chimera -mismatches initial_mutants_rep1 -bwa-alg mem -primer ACCTACCCCNCCGCTCTC -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg initial_mutants_rep1.tpp.cfg python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R1_001.fastq -reads2 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R2_001.fastq -output 10 -mismatches initial_mutants_rep1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg initial_mutants_rep1.tpp.cfg python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr3/LB_culture_rep1_S26_R1_001.fastq.gz -reads2 ./240405_VH00358_89_AAFC5MTM5/kr3/LB_culture_rep1_S26_R2_001.fastq.gz -output LB_culture_rep1 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg LB_culture_rep1.tpp.cfg python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr9/intracellular_mutants_24h_rep1_S29_R1_001.fastq.gz -reads2 ./240405_VH00358_89_AAFC5MTM5/kr9/intracellular_mutants_24h_rep1_S29_R2_001.fastq.gz -output intracellular_mutants_24h_rep1 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg intracellular_mutants_24h_rep1.tpp.cfg python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr8/extracellular_mutants_24h_rep2_S28_R1_001.fastq.gz -reads2 ./240405_VH00358_89_AAFC5MTM5/kr8/extracellular_mutants_24h_rep2_S28_R2_001.fastq.gz -output extracellular_mutants_24h_rep2 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg extracellular_mutants_24h_rep2.tpp.cfg python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./240405_VH00358_89_AAFC5MTM5/kr6/growthout_control_24h_rep2_S27_R1_001.fastq.gz -reads2 ./240405_VH00358_89_AAFC5MTM5/kr6/growthout_control_24h_rep2_S27_R2_001.fastq.gz -output growthout_control_24h_rep2 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689 mv tpp.cfg growthout_control_24h_rep2.tpp.cfg #END cp initial_mutants_rep1.tn_stats initial_mutants_rep1.tn_stats_ #Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):" sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_ sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_ python3 ../parse_tn_stats.py initial_mutants_rep1.tn_stats_ initial_mutants_rep1.tn_stats.xlsx python3 ../parse_tn_stats.py LB_culture_rep1.tn_stats_ LB_culture_rep1.tn_stats.xlsx #calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)" 441057 and 276060 mkdir initial_mutants_rep1_wig mv *.wig initial_mutants_rep1_wig/ zip -r initial_mutants_rep1_wig.zip initial_mutants_rep1_wig/ #The counts-files are too big, not nessasary to send: #~/Tools/csv2xls-0.4/csv_to_xls.py initial_mutants_rep1.tn_stats_ *.counts -d$',' -o initial_mutants_rep1.stats.xls; # contig_1_10 699 411 # contig_1_8 3206 2031 # contig_1_7 3787 2376 # contig_1_6 4604 2871 # contig_1_5 2794 1765 # contig_1_4 83 58 # contig_1_3 2944 1882 # contig_1_2 15446 9678 # contig_1_1 14391 8954 -
delete PCR-duplicate for checking if the template counts in tpp are correct
To address the issue of PCR duplicates in paired-end sequencing data (which you referred to as "PCA-duplicate," but I believe you meant "PCR duplicate"), you can use tools designed for post-alignment processing of SAM/BAM files. These tools identify and remove or mark duplicates where both reads of a pair are identical to another pair in the library, often indicating that they are duplicates resulting from PCR amplification rather than unique sequencing events. Step 0. Install Samtools and Picard Tools for Removing PCR Duplicates: Both Samtools and Picard are widely used for manipulating SAM/BAM files, including removing duplicates. Here's how you can use Picard to remove PCR duplicates: conda install -c bioconda picard samtools Step 1: Convert SAM to BAM and sort the BAM file If your file is in SAM format, you need to convert it to BAM format first using Samtools; PCR duplicate removal requires that the BAM file be sorted by coordinate. This can be done using Samtools: samtools view -Sb initial_mutants_rep1.sam > initial_mutants_rep1.bam samtools sort initial_mutants_rep1.bam -o initial_mutants_rep1_sorted.bam #557566 + 0 read1 #557566 + 0 read2 samtools index initial_mutants_rep1_sorted.bam # contig_1_1 14391 8954 #@SQ SN:gi|420257081|ref|NZ_AKKR01000009.1|contig_1_1 LN:84292 # Extract reads from contig_1_1 samtools view -b initial_mutants_rep1_sorted.bam "gi|420257081|ref|NZ_AKKR01000009.1|contig_1_1" > contig_1_1.bam # Run flagstat on the filtered BAM file samtools flagstat contig_1_1.bam #16589 + 0 read1 #16579 + 0 read2 Step 3: Mark or Remove Duplicates Using Picard Picard Tools can be used to mark duplicates. Here, I'll show you how to remove them: # -Xmx4g between java and -jar # java -jar /usr/local/bin/picard.jar picard MarkDuplicates \ I=contig_1_1.bam \ O=contig_1_1_no_duplicates.bam \ M=marked_dup_metrics.txt \ REMOVE_DUPLICATES=true # ## METRICS CLASS picard.sam.DuplicationMetrics #LIBRARY UNPAIRED_READS_EXAMINED READ_PAIRS_EXAMINED SECONDARY_OR_SUPPLEMENTARY_RDS UNMAPPED_READS UNPAIRED_READ_DUPLICATES READ_PAIR_DUPLICATES READ_PAIR_OPTICAL_DUPLICATES PERCENT_DUPLICATION ESTIMATED_LIBRARY_SIZE #Unknown Library 111 16473 81 111 90 6114 0 0.372629 16270 samtools flagstat contig_1_1_no_duplicates.bam #10405 + 0 read1 #10445 + 0 read2 This command marks duplicates (PCR and optical) and removes them, outputting a file without duplicates. The M option specifies where to write metrics about the duplicates. Step 3 (Alternatively). Using samtools to Remove Duplicates Directly Alternatively, if you prefer a simple and fast tool, samtools has a rmdup utility, but it's less versatile compared to Picard: samtools rmdup -S input_sorted.bam output_no_duplicates.bam Note that samtools rmdup is deprecated in recent versions of samtools because it does not handle all edge cases as well as Picard. Considerations Read Alignment: Ensure that your reads are aligned correctly and that the SAM/BAM files are error-free before duplicate removal. Optical vs. PCR Duplicates: Picard differentiates between PCR duplicates (originating from the same fragment of DNA) and optical duplicates (artifacts from the sequencing platform). Ensure that this distinction is clear if relevant for your analysis. Quality Checks: After removing duplicates, it's wise to perform quality control checks to understand how much of your data was affected by duplicates. -
Check how the failed trimmed reads not contain the the transposon:genomic boundary TAAGAGACAG
#-3.1- find read sequences >VH00358:89:AAFC5MTM5:1:1101:63544:1019_:N:0:TTTCTCTA+CTCGACG ACCTACCCCNCCGCTCTCATCAACCCAATAACGCAGGCAATCAAGCACCCACTGCATCACATAAGGTTGGCTAAGGCGCAATGTATTGCCACAACCGGTCATGTTGTCATATTCACCCTCAGAGGTGAGCCAGTAGTAGCTGGCATTGTCGATCCCACG >VH00358:89:AAFC5MTM5:1:1101:63960:1019_:N:0:TTTCTCTA+CTCGACG CAAAGAGGGGGGCGAAAAGATTTTAAACGATCTTGGCGAAATGAATTTTGAGTTTGTCGTGTGTGACTCACCTGCCGGTATCGAAAGCGGTGCGTTGATGGCACTGTATTTTGCTGACG >VH00358:89:AAFC5MTM5:1:1101:64150:1019_:N:0:TTTCTCTA+CTCGACG TCTTTACCGGGGATGGGACGCAAGATCTGCGCGCACTGGAACCGGCTTATGTTTCCTCCGTTGACAGTGGGAATCTGGCTGGGCATTTGATTGTACTGGCCAATACCTGTGAAGAGTGGGCCGCAGAACCCTTAGCGGCCAACGGGGCCAAGGGATTG >VH00358:89:AAFC5MTM5:1:1101:65400:1019_:N:0:TTTCTCTA+CTCGACG CACCTACCCCCCAGCTCTCATCAACCACAATTGACGCAACATCAGCTGGCGCCATGGCATTAATAACAAACTGGATGAATGGCCCCTGGGATCAACAGCACGAACAAACCGCCGTGTGGTGCCATCAGTTGCGCGCCGAAGGCCATTGACAGAGCACC >VH00358:89:AAFC5MTM5:1:1101:62048:1038_:N:0:TTTCTCTA+CTCGACG CCTACACCCCCGCTCTCATCAACCGCAGCAAAGAAAACGAAAATAATGAAGTCATAAAACTCAAGCGCTCCGCCTAAAGCCGCCAGAGTGAGGGTTTTATAATCTTGCTTATTCAGCCGACGGTTATGAT >VH00358:89:AAFC5MTM5:1:1101:64623:1038_:N:0:TTTCTCTA+CTCGACG CCTACACCCCAGCTCTCATCAACCAATAACCAGTCAACATCACTGACATCATGTTGCTGGCAATATTCCAGCAGTGCATCTGGCGCCCAAAAACGCGCGCGATCCCCTGCTCGCTGACTTAATACTTGAGTGGCGGCCTTGCCCGGCCCAGTAACCCA >VH00358:89:AAFC5MTM5:1:1101:65343:1038_:N:0:TTTCTCTA+CTCGACG ACCTACCACCAAGCTCTCATCAACCTGAATGGATTGAGGGCTACGCTCCCAGATCTGTTCTGCCAATTGGCGCAGATGGTTATCGGTGCAATACAAATGCACAAATGACAAATCGTCTCTTACATCTCAGCTCGATAAACTGCCCTTTGGCATAATGC >VH00358:89:AAFC5MTM5:1:1101:63317:1057_:N:0:GTTCTCTA+CTCGACG GTTGGCGTCCGGCATCTGCATATCACTCATAAAGCGCCTCGATAATCCCACGGATATCCTGGTCGCTAGCTGGGCGCGGGTTGGTGCGCAATGTGAGCTCGGCCAGGGCTGCAGCAATCATATCGGGCAGATGTTGCTGCAATTGGCGCTCATTAACTT >VH00358:89:AAFC5MTM5:1:1101:63430:1057_:N:0:TTTCTCTA+CTCGACG GCACTGGAAGAGCCGACTAGCCTCAATACTCTTGAACTGCTACCGGAATTATTTGCCGCCAATATTGCCTCGGTGAAAATTGAAGGGCGTCAGCGCAGCCCGGCTTATGTCAGCCAGGTGGCGAAAGTGTGGCGGCAGGCAATTGACCGCTATCTGGC >VH00358:89:AAFC5MTM5:1:1101:58299:1076_:N:0:TTTCTCTA+CTCGACG TAAATAGGCCAGACTTGAAATCACACGATCCGGCCAGCGATT #-3.2- generate the genomic sequences blastn -db ../WA-314_m.fna -query initial_mutants_rep1.trimmed1_failed_trim_n20 -out failed_trim_n20_on_yersinia.blastn -evalue 10000 -num_threads 124 -outfmt 6 -max_target_seqs 1 VH00358:89:AAFC5MTM5:1:1101:63544:1019_:N:0:TTTCTCTA+CTCGACG gi|420260421|ref|NZ_AKKR01000094.1|contig_3_51 100.000 141 0 0 19 159 20552 20692 1.20e-70 261 VH00358:89:AAFC5MTM5:1:1101:63960:1019_:N:0:TTTCTCTA+CTCGACG gi|420258256|ref|NZ_AKKR01000038.1|contig_1_38 98.319 119 2 0 1 119 43674 43556 3.14e-55 209 VH00358:89:AAFC5MTM5:1:1101:64150:1019_:N:0:TTTCTCTA+CTCGACG gi|420257402|ref|NZ_AKKR01000018.1|contig_1_12 98.101 158 3 0 1 158 59515 59672 4.26e-75 276 VH00358:89:AAFC5MTM5:1:1101:65400:1019_:N:0:TTTCTCTA+CTCGACG gi|420260713|ref|NZ_AKKR01000106.1|contig_5_2 100.000 57 0 0 18 74 78361 78417 5.91e-24 106 VH00358:89:AAFC5MTM5:1:1101:62048:1038_:N:0:TTTCTCTA+CTCGACG gi|420257081|ref|NZ_AKKR01000009.1|contig_1_1 100.000 110 0 0 21 130 78749 78640 1.62e-53 204 VH00358:89:AAFC5MTM5:1:1101:64623:1038_:N:0:TTTCTCTA+CTCGACG gi|420259377|ref|NZ_AKKR01000067.1|contig_3_23 99.329 149 0 1 10 158 54935 55082 7.13e-73 268 VH00358:89:AAFC5MTM5:1:1101:65343:1038_:N:0:TTTCTCTA+CTCGACG gi|420258810|ref|NZ_AKKR01000051.1|contig_3_3 99.301 143 1 0 16 158 46439 46581 4.29e-70 259 VH00358:89:AAFC5MTM5:1:1101:63317:1057_:N:0:GTTCTCTA+CTCGACG gi|420257889|ref|NZ_AKKR01000029.1|contig_1_25 96.226 159 6 0 1 159 36032 36190 1.20e-70 261 VH00358:89:AAFC5MTM5:1:1101:63430:1057_:N:0:TTTCTCTA+CTCGACG gi|420260713|ref|NZ_AKKR01000106.1|contig_5_2 100.000 158 0 0 1 158 54938 55095 4.23e-80 292 VH00358:89:AAFC5MTM5:1:1101:58299:1076_:N:0:TTTCTCTA+CTCGACG gi|420259608|ref|NZ_AKKR01000075.1|contig_3_31 97.619 42 1 0 1 42 25535 25576 1.04e-14 73.1 samtools faidx WA-314_m.fna "gi|420260421|ref|NZ_AKKR01000094.1|contig_3_51":20534-20692 > genome_rg_for_read1.fasta samtools faidx WA-314_m.fna "gi|420258256|ref|NZ_AKKR01000038.1|contig_1_38":43556-43674 > genome_rg_for_read2.fasta samtools faidx WA-314_m.fna "gi|420257402|ref|NZ_AKKR01000018.1|contig_1_12":59515-59672 > genome_rg_for_read3.fasta samtools faidx WA-314_m.fna "gi|420260713|ref|NZ_AKKR01000106.1|contig_5_2":78344-78501 > genome_rg_for_read4.fasta samtools faidx WA-314_m.fna "gi|420257081|ref|NZ_AKKR01000009.1|contig_1_1":78640-78769 > genome_rg_for_read5.fasta samtools faidx WA-314_m.fna "gi|420259377|ref|NZ_AKKR01000067.1|contig_3_23":54926-55082 > genome_rg_for_read6.fasta samtools faidx WA-314_m.fna "gi|420258810|ref|NZ_AKKR01000051.1|contig_3_3":46424-46581 > genome_rg_for_read7.fasta samtools faidx WA-314_m.fna "gi|420257889|ref|NZ_AKKR01000029.1|contig_1_25":36032-36190 > genome_rg_for_read8.fasta samtools faidx WA-314_m.fna "gi|420260713|ref|NZ_AKKR01000106.1|contig_5_2":54938-55095 > genome_rg_for_read9.fasta samtools faidx WA-314_m.fna "gi|420259608|ref|NZ_AKKR01000075.1|contig_3_31":25535-25576 > genome_rg_for_read10.fasta #-3.3- transposon amplicon start sequence >Transposon_ampli_start ACCTACAACAAAGCTCTCATCAAC CGTGGCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTCAGGGTTGAGATGTGTA TAAGAGACAG #-3.4- generate 10 failed_${read}_group.fasta and multiple align them for read in read1 read2 read3 read4 read5 read6 read7 read8 read9 read10; do cat failed_${read}.fasta genome_rg_for_${read}.fasta transposon_ampli_start.fasta > failed_${read}_group.fasta mafft --adjustdirection --clustalout failed_${read}_group.fasta > failed_${read}_group.aln done cat failed_read1_group.aln failed_read2_group.aln failed_read3_group.aln failed_read4_group.aln failed_read5_group.aln failed_read6_group.aln failed_read7_group.aln failed_read8_group.aln failed_read9_group.aln failed_read10_group.aln > failed_reads_group.aln #-3.5- show the results in checking_failed_reads.pdf -
Source code of parse_tn_stats.py used in the point 1.
import argparse import pandas as pd def parse_tn_stats_detailed(file_path): with open(file_path, 'r') as file: lines = file.readlines() print("Number of lines read:", len(lines)) if lines: print("First few lines:", lines[:10]) metrics_df = pd.DataFrame() current_metric = None metric_data = {} for line in lines: if line.startswith('# ') and not line.startswith('# '): # Ensure it's not a contig data line # Metric definition if current_metric and metric_data: # Save data before starting new metric for contig, value in metric_data.items(): metrics_df.at[contig, current_metric] = value metric_data = {} # Extract the metric name current_metric = line.split(':')[0].strip('# ') print("Processing new metric:", current_metric) # Debug output else: # Data lines for contigs under the current metric parts = line.strip().split(':') contig = parts[0].strip() value = parts[1].strip() try: value = float(value) if value.lower() != 'nan' else pd.NA except ValueError: value = pd.NA # In case of conversion failure metric_data[contig] = value # Capture the last metric data after finishing all lines if current_metric and metric_data: for contig, value in metric_data.items(): metrics_df.at[contig, current_metric] = value metrics_df.fillna(value=pd.NA, inplace=True) return metrics_df def main(): parser = argparse.ArgumentParser(description="Parse detailed TN stats from a file and output to Excel.") parser.add_argument("input_file", help="Input file path for TN stats.") parser.add_argument("output_file", help="Output Excel file path.") args = parser.parse_args() # Parse the detailed metrics from the provided file path detailed_metrics_df = parse_tn_stats_detailed(args.input_file) # Save the DataFrame to an Excel file with pd.ExcelWriter(args.output_file, engine='openpyxl') as writer: detailed_metrics_df.to_excel(writer, sheet_name='Detailed Metrics') print(f"Final DataFrame saved to Excel: {args.output_file}") if __name__ == "__main__": main()
Tn-seq data analysis
Genome-wide identification of virulence-associated genes in Staphylococcus aureus using Transposon insertion-site deep sequencing
5.7 Mutagenesis in S. aureus
5.7.1 Generation of random Transposon mutant pools in S. aureus
- Pooled mariner transposon mutant libraries were generated as previously described 246.
- Briefly, 6850 transformed with plasmid pBTn was revived on TSA Cm 10 plates.
- An overnight culture was prepared by picking a colony and resuspending in broth with Cm 10 and without Xylose, followed by incubation at 30°C with agitation at 180 rpm.
- The culture was diluted in ratio 1:100 in fresh broth containing 0.5% Xylose, Cm 10 and Erm 5, followed by overnight incubation at 30°C with agitation at 180 rpm.
- This was done in 10 replicates simultaneously. The cultures were again diluted in ratio 1:100 in fresh broth containing 0.5% Xylose and Erm 5, followed by overnight incubation at 42°C with agitation at 180 rpm. This step was repeated two more times.
- All cultures were combined and centrifuged at 3000 xg for 10 minutes.
- The pellets were mixed with fresh broth so that the OD600nm was 10.0.
- Stocks were prepared as previously described (See Section 5.1.2). Successful transposition and subsequent loss of plasmid due to temperature elevation was assessed by enumeration of Erm resistant but Cm sensitive bacteria.
- A trial was made with more than one medium and more than 3 temperatures elevation steps to assess maximum efficiency of transposition (See Section 2.1.1).
- Colonies were randomly chosen from Erm 5 plates and tested for insertion in the genome by Arbitrary PCR (See Section 5.6.1) followed by
5.9 High throughput analysis of nucleic acids
5.9.1 Transposon insertion-site deep sequencing (Tn-seq)
- Chromosomal DNA was isolated from pooled mariner transposon mutant libraries including inocula and output harvested from various conditions, by the method described in Section 5.5. Fragment DNA libraries were prepared and sequenced using the following steps.
5.9.1.1 Fragmentation of S. aureus genomic DNA
- Hydrodynamic shearing of genomic DNA was carried out by application of ultrasonics on a Bioruptorr instrument.
- 1 ml of DNA dissolved in water were taken in 15 ml polystyrene tubes and sonicated for 10 cycles, with the following specifications.
- Ultrasonic wave frequency: 20kHz, wave power: H i.e. 320W, time: 1 minute/cycle (30 seconds ’ON’ and 30 seconds ’OFF’).
- Fragmentation was carried out at 4°C to avoid shearing by the heat generated from the sonication probe.
- This setting consistently resulted in fragments sizes between 100 to 500 nucleotides.
- The size range was analyzed by agarose gel electrophoresis and on Agilent 2100 Bioanalyzer instrument.
5.9.1.2 End-repair and size selection of DNA fragments
- 5 μg of fragmented DNA from each sample were repaired using the commercially available NEBNextr End repair module following manufacturer’s instructions in a volume of 100 μl.
- Fragments within the size range of 200-300 nucleotides were selected using a gel-free double-Solid Phase Reversible Immobilization (dSPRI), method G, as previously described using AMPure XP beads in three steps.
- Step 1: Immediately after end repair, the samples were mixed with 0.9X volume of beads and incubated at room temperature for 20 minutes. The beads were separated using a magnetic rack and the residual solution was transfered to a fresh microfuge tube.
- Step 2: The DNA solution was subsequently mixed with 0.11X volume of beads and incubated at room temperature for 7 minutes. The beads were again separated and the residual solution was disposed.
- Step 3: The beads were washed twice with 70% Ethanol for 30 seconds without removing tubes from the magnetic rack.
- The beads were air-dried for 2-3 minutes and eluted in 42 μl of sterile nuclease-free water.
- If required, the size range was confirmed by agarose gel electrophoresis.
5.9.1.3 dA-tailing of blunt end DNA fragments
- After the correct size of DNA fragments were obtained, NEBNextr dA-Tailing module was used to add non-templated deoxy-Adenosine monophosphate (dAMP) molecules to the 3‘ ends, according to manufacturer’s instructions.
- The DNA fragments were purified using AMPure XP beads and eluted into 30 μl of sterile nuclease-free water.
5.9.1.4 Generation and ligation of adapters to DNA fragments
- For generation of multiplexing dsDNA adapters compatible with Illuminar sequencing platform, two separate oligonucleotides were used.
- MultiPlex-Y-Adapt_f with a 5′ phosphorylation and MultiPlex-Y-Adapt_r with a 3′ terminal phosphorothioate linkage, were mixed in equimolar concentrations in 1X Oligo-annealing buffer.
- The reaction mixture was heated to 94°C for 5 minutes followed by gradual cooling to room temperature and finally incubated on ice.
- Adaptors were ligated to dA-tailed fragments overnight at 16°C using T4 DNA ligase in the presence of 1X T4 ligase buffer and 50% Polyethylene glycol (PEG), added to the previous reaction from Section 5.9.1.3. Upon completion, DNA fragments were purified with AMPure XP beads and eluted in 25 μl of sterile nuclease-free water.
5.9.1.5 Massively parallel sequencing on Illuminar platform
- Before sequencing transposon ends were enriched by aforementioned PCR steps (See Section 5.6.1), purified with AMPure XP beads and checked for quality (See Section 5.5.7).
- The resulting DNA fragment libraries were sequenced on the Illuminar Hi-Seq 2500 platform obtaining 10-30 million single reads per sample with indices, using the transposon-specific oligonucleotide primer Himar1-Seq.
5.9.1.6 Tn-seq data analysis
- Sequencing files were obtained and was processed and analyzed, as previously described 301,326 .
- In brief, Illuminar adapter sequences were removed using cutadapt version 1.2.1 327.
- The sequence reads were checked for the nucleotide pattern ‘CAACCTGT’ originating from the transposon Inverted Terminal Repeats (ITR). [In my case ‘TAAGAGACAG’]
- Only reads containing this specific sequence with an allowance of one mismatch or gap and with minimum length of 16 nucleotides were used for further analyses. [In my case 20]
- Further, the reads were mapped to the Staphylococcus aureus 6850 genome (GenBank accession CP006706) using Bowtie 2 algorithm version 2.1.0 328.
- To identify transposon insertion sites (TIS), the aligned start positions of mapped reads were extracted and each position on the genome, covered by at least one alignment start, was annotated as TIS.
- The genomic position was adjusted strand-specifically to account for the 1 nucleotide shift of the reads mapping on the positive or negative strand.
- Statistical analysis of enriched or depleted reads from each TIS, was performed using DESeq2 version 1.6.2 261.
- The HeLa infection experiment was modelled as a time course including a technical replicate with the input libraries as time t0.
- For the mouse lung infection experiment, the 3 output libraries were compared to the input libraries.
- Genes with very low mean normalized read depth (mnrd) <4 were excluded in the HeLa experiment and those with mnrd 8 were excluded from the animal experiments.
- The P-values were corrected for multiple testing and the TIS with adjusted P-value <= 0.05 were considered as significantly increased or decreased.
5.9.2 RNA deep sequencing
- RNA were isolated as previously described (See Section 5.5.5), followed by removal of DNA using DNase I enzyme (See Section 5.5.6).
- Quantity and quality of RNA was determined by spectrophotometric method using Nanodrop and 1.8% agarose gel containing formamide. mRNA enrichment was performed by Ribodepletion Kits followed by library Preparation for Illuminar .
- Depletion of processed transcripts, were performed by using Terminal 5’-phosphate-dependent exonuclease (TEX) as previously described (Sharma 2010).
- Briefly, RNA samples were poly(A)-tailed using poly(A) polymerase.
- 5’-triphosphates were removed by treatment with tobacco acid pyrophosphatase (TAP).
- RNA adapters were ligated to the 5’-phosphate ends and first-strand cDNA were generated using an oligo(dT)-primer and M-MLV reverse transcriptase.
- High fidelity DNA polymerase cDNA was amplified by PCR. cDNA were sequenced on Illuminar HiSeq platform2 , yielding 100 bp paired end reads.
- Adapters removed and trimmed to 70 bp using Trimmomatic (Bolger 2014) and only reads exceeding a mean base quality 5 within all sliding windows of 5bp were mapped to the S. aureus USA300 FPR3757 genome (NCBI accession NC_007793.1), using Bowtie2 (25).
- Only paired and concordant alignments were considered further, yielding at least 12 million uniquely mapped read pairs per replicate. A total of 2693 coding and non-coding transcripts were identified for further analysis.
- Differential transcript abundance analysis was performed using the DESeq2 package v.1.5.9 (12) in R.
[301] Natural mutations in a staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Proceedings of the National Academy of Sciences, 2016. [326] Transcriptional landscape and essential genes of neisseria gonorrhoeae. Nucleic acids research, 42(16):10579–10595, 2014. [327] Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal, 17(1): pp–10, 2011. [328] Fast gapped-read alignment with bowtie 2. Nature methods, 9(4):357–359, 2012.
How does the adapter in Illumina sequencing work?
Why are adapter sequences trimmed from only the 3′ ends of reads? https://support.illumina.com.cn/bulletins/2016/04/adapter-trimming-why-are-adapter-sequences-trimmed-from-only-the–ends-of-reads.html
Expands one nucleic base at a time https://www.researchgate.net/figure/Illumina-sequencing-process-A-DNA-library-Breaks-the-genome-DNA-to-form-DNA_fig3_357155980
In Illumina sequencing, the barcode (also known as the index) is indeed a critical part of the sequencing process because it allows for the identification and demultiplexing of multiple samples that are sequenced together in the same run.
Here’s how it works:
-
Adapter Ligation: First, adapters are ligated to the fragmented DNA. These adapters contain the sequences for P5 and P7 priming sites, necessary for flow cell attachment and the initiation of the sequencing reaction.
-
Index Sequences: The adapters also include index sequences (barcodes). In the case of dual-indexing, one index (Index 1) is on the adapter ligated at the P7 end, and another index (Index 2) is on the adapter ligated at the P5 end. These indexes are unique to each sample.
-
Sequencing Initiation: Sequencing begins with the binding of sequencing primers to their complementary sites on the adapters—not directly from the index sequences. However, the index sequences are read during specific additional sequencing reactions:
- For Read 1, sequencing starts from the P5 end.
- If performing paired-end sequencing, after Read 1 is complete, the machine performs a read of the Index 1 sequence.
- Then, the flow cell is reconfigured to sequence Read 2 from the P7 end.
- Finally, if dual-indexing, the Index 2 sequence is read.
-
Index Reading (Read1 Primer and i7 Index Primer): The indexes are not part of the main sequence reads (Read 1 or Read 2) but are read in separate, dedicated sequencing reactions using specific index primers after the completion of the standard sequencing cycles.
The crucial point is that the sequencing of the index sequences happens after the main DNA fragment has been sequenced, during dedicated index read cycles. The readout of the indexes is integral to the sequencing run and allows the software to assign each sequence to the correct sample in the analysis phase, enabling the pooling of multiple samples in a single sequencing run. This process is called demultiplexing.
Tn5 adapter https://teichlab.github.io/scg_lib_structs/methods_html/plate_and_piATAC-seq.html
Y-shaped-adaptors https://www.researchgate.net/figure/DNA-template-ligation-with-Y-shaped-adaptors-Blunt-ended-ds-DNA-templates-5_fig2_323640739
(0) Final library structure:
5'- AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGXXXXXXXX...XXXXXXXXCTGTCTCTTATACACATCTCCGAGCCCACGAGACNNNNNNNNATCTCGTATGCCGTCTTCTGCTTG
TTACTATGCCGCTGGTGGCTCTAGATGTGNNNNNNNNAGCAGCCGTCGCAGTCTACACATATTCTCTGTCXXXXXXXX...XXXXXXXXGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTGNNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
Illumina P5 i5 s5 ME cDNA ME s7 i7 Illumina P7
Library sequencing:
(1) Add read 1 sequencing primer to sequence the first read (bottom strand as template):
Primer1
5'- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG|--READ1---->
3'- TTACTATGCCGCTGGTGGCTCTAGATGTGNNNNNNNNAGCAGCCGTCGCAGTCTACACATATTCTCTGTCXXXXXXXX...XXXXXXXXGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTGNNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
(2) Add index 1 sequencing primer to sequence the first index (i7) (bottom strand as template, 8 cycles):
5'- CTGTCTCTTATACACATCTCCGAGCCCACGAGAC------>
3'- TTACTATGCCGCTGGTGGCTCTAGATGTGNNNNNNNNAGCAGCCGTCGCAGTCTACACATATTCTCTGTCXXXXXXXX...XXXXXXXXGACAGAGAATATGTGTAGAGGCTCGGGTGCTCTGNNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5'
(3) Cluster regeneration, add Index 2 sequencing primer to sequence the second index (i5) (top strand as template, 8 cycles. Single cells can be identified as the combination of i5 and i7):
5'- AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGXXXXXXXX...XXXXXXXXCTGTCTCTTATACACATCTCCGAGCCCACGAGACNNNNNNNNATCTCGTATGCCGTCTTCTGCTTG
<-------AGCAGCCGTCGCAGTCTACACATATTCTCTGTC -5'
(4) Add read 2 sequencing primer to sequence the second read (top strand as template):
5'- AATGATACGGCGACCACCGAGATCTACACNNNNNNNNTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGXXXXXXXX...XXXXXXXXCTGTCTCTTATACACATCTCCGAGCCCACGAGACNNNNNNNNATCTCGTATGCCGTCTTCTGCTTG
<----READ2--|GACAGAGAATATGTGTAGAGGCTCGGGTGCTCTG -5'
Primer2Tarifvertrag TV-Ärzte/VKA
https://www.vka.de/verband/unsere-mitgliedverbaende https://www.av-hamburg.de/ https://www.av-hamburg.de/fileadmin/dokumente/tarifvertraege/aktuell/entgelttabellen/Allgemeine_Tabelle_01.03.2024_2.pdf https://www.av-hamburg.de/fileadmin/dokumente/mitglieder/AV14.26.2401_Namensliste_Mitglieder_20240122_01.pdf https://www.uke.de/karriere/das-uke-als-arbeitgeber/verguetung-tarifvertraege/index.html https://www.berufe-sh.de/informationen/gehaltstabellen.html https://www.personalrat.uni-freiburg.de/aktuelles/gehaltstabellen
Hier sind einige Beispiele für Krankenhäuser in Deutschland, die unter den Tarifvertrag TV-Ärzte/VKA fallen. Diese sind typischerweise kommunale Krankenhäuser, deren Träger Mitglieder der Vereinigung der kommunalen Arbeitgeberverbände (VKA) sind:
- Städtisches Klinikum München – Eines der größten kommunalen Krankenhäuser in Deutschland.
- Klinikum Stuttgart – Das größte Krankenhaus der Region Stuttgart, das von der Stadt betrieben wird.
- Klinikum Dortmund – Ein großes kommunales Krankenhaus in Dortmund.
- Städtische Kliniken Frankfurt – Eine Gruppe von Krankenhäusern unter der Trägerschaft der Stadt Frankfurt.
- Klinikum Karlsruhe – Das städtische Krankenhaus in Karlsruhe.
- Städtisches Klinikum Braunschweig – Ein großes Krankenhaus in Niedersachsen.
- Klinikum Nürnberg – Ein großes kommunales Krankenhaus in Bayern.
- Klinikum Bremen-Mitte – Teil des kommunalen Klinikverbunds Gesundheit Nord in Bremen.
- Klinikum Oldenburg – Ein kommunales Krankenhaus in Niedersachsen.
- Städtisches Klinikum Magdeburg – Das kommunale Krankenhaus in der Stadt Magdeburg.
- Universitätsklinikum Hamburg-Eppendorf (UKE) Körperschaft des öffentlichen Rechts
NAMENSLISTE DER MITGLIEDER DER AVH (Auszug, Stand 22. Januar 2024)
- ab ausblick hamburg ggmbh www.ausblick-hamburg.de
- AHS Hamburg Aviation Handling Services GmbH www.ahs-de.com
- Alida Schmidt-Stiftung www.alida.de
- Ambulante Psychosoziale Dienste
- Lydia Buchfink GmbH & Co. KG
- www.lydiabuchfink.de
- Arbeit und Leben DGB/VHS Hamburg e.V. www.hamburg.arbeitundleben.de
- Archäologisches Museum Hamburg
- und Stadtmuseum Harburg
- Stiftung öffentlichen Rechts
- www.amh.de
- ASB Arbeiter-Samariter-Bund Sozialeinrichtungen
- (Hamburg) GmbH
- www.asb-hamburg.de
- Asklepios Kliniken Hamburg GmbH www.asklepios.com
- Asklepios Westklinikum Hamburg GmbH www.asklepios.com/hamburgrissen
- Ballin Stiftung e.V. www.ballin.hamburg
- BBW Berufsbildungswerk Hamburg gGmbH www.bbw-hamburg.de
- Bernhard-Nocht-Institut für Tropenmedizin
- Stiftung öffentlichen Rechts
- www.bni-hamburg.de
- BFW Berufsförderungswerk Hamburg gGmbH www.bfw-hamburg.de
- BTZ Berufliches Trainingszentrum Hamburg gGmbH www.btz-hamburg.de
- Bücherhallen Hamburg
- Stiftung Hamburger Öffentliche Bücherhallen
- www.buecherhallen.de
- BdB Bundesverband der Berufsbetreuer/innen e.V.
- (Gast)
- www.bdb-ev.de
- Compass Sozialpsychiatrische Dienste GmbH
- (Gast)
- www.compass-hamburg.de
- Dataport – Anstalt des öffentlichen Rechts –
- (Gast)
- www.dataport.de
- Deutsches Elektronen-Synchrotron DESY
- Stiftung bürgerlichen Rechts
- www.desy.de
- Deutsches Maritimes Zentrum e.V.
- (Gast)
- www.dmz-maritim.de
- Deutsches Schauspielhaus in Hamburg
- Neue Schauspielhaus GmbH
- www.schauspielhaus.de
- Diesterweg-Stiftung
- Stiftung bürgerlichen Rechts
- (Gast)
- www.diesterweg-stiftung.de
- Elbe-Werkstätten GmbH www.elbe-werkstaetten.de
- Elbkinder KITA Hamburg Servicegesellschaft mbH
- Elbkinder – Vereinigung Hamburger
- Kindertagesstätten gGmbH
- www.elbkinder-kitas.de
- Elbphilharmonie und Laeiszhalle
- Betriebsgesellschaft mbH
- www.elbphilharmonie.de
- EEW Energy from Waste Stapelfeld GmbH www.eew-energyfromwaste.com
- European X-Ray Free-Electron Laser Facility
- (European XFEL) GmbH
- (Gast)
- www.xfel.eu
- Flüchtlingszentrum Hamburg
- Zentrale Information und Beratung
- für Flüchtlinge gGmbH
- (Gast)
- www.fz-hh.de
- Flutopfer-Stiftung von 1962 www.alida.de
- f & w fördern und wohnen AöR www.foerdernundwohnen.de
- Forschungsstelle für Zeitgeschichte
- in Hamburg – FZH –
- Stiftung bürgerlichen Rechts
- www.zeitgeschichte-hamburg.de
- Freie und Hansestadt Hamburg www.hamburg.de/personalamt
- GBI Großhamburger Bestattungsinstitut rV www.gbi-hamburg.de
- Gesellschaft zur Beseitigung
- von Sonderabfällen mbH
- www.sad-rondeshagen.de
- GIGA German Institute of Global and Area Studies
- Stiftung des bürgerlichen Rechts
- www.giga-hamburg.de
- GKL Gemeinsame Klassenlotterie der Länder
- Anstalt öffentlichen Rechts
- (Gast)
- www.gkl.org
- GMH | Gebäudemanagement Hamburg GmbH www.gmh-hamburg.de
- Grone Service- und Verwaltungsgesellschaft mbH www.grone.de
- HADAG Seetouristik und Fährdienst AG www.hadag.de
- HADAG Verkehrsdienste GmbH www.hadag.de
- Hamburg Airport
- Flughafen Hamburg GmbH
- www.hamburg-airport.de
- Hamburg Institute for Advanced Study e.V. (HIAS) www.hias-hamburg.de
- Hamburg Invest Entwicklungsgesellschaft
- mbH & Co. KG
- www.hamburg-invest.com
- Hamburg Kreativ Gesellschaft mbH
- (Gast)
- www.kreativgesellschaft.org
- Hamburg Marketing GmbH www.marketing.hamburg.de
- Hamburg Messe und Congress GmbH www.hamburg-messe.de
- Hamburg Port Authority AöR www.hamburg-port-authority.de
- Hamburg Tourismus GmbH www.hamburg-tourismus.de
- hamburger arbeit GmbH www.hamburger-arbeit.de
- Hamburger Arbeitsassistenz gGmbH www.hamburger-arbeitsassistenz.de
- Hamburger Blindenstiftung
- Stiftung privaten Rechts
- www.blindenstiftung.de
- Hamburger Friedhöfe – AöR – www.friedhof-hamburg.de
- Hamburger Kind – Bildung und Betreuung gGmbH www.hamburgerkind.de
- Hamburger Krematorium GmbH www.krematorium-hamburg.de
- Hamburger Kunsthalle
- Stiftung öffentlichen Rechts
- www.hamburger-kunsthalle.de
- Hamburger Lebenshilfe-Werk gGmbH www.lebenshilfe-werk-ggmbh.de
- Hamburger Stadtentwässerung AöR www.hamburgwasser.de
- Hamburgische Staatsoper GmbH www.staatsoper-hamburg.de
- Handwerkskammer Hamburg
- Körperschaft öffentlichen Rechts
- (Gast)
- www.hwk-hamburg.de
- HEG Hamburger Entsorgungsgesellschaft mbH www.heg-hamburg.de
- Helmholtz-Zentrum hereon GmbH www.hereon.de
- HIW Hamburg Invest
- Wirtschaftsförderungsgesellschaft mbH
- www.hamburg-invest.com
- HVF Hamburgischer Versorgungsfonds AöR www.hvf.hamburg.de
- Institut für die Geschichte der deutschen Juden
- Stiftung des bürgerlichen Rechts
- (Gast)
- www.igdj-hh.de
- Institut für Friedensforschung
- und Sicherheitspolitik – IFSH –
- an der Universität Hamburg
- Stiftung des bürgerlichen Rechts
- www.ifsh.de
- ipb – Institut für Innovation und Praxistransfer
- in der Betreuung gGmbH
- (Gast)
- www.ipb-weiterbildung.de
- ISZ Immobilien Service Zentrum GmbH
- Johann Daniel Lawaetz-Stiftung www.lawaetz.de
- Jugendhilfe e.V.
- (Gast)
- www.jugendhilfe.de
- Kampnagel Internationale Kulturfabrik GmbH www.kampnagel.de
- Kassenärztliche Vereinigung Hamburg (KVH)
- Körperschaft des öffentlichen Rechts
- (Gast)
- www.kvhh.de
- Leben mit Behinderung Hamburg
- Sozialeinrichtungen gGmbH
- www.lmbhh.de
- Leben mit Behinderung Hamburg
- Elternverein e.V.
- www.lmbhh.de
- Leibniz-Institut für Virologie (LIV)
- Stiftung bürgerlichen Rechts
- www.leibniz-liv.de
- Logistik-Initiative Hamburg Management GmbH www.hamburg-logistik.net
- LOTTO Hamburg GmbH
- (Gast)
- www.lotto-hh.de
- LungenClinic Grosshansdorf GmbH www.lungenclinic.de
- MARKK Museum am Rothenbaum –
- Kulturen und Künste der Welt
- Stiftung öffentlichen Rechts
- www.markk-hamburg.de
- Medienanstalt Hamburg / Schleswig-Holstein
- (MA HSH) AöR
- www.ma-hsh.de
- Museum für Kunst und Gewerbe Hamburg
- Stiftung öffentlichen Rechts
- www.mkg-hamburg.de
- NMS New Mobility Solutions Hamburg GmbH www.new-mobility-solutions.de
- Norddeutsche Hörbücherei e.V. www.blindenbuecherei.de
- Olympiastützpunkt Hamburg / Schleswig-Holstein e.V.
- (Gast)
- www.osphh-sh.de
- PepKo Perspektiv-Kontor Hamburg gGmbH www.pepko-hamburg.de
- projekt märz Partnerschaftsgesellschaft www.ambulante-sozialpsychiatrie.
- hamburg
- SAEMS Special Airport Equipment and Maintenance
- Services GmbH & Co. KG
- Schulservice Hamburg
- Gesellschaft für Facility Management mbH
- servTEC
- Hamburg Wasser Service und Technik GmbH
- www.servTEC.de
- SGG Städtische Gebäudeeigenreinigung GmbH
- Sprungbrett e.V.
- (Gast)
- www.sprungbrett-bergedorf.de
- Stadtreinigung Hamburg AöR www.srhh.de
- Statistisches Amt
- für Hamburg und Schleswig-Holstein AöR
- www.statistik-nord.de
- Stiftung Bürgerhaus Wilhelmsburg www.buewi.de
- Stiftung Centralbibliothek für Blinde www.blindenbuecherei.de
- Stiftung Das Rauhe Haus
- (Gast)
- www.rauheshaus.de
- Stiftung Grone-Schule www.grone.de
- Stiftung Hamburger Gedenkstätten und Lernorte
- zur Erinnerung an die Opfer der NS-Verbrechen
- www.gedenkstaetten-hamburg.de
- Stiftung Historische Museen Hamburg www.shmh.org
- Stilbruch Betriebsgesellschaft mbH www.stilbruch.de
- Studierendenwerk Hamburg AöR www.studierendenwerk-hamburg.de
- Thalia Theater GmbH www.thalia-theater.de
- TuTech Innovation GmbH www.tutech.de
- Universitätsklinikum Hamburg-Eppendorf (UKE)
- Körperschaft des öffentlichen Rechts
- www.uke.de
- Verbraucherzentrale Hamburg e. V. www.vzhh.de
- Wilhelm Carstens Gedächtnis-Stiftung www.alida.de
- …
Clinical metagenomics [Talks for Shenzhen and so on]
https://www.nature.com/articles/s41576-019-0113-7
Microbiome: The entirety of organisms that colonize individual sites in the human body.
Microarrays: Commonly referred to as ‘chips’, these platforms consist of spots of DNA fragments, antibodies or proteins printed onto surfaces, enabling massive multiplexing of hundreds to thousands of targets.
Reads: In DNA sequencing, reads are inferred sequences of base pairs corresponding to part of or all of a single DNA fragment.
Metagenomic NGS (mNGS): A shotgun sequencing approach in which all genomic content (DNA and/or RNA) of a clinical or environmental sample is sequenced.
Transmission network analysis: The integration of epidemiological, laboratory and genomic data to track patterns of transmission and to infer origin and dates of infection during an outbreak.
Precision medicine: An approach to medical care by which disease treatment and prevention take into account genetic information obtained by genomic or molecular profiling of clinical samples.
Reference standards: In laboratory test development, well-characterized, standardized and validated reference materials or databases that enable measurement of performance characteristics of an assay, including sensitivity, specificity and accuracy.
Latex agglutination: A clinical laboratory test for detection of a specific antibody in which the corresponding antigen is adsorbed on spherical polystyrene latex particles that undergo agglutination in the presence of the antibody.
Seroconversion: The development of detectable antibodies in the blood that are directed against an infectious agent, such as HIV-1, after which the infectious disease can be detected by serological testing for the antibody. 机体的免疫系统在受到抗原(包括细菌、病毒、甚至自身肿瘤细胞等)刺激后会产生抗体,我们从免疫细胞(B细胞)开始产生抗体算起,将血清中无法检测到抗体至能够检测到抗体的这个“转换点”定义为血清转换。
Library: In DNA sequencing, a collection of DNA fragments with known adapter sequences at one or both ends that is derived from a single clinical or environmental sample.
Sanger sequencing: A classical method of DNA sequencing based on selective incorporation of chain-terminating dideoxynucleotides developed by Frederick Sanger and colleagues in 1977; now largely supplanted by next-generation sequencing.
Subtyping (ST): In microbiology, refers to the identification of a specific genetic variant or strain of a microorganism (for example, virus, bacterium or fungus), usually by sequencing all or part of the genome.
Liquid biopsy: The detection of molecular biomarkers from minimally invasive sampling of clinical body fluids, such as DNA sequences in blood, for the purpose of diagnosing disease.
Spike-in: In laboratory test development, refers to the use of a nucleic acid fragment or positive control microorganism that is added to a negative sample matrix (for example, plasma from blood donors) or clinical samples and that serves as an internal control for the assay.
No-template control: In PCR or sequencing reactions, a negative control sample in which the DNA or cDNA is left out, thus monitoring for contamination that could produce false-positive results.
Biorobots: The automated instrumentation in the clinical laboratory that enables parallel processing of many samples at a time.
Point-of-care: Refers to diagnostic testing or other medical procedures that are done near the time and place of patient care (for example, at the bedside, in an emergency department or in a developing-world field laboratory).
Cluster density: On Illumina sequencing systems, a quality control metric that refers to the density of the clonal clusters that are produced, with each cluster corresponding to a single read. An optimal cluster density is needed to maximize the number and accuracy of reads generated from a sequencing run.
Q-score: A quality control metric for DNA sequencing that is logarithmically related to the base calling error probabilities and serves as a measurement of read accuracy.
Proficiency testing: A method for evaluating the performance of individual laboratories for specific laboratory tests using a standard set of unknown samples that permits interlaboratory comparisons.
Nanopore sequencing: A sequencing method in which DNA or RNA molecules are transported through miniature pores by electrophoresis. Sequencing reads are generated by measurement of transient changes in ionic current as the molecule passes through the pore.
Box 1 | Where is the signal — cellular or cell-free DNA? Metagenomic sequencing for clinical diagnostic purposes typically uses a shotgun approach by sequencing all of the DNA and/or RNA in a clinical sample. Clinical samples can vary significantly in their cellularity, ranging from cell-free fluids (that is, plasma, bronchoalveolar lavage fluid or centrifuged cerebrospinal fluid) to tissues. In the next-generation sequencing (NGS) field, there is great interest in the use of liquid biopsies from cell-free DNA (cfDNA) extracted from body fluids, such as plasma, to identify chromosomal or other genetic mutations and thus diagnose malignancies in the presymptomatic phase123. Similarly, cfDNA analysis has been useful for non-invasive prenatal testing applications, such as for the identification of trisomy 21 (ref.124 ). One study has described the potential utility of cfDNA analysis in diagnosing invasive fungal infection in cases where biopsy is not possible57. Another advantage to cfDNA analysis is the higher sensitivity of metagenomic sequencing owing to less cellular background from the human host. However, limitations of cfDNA analysis may include decreased sensitivity for detection of predominantly intracellular pathogens, such as human T cell lymphotropic virus, Rickettsia spp. and Pneumocystis jirovecii, and loss of the ability to interrogate cellular human host responses with RNA sequencing.
Box 2 | Nanopore sequencing
-
Abstract
- Clinical metagenomic next-generation sequencing (mNGS), the comprehensive analysis of microbial and host genetic material (DNA and RNA) in samples from patients, is rapidly moving from research to clinical laboratories.
- This emerging approach is changing how physicians diagnose and treat infectious disease, with applications spanning a wide range of areas, including antimicrobial resistance [x], the microbiome [x], human host gene expression (transcriptomics) [x] and oncology [x].
- Here, we focus on the challenges of implementing mNGS in the clinical laboratory and address potential solutions for maximizing its impact on patient care and public health.
-
Introduction
-
The field of clinical microbiology comprises both diagnostic microbiology, the identification of pathogens from clinical samples to guide management and treatment strategies for patients with infection, and public health microbiology, the surveillance and monitoring of infectious disease outbreaks in the community.
-
Traditional diagnostic techniques in the microbiology laboratory include growth and isolation of microorganisms in culture, detection of pathogen-specific antibodies (serology) or antigens and molecular identification of microbial nucleic acids (DNA or RNA), most commonly via PCR.
-
[Disadvantage] While most molecular assays target only a limited number of pathogens using specific primers or probes, metagenomic approaches characterize all DNA or RNA present in a sample, enabling analysis of the entire microbiome as well as the human host genome or transcriptome in patient samples.
-
Metagenomic approaches have been applied for decades to characterize various niches, ranging from marine environments1 to toxic soils2 to arthropod (节肢动物的) disease vectors 3,4 to the human microbiome5,6.
-
These tools have also been used to identify infections in ancient remains7, discover novel viral pathogens 8 [Viral pathogen discovery] and characterize the human virome in both healthy and diseased states9–11 and for forensic applications12.
-
The capacity to detect all potential pathogens — bacteria, viruses, fungi and parasites — in a sample and simultaneously interrogate host responses has great potential utility in the diagnosis of infectious disease.
-
Metagenomics for clinical applications derives its roots from the use of microarrays in the early 2000s13,14.
-
Some early successes using this technology include the discovery of the SARS coronavirus15, gene profiling of mutations in cancer16 and in-depth microbiome analysis of different sites in the human body17.
-
However, it was the advent of next-generation sequencing (NGS) technologies in 2005 that jump-started the metagenomics field18.
-
For the first time, millions to billions of reads could be generated in a single run, permitting analysis of the entire genetic content of a clinical or environmental sample.
-
The proliferation of available sequencing instruments and exponential decreases in sequencing costs over the ensuing decade drove the rapid adoption of NGS technology.
-
To date, several studies have provided a glimpse into the promise of NGS in clinical and public health settings.
-
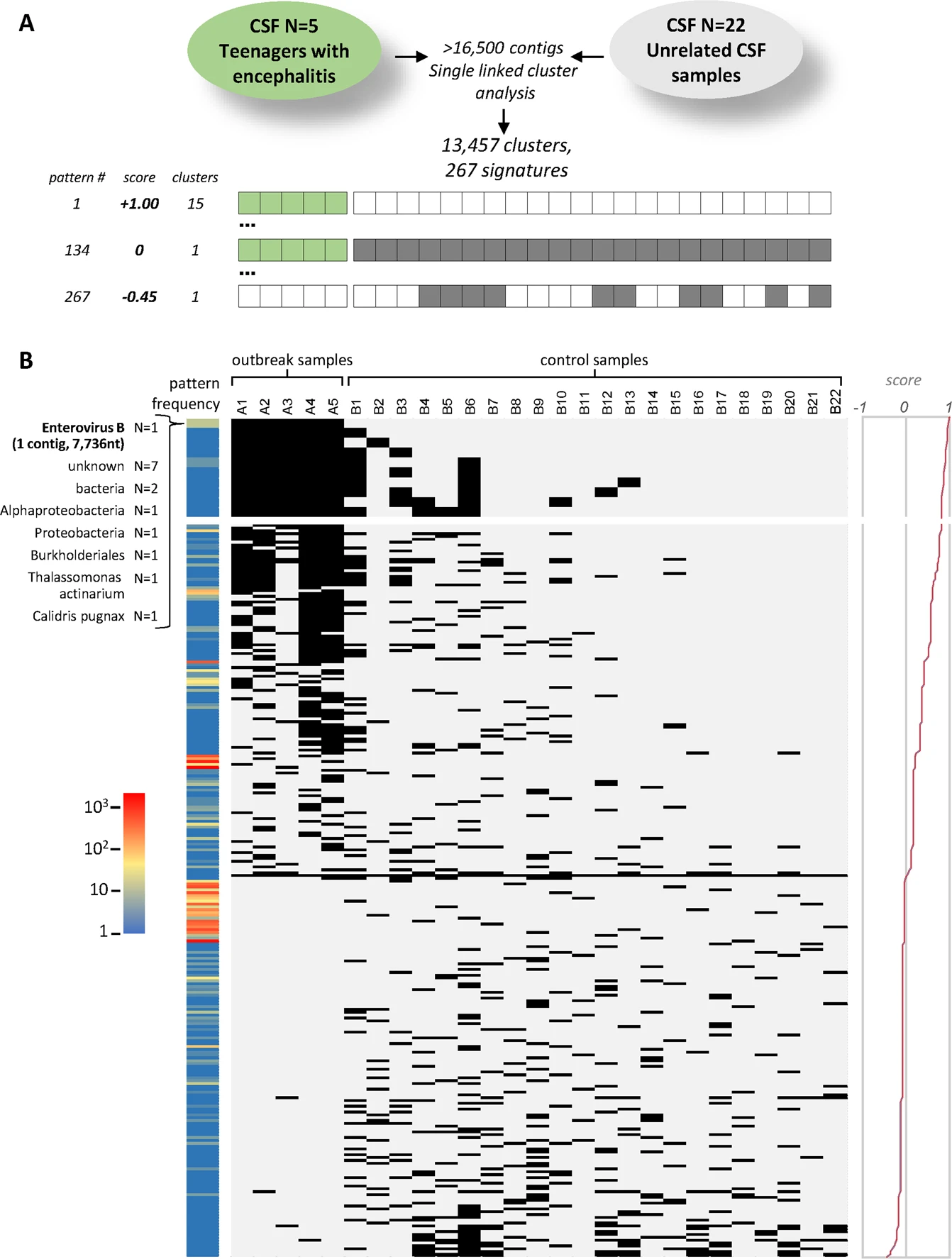
For example, NGS was used for the clinical diagnosis of neuroleptospirosis in a 14-year-old critically ill boy with meningoencephalitis19 [–>The enterovirus example in DAMIAN]; this case was the first to demonstrate the utility of metagenomic NGS (mNGS) in providing clinically actionable information, as successful diagnosis prompted appropriate targeted antibiotic treatment and eventual recovery of the patient.
-
Examples in public health microbiology include the use of NGS, in combination with transmission network analysis20 [Integration of Sequencing and Epidemiologic Data for Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infections in a Tertiary-Care Hospital], to investigate outbreaks of the Escherichia coli strain O104:H4 (ref. 21) and for surveillance of antimicrobial resistance in the food supply by bacterial whole-genome sequencing22.
-
Increasingly, big data provided by mNGS is being leveraged for clinical purposes, including characterization of antibiotic resistance directly from clinical samples23 and analysis of human host response (transcriptomic) data to predict causes of infection and evaluate disease risk24,25.
-
Thus, mNGS can be a key driver for precision diagnosis [What is the exact definition of precision diagnosis?] of infectious diseases, advancing precision medicine [precision diagnosis–>precision medicine] efforts to personalize patient care in this field.
-
Despite the potential and recent successes of metagenomics, clinical diagnostic applications have lagged behind research advances owing to a number of factors.
-
[Factor 1] A complex interplay of microbial and host factors influences human health, as exemplified by the role of the microbiome in modulating host immune responses26, and it is often unclear whether a detected microorganism is a contaminant, colonizer or bona fide [真实地] pathogen.
-
[Factor 2] Additionally, universal reference standards and proven approaches to demonstrate test validation, reproducibility and quality assurance for clinical metagenomic assays are lacking.
-
Considerations of cost, reimbursement, turnaround time, regulatory considerations and, perhaps most importantly, clinical utility also remain major hurdles for the routine implementation of clinical mNGS in patient care settings 27.
-
We review here the various applications of mNGS currently being exploited in clinical and public health settings.
-
We discuss the challenges involved in the adoption of mNGS in the clinical laboratory, including validation and regulatory considerations that extend beyond its initial development in research laboratories, and propose steps to overcome these challenges.
-
Finally, we envisage future directions for the field of clinical metagenomics and anticipate what will be achievable in the next 5 years.
-
-
Applications of clinical metagenomics
- To date, applications of clinical metagenomics have included infectious disease diagnostics for a variety of syndromes and sample types, microbiome analyses in both diseased and healthy states, characterization of the human host response to infection by transcriptomics and the identification of tumour-associated viruses and their genomic integration sites (Fig. 1; Table 1).
- Aside from infectious disease diagnostics, adoption of mNGS in clinical laboratories has been slow, and most applications have yet to be incorporated into routine clinical practice.
- Nonetheless, the breadth and potential clinical utility of these applications are likely to transform the field of diagnostic microbiology in the near future.
3.1. [TODO] Make a similar table as Table 1 for my own projects
- Sequencing method Clinical sample type Potential clinical indications Clinical test available? Refs
- Infectious disease diagnosis — targeted analyses: 1 or 2 papers
- Infectious disease diagnosis — untargeted analyses: 1 or 2 papers
- Microbiome analyses: 1 or 2 papers
- Human host response analyses: RNAseq data 1 or 2 papers
- [Optional] Oncological analyses3.2. Applications of clinical metagenomics | Infectious disease diagnosis | Introduction
- The traditional clinical paradigm for diagnosis of infectious disease in patients, applied for more than a century, involves a physician formulating a differential diagnosis and then ordering a series of tests (generally ‘one bug, one test’) in an attempt to identify the causative agent.
- The spectrum of conventional testing for pathogens in clinical samples ranges from the identification of microorganisms growing in culture (for example, by biochemical phenotype testing or matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry), the detection of organism-specific biomarkers (such as antigen testing by latex agglutination or antibody testing by enzyme-linked immunosorbent assay (ELISA)) or nucleic acid testing by PCR for single agents to multiplexed PCR testing using syndromic panels.
- These panels generally include the most common pathogens associated with a defined clinical syndrome, such as meningitis (脑膜炎) and encephalitis [ensefәˊlaitis], acute respiratory infection, sepsis or diarrhoeal disease 28–31.
- Molecular diagnostic assays provide a fairly cost-effective and rapid (generally <2 hours of turnaround time) means to diagnose the most common infections.
- However, nearly all conventional microbiological tests in current use detect only one or a limited panel of pathogens at a time or require that a microorganism be successfully cultured from a clinical sample.
- By contrast, while NGS assays in current use cannot compare with conventional tests with respect to speed — the sequencing run alone on a standard Illumina instrument takes >18 hours — mNGS enables a broad range of pathogens — viruses, bacteria, fungi and/or parasites — to be identified from culture or directly from clinical samples on the basis of uniquely identifiable DNA and/or RNA sequences32.
- Another key advantage of NGS approaches is that the sequencing data can potentially be leveraged for additional analyses beyond the mere identification of a causative pathogen, such as microbiome characterization and parallel analyses of human host responses through transcriptome profiling by RNA sequencing (RNA-seq).
- Thus, the clinical utility of NGS in diagnosis may be in the most difficult-to-diagnose cases or for immunocompromised patients, in whom the spectrum of potential pathogens is greater.
- Eventually, mNGS may become cost competitive with multiplexed assays or used as an upfront ‘rule out’ assay to exclude infectious aetiologies.
- Of course, detection of nucleic acids, either by multiplex PCR panels or NGS, does not by itself prove that an identified microorganism is the cause of the illness, and findings have to be interpreted in the clinical context.
- In particular, discovery of an atypical or novel infectious agent in clinical samples should be followed up with confirmatory investigations such as orthogonal testing of tissue biopsy samples and demonstration of seroconversion or via the use of cell culture or animal models, as appropriate8, to ascertain its true pathogenic potential.
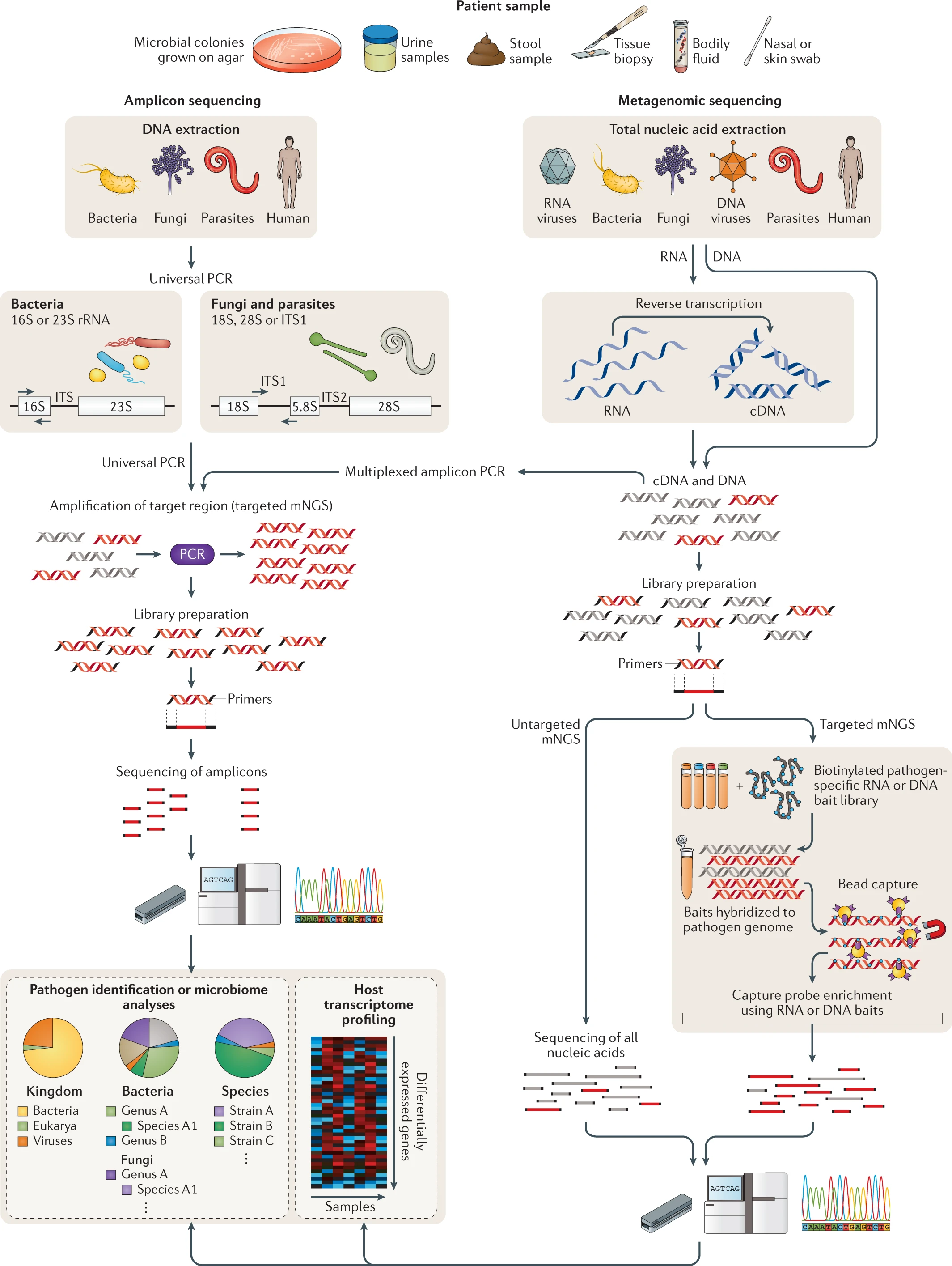
- * NGS of clinical samples as performed in either research or clinical laboratories involves a number of steps, including nucleic acid extraction, enrichment for DNA and/or RNA, library preparation, PCR amplification (if needed), sequencing and bioinformatics analysis (Fig. 2) .
- Any body fluid or tissue yielding sufficient nucleic acid is amenable to NGS analysis, which can either be targeted, that is, enriching individual genes or genomic regions, or untargeted, as is the case for metagenomic ‘shotgun’ approaches (Fig. 2).
- The details for the specific steps vary by laboratory and are described extensively elsewhere33–37.3.3. * Applications of clinical metagenomics | Infectious disease diagnosis | Targeted NGS analyses
- 多重引物PCR(Multiplexed amplicon PCR)和通用PCR(Universal PCR)是两种不同的聚合酶链反应(PCR)技术,它们在设计和应用上有所区别:
- 多重引物PCR:
- 多重引物PCR是一种可以同时扩增多个目标DNA片段的PCR技术。在这种方法中,多对特异性引物在一个反应体系中使用,允许同时检测多个基因序列。
- 这种方法常用于病原体检测、遗传标记分析和多基因疾病的研究。例如,它可以在一个实验中同时检测多种病毒或细菌的存在。
- 多重引物PCR需要精确设计引物,以避免引物之间的交叉反应和非特异性扩增。
- 通用PCR:
- 通用PCR指使用一对引物针对广泛的物种或多个样本中的相同基因区域进行扩增的PCR技术。这对引物通常针对高度保守的DNA序列,使其能够扩增来自不同物种的相似序列。
- 通用PCR常用于物种鉴定、系统发育分析和环境样本的微生物多样性研究。例如,通过通用PCR可以从多个生物样品中扩增相同的微生物16S rRNA基因区域,以分析不同样品中的微生物组成。
- 通用PCR较为简单,但其特异性较低,可能会扩增到非目标序列。
- 总的来说,多重引物PCR适用于需要同时检测多个特定目标的情况,而通用PCR更适合于需要从广泛样本中扩增相同基因区域的应用。两者各有优势和局限性,选择哪种技术取决于实验的具体需求和目的。
- Universal primers refer to the primers used in universal PCR. Universal PCR is a type of polymerase chain reaction (PCR) where the same set of primers is used to amplify DNA from different species or sources. These primers are designed to target conserved regions of DNA that are similar across various organisms, making them useful for a wide range of applications, including taxonomic classification, phylogenetic studies, and environmental biodiversity assessments. The universal nature of these primers allows for the amplification of a specific gene or DNA segment from a broad array of species within a single PCR assay.
- Targeted approaches have the benefit of increasing the number and proportion of pathogen reads in the sequence data.
- This step can increase the detection sensitivity for microorganisms being targeted, although it limits the breadth of potential pathogens that can be identified.
- * An example of a targeted approach is the use of highly conserved primers for universal PCR amplification and detection of all microorganisms corresponding to a specific type from clinical samples, such as 16S ribosomal RNA (rRNA) gene amplification for bacteria38,39 and 18S rRNA and internal transcribed spacer (ITS) gene amplification for fungi40 (Fig. 2).
- Previously, such approaches were followed by Sanger sequencing of the resulting PCR amplicon to identify the pathogen and make a diagnosis; now, this step is commonly accomplished using NGS.
- Universal PCR for detection of bacteria and fungi has now been adopted in many hospital laboratories and has increased the number and proportion of infectious diagnoses39,41, although the technique is limited by the breadth of detection (that is, bacteria or fungi only or even a more limited range of targets, such as mycobacteria only, depending on the primer sets used) and by concerns regarding sensitivity42.
- * Another example of a targeted NGS approach is the design of primers tiled across the genome to facilitate PCR amplification and amplicon NGS for recovery of viral genomes directly from clinical samples43 .
- This method has been used to track the evolution and spread of Zika virus (ZIKV) in the Americas44–46 and of Ebola virus in West Africa47, with some demonstrations of real-time monitoring having an impact on public health interventions.
- * Another targeted approach is capture probe enrichment, whereby metagenomic libraries are subjected to (使遭受) hybridization using capture ‘bait’ probes48.
- These probes are generally 30–120 bp in length, and the number of probes can vary from less than 50 to more than 2 million 49–52.
- Although this enrichment method has been shown to increase the sensitivity of metagenomic detection in research settings, especially for viruses, it has yet to be used routinely for clinical diagnosis.
- A promising application of this approach may be the enrichment of clinical samples for characterization of antibiotic resistance23, a considerable problem in hospitals and the primary focus of the US National Action Plan for Combating Antibiotic-Resistant Bacteria53.
- * However, drawbacks of capture probe enrichment, compared with untargeted approaches for infectious disease diagnosis, include a bias towards targeted microorganisms, added steps, increased costs and long hybridization times (24–48 hours) as a result of the additional processing needed for maximal efficiency.3.4. Applications of clinical metagenomics | Infectious disease diagnosis | Untargeted metagenomic NGS analyses
- Untargeted shotgun mNGS analyses forego (放弃) the use of specific primers (namely using Universal Primer in Amplicon sequencing) or probes (namely using baits in targeted mNGS) 54.
- Instead, the entirety of the DNA and/or RNA (after reverse transcription to cDNA) is sequenced.
- *(Can refer to the project of Holger and Anna) With pure cultures of bacteria or fungi, mNGS reads can be assembled into partial or complete genomes.
- (We have the method 3.3. targeted capture probe for the application here) These genome sequences are then used for subtyping and/or monitoring hospital outbreaks in support of infection control and/or public health surveillance efforts. - For example, a seminal study described theuse of whole-genome sequencing of multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae to track the origin and evolution of a hospital outbreak55.
- (We have the method 3.3. targeted capture probe for the application here) This study demonstrated for the first time the high-resolution mapping of likely transmission events in a hospital, some of which were unexpected on the basis of initial epidemiological data, and also identified putative resistance mutations in emerging resistant strains.
- The integration of genomic and epidemiological datayielded actionable insights that would have been useful for curbing transmission.
- Untargeted mNGS of clinical samples is perhaps the most promising approach for the comprehensive diagnosis of infections.
- In principle, nearly all pathogens, including viruses, bacteria, fungi and parasites, can be identified in a single assay56.
- mNGS is a needle-in-a-haystack endeavour, as only a small proportion (typically <1%) of reads are non-human, of which only a subset may correspond to potential pathogens.
- A limitation of mNGS is that the sensitivity of the approach is critically dependent on the level of background.
- Tissues, for example, have increased human host background relative to cell-free body fluids, resulting in a reduced number and proportion of microbial reads and hence a decrease in mNGS sensitivity33,36,37.
- Moreover, defining specific microbial profiles that are diagnostic or predictive of disease development can be difficult, especially from nonsterile sites that harbour a complex microbiome, such as respiratory secretions or stool6.
- Nevertheless, several groups have successfully validated mNGS in Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratories for the diagnosis of infections, including meningitis (脑膜炎) or encephalitis (脑炎)36,37, sepsis33,57 and pneumonia58, and these assays are now available for clinical reference testing of patients.3.5. Applications of clinical metagenomics | Clinical microbiome analyses
- Many researchers now use mNGS instead of targeted sequencing of the 16S rRNA gene for in-depth characterization of the microbiome59.
- There is growing public awareness of the microbiome and its likely involvement in both acute and chronic disease states60.
- However, no microbiome-based tests have been clinically validated for the diagnosis or treatment of disease, in part owing to an incomplete understanding of the complexity of the microbiome and its role in disease pathogenesis.
- One future clinical application of microbiome analysis may be in the management and treatment of Clostridium difficile-associated disease.
- C. difficile is an opportunistic bacterium that can infect the gut, resulting in the production of toxins that can cause diarrhoea, dehydration, sepsis and death.
- C. difficile infection occurs only in the setting of a microbiome that is altered by factors such as exposure to broad-spectrum antibiotics or recent gastrointestinal surgery61.
- The importance of the microbiome in C. difficile infection is underscored by the 80–90% effectiveness of faecal stool transplantation in treating and potentially curing the disease62,63.
- The use of mNGS to characterize the microbiome in multiple studies has facilitated the development of bacterial probiotic mixtures that can be administered as pills for prophylaxis or treatment of C. difficile-associated disease (Fig. 1B).
- Another potential application of the microbiome is in the analysis of bacterial diversity, which can provide clues as to whether a patient’s illness is infectious or non-infectious.
- For example, a study of mNGS for the identification of respiratory pathogens in patients with pneumonia found that individuals with culture-proven infection had significantly less diversity in their respiratory microbiome25.
- Alterations of the microbiome, known as dysbiosis, have also been shown to be related to obesity, diabetes mellitus and inflammatory bowel disease64, and manipulation of the microbiome may be a pathway to treating these pathological conditions.3.6. Applications of clinical metagenomics | Human host response analyses
- Clinical mNGS typically focuses on microbial reads; however, there is a complementary role for the analysis of gene expression in studying human host responses to infection65 (Fig. 1c).
- mNGS of RNA libraries used for the detection of pathogens such as RNA viruses in clinical samples incidentally produces host gene expression data for transcriptome (RNA-seq) analyses66.
- Although RNA-seq analyses are commonly performed on whole blood or peripheral blood mononuclear cell (PBMC) samples, any body fluid or tissue type is potentially amenable to these analyses.
- Classification of genes by expression profiling using RNA-seq has been used to characterize several infections, including staphylococcal bacteraemia67, Lyme disease68, candidiasis69, tuberculosis (discriminating between latent and active disease risk)70–72 and influenza73–75.
- Machine-learning-based analyses of RNA-seq data have been used for cancer classification76, and translation of these approaches may be promising for infectious diseases.
- Panels containing a limited number of host biomarkers are being developed as diagnostic assays for influenza77, tuberculosis70 and bacterial sepsis 78.
- Although no RNA-seq-based assay has been clinically validated to date for use in patients, the potential clinical impact of RNA-seq analyses is high.
- Interrogation of RNA reads from microorganisms corresponding to active microbial gene expression might enable the discrimination between infection versus colonization 25 and live (viable) versus dead organisms79.
- Moreover, RNA-seq analyses of the human host can be used to identify novel or underappreciated host–microbial interactions directly from clinical samples, as previously shown for patients with Lyme disease68, dengue 80 or malaria81.
- RNA-seq may be particularly useful in clinical cases in which the causative pathogen is only transiently present (such as early Lyme disease82 or arboviral infections, including West Nile virus83 or ZIKV84); analogous to serologic testing, indirect diagnosis of infections may be possible on the basis of a pathogen-specific human host response.
- Analysis of pathogen-specific host responses may also be useful in discriminating the bona fide causative pathogen or pathogens in a complex clinical metagenomic sample, such as a polymicrobial abscess or respiratory fluid25.
- * Yet another promising application of RNA-seq is in discriminating infectious versus non-infectious causes of acute illness25.
- If an illness is judged more likely to be non-infectious (for example, an autoimmune disease) on the basis of the host response, for example, clinicians may be more willing to discontinue antibiotics and treat the patient aggressively with steroids and other immunosuppressive medications.
- As large-scale sequencing data continue to be generated, perhaps driven by routine clinical mNGS testing, secondary mining of human reads might improve the accuracy of clinical diagnoses by incorporating both microbial and host gene expression data.3.7. Applications of clinical metagenomics | Applications in oncology
- In oncology, whole-genome or directed NGS approaches to identify mutated genes can be used to simultaneously uncover viruses associated with cancer (that is, herpesviruses, papillomaviruses and polyomaviruses) and/or to gather data on virus–host interactions85.
- For example, mNGS was critical in the discovery of Merkel cell polyomavirus (Fig. 1d), now believed to be the cause of Merkel cell carcinoma, a rare skin cancer seen most commonly in elderly patients86.
- To date, the US Food and Drug Administration (FDA) has approved the clinical use of two NGS panels testing for actionable genomic aberrations in tumour samples 87. - Detection of reads corresponding to both integrated and exogenous viruses in these samples would be possible with the addition of specific viral probes to the panel or accomplished incidentally while sequencing the whole tumour genome or exome.
- Additional knowledge of integrated or active viral infections in cancers and their involvement in signalling pathways may inform preventive and therapeutic interventions with targeted antiviral and/or chemotherapeutic drugs88, as evidenced by the decreased risk of hepatitis C virus-associated hepatocellular carcinoma after treatment with direct-acting antiviral agents89.
- In the future, mNGS of cell-free DNA from liquid biopsy samples (for example, plasma) might be leveraged for the simultaneous identification of early cancer and diagnosis of infection in immunocompromised patients (Box 1).-
[OPTIONAL, or short with 2-3 slides, more technically, e.g. 4.1-4.4] Clinical implementation of metagenomic NGS
Implementation of mNGS in the clinical laboratory is a complex endeavour that requires customization of research protocols using a quality management approach consistent with regulatory standards 90. Library prepara tion reagents, sequencing instrumentation and bioin formatics tools are constantly changing in the research environment. However, in the clinical laboratory, assays need to be implemented following standardized (locked- down) protocols. Changes made to any component of the assay need to be validated and shown to have acceptable performance before testing in patients. Periodic updates and repeat validation studies are performed as deemed necessary to incorporate interim technological advances in NGS reagents, protocols and instrumentation. Metagenomic methods for pathogen detection pres ent a particularly challenging scenario for clinical vali dation (Fig. 3), as it is not practical to test an essentially unlimited number of different organisms for the assay to be considered validated. Although the FDA has pro vided general guidelines for clinical validation of NGS infectious disease testing91, there are no definitive reco mmendations for the clinical implementation of mNGS testing, nor is there mention of specific requirements. However, a best-practice approach can be taken that includes failure-mode analysis and evaluations of per formance characteristics using representative organ isms with ongoing assay monitoring and independent confirmation of unexpected results.
4.1. Clinical implementation of metagenomic NGS | Sensitivity and enrichment or depletion methods
Sensitivity and enrichment or depletion methods
A key limitation of mNGS is its decreased sensitivity with
high background, either predominantly from the human
host (for example, in tissue biopsies) or the microbiome
(for example, in stool). The background can be clini
cally relevant as the pathogen load in infections, such as
Shigella flexneri in stool from patients with diarrhoea92 or
ZIKV in plasma from patients with vector-borne febrile
illness93, can be very low (<103 copies per ml).
Host depletion methods for RNA libraries have been
developed and shown to be effective, including DNase I
treatment after extraction to remove residual human
background DNA94; the use of RNA probes followed
by RNase H treatment95; antibodies against human and
mitochondrial rRNA (the most abundant host RNA
types in clinical samples)96; and/or CRISPR–Cas9-based
approaches, such as depletion of abundant sequences by
hybridization97.
Unfortunately, there are no comparably effective
parallel methods for DNA libraries. Limited enrich
ment in the 3–5 times range can be achieved with
the use of antibodies against methylated human host
DNA98, which enriches microbial reads owing to the
lack of methylated DNA in most pathogen genomes.
Differential lysis of human cells followed by degrada
tion of background DNA with DNase I — thus retain
ing and enriching for nucleic acid from organisms with
cell walls, which include some bacteria and fungi — has
been shown to provide substantial microbial enrichment
of up to 1,000 times94,99,100. However, the performance of
differential lysis methods can be limited by a number
of factors. These limitations include potential decreased
sensitivity for microorganisms without cell walls, such
as Mycoplasma spp. or parasites; a possible paradoxi
cal increase in exogenous background contamination
by use of additional reagents101; and the inability to
detect free nucleic acid from dead organisms that are
lysed in vivo by human host immune cells or antibiotic
treatment. The importance of retaining the ability for
cell-free DNA detection from culture-negative samples
from dead organisms is also why incorporation of a
propidium monoazide treatment step to select for DNA
from live organisms may not be clinically useful as an
enrichment method for mNGS102 . In general, both the
differential lysis and propidium monoazide approaches
would also be cumbersome to implement in a highly
reproducible fashion, which is needed for clinical
laboratory validation.
To some extent, the human host background limi
tation may be overcome with brute force, made possi
ble by the increasing capacities of available sequencers.
For instance, an astrovirus was detected in a child with
encephalitis by ultradeep sequencing of brain tissue,
yielding only 1,612 reads out of ~134 million (0.0012%)
sequences103. Yet another approach to improve sensitiv
ity is to leverage a hybrid method for enrichment, such
as metagenomic sequencing with spiked primers46 .
Combining targeted with untargeted sequencing, the
method uses variably sized panels (100–10,000) of short
primers that are added (‘spiked’) into reaction mixtures
to enrich for specific target organisms while retaining
the breadth of metagenomic sequencing for off-target
organisms. When spiked at the reverse transcription
step, a panel of ZIKV-specific primers was found to
increase the number of ZIKV reads by more than ten
fold without appreciably decreasing broad metagenomic
sensitivity for other pathogens, enabling whole-genome
viral sequencing to characterize ZIKV spread from
Brazil into Central America and Mexico46.4.2. Clinical implementation of metagenomic NGS | Laboratory workflow considerations
The complexity of mNGS analysis requires highly
trained personnel and extreme care in sample handling
to avoid errors and cross-contamination. Even miniscule
amounts of exogenous DNA or RNA introduced during
sample collection, aliquoting, nucleic acid extraction,
library preparation or pooling can yield a detectable
signal from contaminating reads. In addition, labora
tory surfaces, consumables and reagents are not DNA
free. A database of background microorganisms com
monly detected in mNGS data and arising from nor
mal flora or laboratory contamination101,104 typically
needs to be maintained for accurate mNGS analyses.
Microorganisms on this list are either not reported or
will require higher thresholds for reporting if they are
clinically significant organisms.
Clinical laboratory operations are characterized by
a defined workflow with scheduled staffing levels and
are less amenable to on-demand testing than those of
research laboratories. As samples are typically handled in
batches, the frequency of batch analysis is a major deter
minant of overall turnaround time. Unless fully auto
mated sample-handling systems are readily available,
wet lab manipulations for mNGS require considerable
hands-on time to perform, as well as clinical staff who
are highly trained in molecular biology procedures.
There are ergonomic concerns with repetitive tasks
such as pipetting, as well as potential for inadvertent
sample mix-up or omission of critical steps in the work
flow. Maintaining high quality during complex mNGS
procedures can be stressful to staff, as slight deviations in
sample handling can lead to major changes in the results
generated. Separating the assay workflow into multiple
discrete steps to be performed by rotating shifts can be
helpful to avoid laboratory errors.4.3. Clinical implementation of metagenomic NGS | Reference standards
Well-characterized reference standards and controls areneeded to ensure mNGS assay quality and stability overtime. Most available metagenomic reference materialsare highly customized to specific applications (for example, ZymoBIOMICS Microbial Community Standardfor microbiome analyses and bacterial and fungal metagenomics105) and/or focused on a more limited spectrum of organisms (for example, the National Instituteof Standards and Technology (NIST) reference materialsfor mixed microbial DNA detection, which contain onlybacteria106). Thus, these materials may not be applicableto untargeted mNGS analyses.
Custom mixtures consisting of a pool of microorganisms (mock microbial communities) or theirnucleic acids can be developed as external controls toestablish limits of detection for mNGS testing. Internalspike-in control standards are available for other NGS
applications such as transcriptome analysis by RNA-seq, with External RNA Controls Consortium (ERCC)RNA standards composed of synthetic RNA oligonucleotides spanning a range of nucleotide lengths andconcentrations 107. The complete set or a portion ofthe ERCC RNA standards (or their DNA equivalents)can be used as spike-in internal controls to controlfor assay inhibition and to quantify titres of detectedpathogens by standard curve analysis108. Nonetheless,
the lack of universally accepted reference standards formNGS makes it difficult to compare assay performancesbetween different laboratories. There is a critical needfor standardized reference organisms and genomicmaterials to facilitate such comparisons and to defineoptimal analysis methods.4.4. Clinical implementation of metagenomic NGS | Bioinformatics challenges
User-friendly bioinformatics software for analysis ofmNGS data is not currently available. Thus, customizedbioinformatics pipelines for analysis of clinical mNGSdata56,109–111 still require highly trained programming staffto develop, validate and maintain the pipeline for clinicaluse. The laboratory can either host computational servers locally or move the bioinformatics analysis and datastorage to cloud platforms. In either case, hardware andsoftware setups can be complex, and adequate measuresmust be in place to protect confidential patient sequence
data and information, especially in the cloud environment.Storage requirements for sequencing data can quicklybecome quite large, and the clinical laboratory must decideon the quantity, location and duration of data storage.
Bioinformatics pipelines for mNGS analysis use anumber of different algorithms, usually developed forthe research setting and constantly updated by software developers. As for wet lab procedures, it is usuallynecessary to make custom modifications to the pipelinesoftware and then lock down both the software and reference databases for the purposes of clinical validation112.
A typical bioinformatics pipeline consists of a series of
analysis steps from raw input FASTQ files including
quality and low-complexity filtering, adaptor trimming,
human host subtraction, microorganism identification
by alignment to reference databases, optional sequence
assembly and taxonomic classification of individual
reads and/or contiguous sequences (contigs) at levels
such as family, genus and species (Fig. 4). Each step in
the pipeline must be carefully assessed for accuracy and
completeness of data processing, with consideration for
propagation of errors. Sensitivity analyses should be
performed with the inclusion of both in silico data and
data generated from clinical samples. Customized data
sets can be prepared to mimic input sequence data and
expand the range of microorganisms detected through in
silico analysis37. The use of standardized reference mate
rials and NGS data sets is also helpful in comparative
evaluation of different bioinformatics pipelines105.
Additionally, public databases for microbial reference
genomes are being continuously updated, and laborato
ries need to keep track of the exact versions used in addi
tion to dealing with potential misannotations and other
database errors. Larger and more complete databases
containing publicly deposited sequences such as the
National Center for Biotechnology Information (NCBI)
Nucleotide database are more comprehensive but also
contain more errors than curated, more limited data
bases such as FDA-ARGOS91,113 or the FDA Reference
Viral Database (RVDB) 114. A combined approach that
incorporates annotated sequences from multiple data
bases may enable greater confidence in the sensitivity
and specificity of microorganism identification.
Performance validation and verification for bioinfor
matics analysis constitute a time-consuming endeavour
and include analysis of control and patient data sets and
comparisons, with orthogonal clinical testing to deter
mine the accuracy of the final result36. Establishing
thresholds enables separation of true-positive matches
from the background, and these thresholds can incor
porate metrics such as the number of sequence reads
aligning to the detected microorganism, normalized to
reads per million, external no-template control samples or
internal spike-in material; the number of nonoverlapping
genomic regions covered; and the read abundance in clin
ical samples relative to negative control samples (to avoid
reporting of contaminant organisms). Receiver–operator
curve (ROC) analysis is a useful tool to determine opti
mal threshold values for a training set of clinical samples
with known results, with verification of pre-established
thresholds using an independent validation set36.
As in the wet lab workflow, analysis software and ref
erence databases should ideally be locked down before
validation and clinical use. Many laboratories maintain
both production and up-to-date development versions
of the clinical reference database (for example, the NCBI
nucleotide database is updated every 2 weeks), with the
production database being updated at regular, prespec
ified intervals. Standardized data sets should be used to
verify the database after any update and to ensure that
assay results are accurate and reproducible, as errors
can be introduced from newly deposited sequences and
clinical metadata.4.5. Clinical implementation of metagenomic NGS | Cost considerations
Although there have been substantial cost reductions in
the generation of sequence data, the overall per-sample
reagent cost for sequencing remains fairly high. Most lab
oratories lack the robotic equipment and established
automated protocols to multiplex large numbers
of patient samples in a single run. Thus, the majority of
library preparation methods for mNGS are performed
manually and hence incur considerable staff time. The
additional resources needed to run and maintain a
bioinformatics analysis pipeline are also considerable,
and steps taken to ensure regulatory oversight can add
notably to costs as well. This leads to an overall cost
of several hundreds to thousands of dollars per sam
ple analysed, which is higher than that for many other
clinical tests.
Technical improvements in hardware are needed
for mNGS sample processing to increase throughput
and to reduce costs. As NGS procedures become more
standardized, there has been a drive towards increasing
automation with the use of liquid-handling biorobots115.
Typically, two biorobots are needed for clinical mNGS
for both the pre-amplification and post-amplification
steps to avoid PCR amplicon cross-contamination.
Increased multiplexing is also possible with the greatly
enhanced output from the latest generation of sequenc
ers, such as the Illumina NovaSeq instruments. However,
a potential limitation with running larger numbers of
samples per run is longer overall turnaround times for
clinical use owing to the requirement for batch pro
cessing as well as sample workflow and computational
analysis considerations. Additionally, high-throughput
processing of clinical samples for NGS may only be
possible in reference laboratories. The development of
microfluidic devices for NGS sample library preparation,
such as VolTRAX116, could eventually enable clinicians
to use mNGS more widely in hospital laboratories or
point-of-care settings.4.6. Clinical implementation of metagenomic NGS | Regulatory considerations
Clinical laboratories are highly regulated, and general
laboratory and testing requirements apply to all mole
cular diagnostic assays reported for patient care 90 .
Quality control is paramount, and methods must be
developed to ensure analytic accuracy throughout the
assay workflow. Important quality control steps can
include initial sample quality checks, library param
eters (concentration and size distribution), sequence
data generation (cluster density and Q-score), recovery of
internal controls and performance of external controls.
Validation data generated during assay development and
implementation should be recorded and made availa
ble to laboratory inspectors (for laboratory-developed
tests) or submitted to regulatory agencies, such as the
FDA in the USA or the European Medicines Agency
(EMA) in Europe, for approval.
Ongoing monitoring is particularly important for
mNGS assays to verify acceptable performance over
time and to investigate atypical findings36. Monitoring is
accomplished using sample internal controls, intra-run
control samples, swipe tests for contamination and perio
dic proficiency testing. Unexpected or unusual results are
further investigated by reviewing patients’ clinical charts
or by confirmatory laboratory testing using orthogonal
methods. Identification of microorganisms that have
not been identified before in the laboratory should be
independently confirmed, usually through clinical ref
erence or public health laboratory testing. Atypical or
novel organisms should be assessed for their clinical
significance, and these findings should be reported and
discussed with health-care providers, with consideration
for their potential pathogenicity and for further testing
and treatment options. Clinical microbial sequencing
boards, modelled after tumour boards in oncology, can
be convened via real-time teleconferencing to discuss
mNGS results with treatment providers in clinical con
text (Fig. 3). Detection of microorganisms with public
health implications such as Sin Nombre hantavirus or
Ebola virus should be reported, as appropriate, to the
relevant public health agencies.-
Conclusions and future perspectives
Technological advancements in library preparation methods, sequence generation and computational bio informatics are enabling quicker and more comprehen sive metagenomic analyses at lower cost. Sequencing technologies and their applications continue to evolve. Real-time sequencing in particular may be a game- changing technology for point-of-care applications in clinical medicine and public health, as laboratories have begun to apply these tools to diagnose atypical infec tions and track pathogen outbreaks, as demonstrated by the recent deployment of real-time nanopore sequencing for remote epidemiological surveillance of Ebola44 and ZIKV44,45, and even for use aboard the International Space Station117 (Box 2). Nonetheless, formidable challenges remain when implementing mNGS for routine patient care. In par ticular, sensitivity for pathogen detection is decreased in clinical samples with a high nucleic acid background or with exceedingly low pathogen titres; this concern is only partially mitigated by increasing sequencing depth per sample as costs continue to drop. As a comprehen sive direct detection method, mNGS may eventually replace culture, antigen detection and PCR methods in clinical microbiology, but indirect approaches such as viral serological testing will continue to play a key part in the diagnostic work-up for infections27, and functional assays such as culture and phenotypic susceptibility test ing will likely always be useful for research studies. In summary, while current limitations suggest that mNGS is unlikely to replace conventional diagnostics in the short term, it can be a complementary, and perhaps essential, test in certain clinical situations. Although the use of mNGS for informing clinical care has been demonstrated in multiple case reports and small case series118, nearly all studies have been retro spective, and clinical utility has yet to be established in a large-scale prospective clinical trial. Prospective clinical studies will be critical to understand when to perform mNGS and how the diagnostic yield compares with that of other methods. For example, the mNGS transcrip tomic approach might enable effective treatment triage, whereby antimicrobials are only needed for patients showing an ‘infectious profile’ of gene expression and those with a ‘non-infectious profile’ can be treated for other causes. In particular, prospective clinical trial and economic data showing the cost-effectiveness of these relatively expensive tests in improving patient outcomes are needed to justify their use. These data will also sup port a pathway towards regulatory approval and clini cal reimbursement. High-quality evidence that clinical metagenomic assays are effective in guiding patient management will require protocols that minimize potential assay and patient selection bias and compare relevant health outcomes using data sets generated from large patient cohorts119,120. We predict that, over the next 5 years, prospective clinical trial data evaluating the clinical utility and cost- effectiveness of mNGS will become available; overall costs and turnaround time for mNGS will continue to drop; other aspects of mNGS beyond mere identifica tion, such as incorporation of human host response and microbiome data, will prove clinically useful; robotic sample handling and microfluidic devices will be devel oped for push-button operation; computational analysis platforms will be more widely available, both locally and on the cloud, obviating the need for dedicated bioinfor matics expertise; and at least a few mNGS-based diag nostic assays for infectious diseases will attain regulatory approval with clinical reimbursement. We will witness the widespread democratization of mNGS as genomic analyses become widely accessible not only to physicians and researchers but also to patients and the public via crowdsourcing initiatives121,122 . Furthermore, in a world with constantly emerging pathogens, we envisage that mNGS-based testing will have a pivotal role in monitor ing and tracking new disease outbreaks. As surveillance networks and rapid diagnostic platforms such as nano pore sequencing are deployed globally, it will be possi ble to detect and contain infectious outbreaks at a much earlier stage, saving lives and lowering costs. In the near future, mNGS will not be a luxury but a necessity in the clinician’s armamentarium as we engage in the perpetual fight against infectious diseases. -
Fig. 1 for chapter 4 | Clinical applications of metagenomic sequencing (USING: Overview of applications of clinical metagenomics:
-
Infectius disease diagnostics (untargeted analyses using metagnenomic sequencing using DAMIAN): DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples, explain the cohort samples! will be further developed!
-
With the methods, we can only assemble a small part or a short contig of virus or bacteria. However, if we want to know if want to compare two different isolates, we need generally the complete sequences of virus, we can use the targeted capture sequencing!
-
Infectius disease diagnostics (targeted analyses using capture probe enrichment) Paper: Target capture sequencing reveals a monoclonal outbreak of respiratory syncytial virus B infections among adult hematologic patients
-
Microbiome analyses (Metagenomic sequencing using 16S Amplicon sequencing or Unbiased shotgun metagenomics?) • Unbiased shotgun metagenomics • Amplicon metagenomics Fragment DNA and sequence PCR amplify a gene of interest randomly Tells you what types of organisms there are Bacteria/Archaea (16S rRNA), Microbial Unexpected Viral Euks (18S rRNA), Fungi (ITS), #DELETE “Virus (no Infection good marker) —-> • Targeted analyses using capture probe enrichment”
-
Human host response analyses (RNA sequencing)
-
[TODO]: based on the Figure 2, make shorter slide (Overview.png)
- Project1: mark the keywords in the plots with highlighted color e.g. with green: Amplicon sequencing + Bacteria + microbiome analyses; Changes in the composition of the upper respiratory tract microbial community in granulomatosis with polyangiitis; Fig. 1. Alpha and beta diversity of nasal samples from patients with GPA and RA and healthy controls. (Figure1.jpg)
- Project2: keywords: Metagenomic sequencing + Targeted mNGS + microbiome analyses (monitoring hospital outbreaks); Target capture sequencing reveals a monoclonal outbreak of respiratory syncytial virus B infections among adult hematologic patients
- Project 3: keywords: Metagenomic sequencing + Untargeted mNGS + Pathogen identification; DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples in the example enterovirus detection Figure7.png.
- Project 4: keywords: Metagenomic sequencing + Untargeted mNGS + microbiome analyses; (Can refer to the project of Holger and Anna) With pure cultures of bacteria or fungi, mNGS reads can be assembled into partial or complete genomes; Based on the provided description, the project does indeed relate to microbiome analyses; Genomics of Invasive Cutibacterium acnes Isolates from Deep-Seated Infections; C.acnes_Figure1.jpg
- Future project 5: keywords: Metagenomic sequencing + Untargeted mNGS + Host transcriptome profiling: Flowchart3.png.
-
A
-
Applications in infectious disease diagnostics include direct identification of microorganisms from primary clinical samples (part Aa);
-
antimicrobial resistance prediction by characterization of resistance genes (part Ab);
-
detection of species-level or strain-level virulence determinants, such as secretion of specific endotoxins or exotoxins (part Ac);
-
and antiviral resistance prediction (part Ad). As shown for HIV-1, recovery of the complete viral genome from a patient sample by metagenomic next-generation sequencing (mNGS) (part Ad, graph) facilitates sequence analysis to predict susceptibility [sәseptәˊbiliti] or resistance to antiretroviral drugs (part Ad, bar plot); [????] the susceptibility profile for the analysed strain (black bars) predicts resistance to the non-nucleoside reverse transcriptase inhibitor (NNRTI) class of drugs (denoted by an asterisk), as opposed to nucleoside reverse transcriptase inhibitors (NRTIs) or protease inhibitors (PIs).
-
B
-
Microbiome analyses can inform disease prognosis in acute and chronic disease states and underlie the development of probiotic therapies. Coloured bars represent individual microbiota species. A reduction in species diversity is seen in dysbiosis (an unhealthy state), such as present in patients with Clostridium difficile-associated disease. Stool from healthy individuals can be harvested to treat patients with C. difficile infection by faecal stool transplantation or as orally administered encapsulated faecal pills. Alternatively, synthetic stool generated from microbiota species observed in healthy individuals can be used as probiotics to treat patients. In addition to C. difficile infection, chronic diseases such as obesity, inflammatory bowel disease and diabetes mellitus are potential targets for probiotic therapy.
-
C RNA-sequencing-based transcriptomics can improve the diagnosis of infectious and non-infectious conditions on the basis of the human host response. Host transcriptomic profiling by NGS can enable the construction of a classifier metric to discriminate between patients with infection (red bars) from uninfected patients (blue bars) with high accuracy (part Ca).
-
Metric scores above the dotted line indicate infection, whereas scores below the dotted line indicate absence of infection; the overall accuracy of the classifier metric shown is 83%. Cluster heat map analysis identifies individual, differentially expressed host genes associated with infection (genes A–F) versus those associated with no infection (genes G–L) (part Cb).
-
D
-
Sequencing of viral tumours or liquid biopsy analyses in oncology can be used for simultaneous pathogen detection and characterization of host genetic mutations.
-
mNGS can be used to detect Merkel cell polyomavirus, the virus associated with the development of Merkel cell carcinoma.
-
Simultaneous sequencing of host DNA can identify mutations that arise from integration of the viral genome containing the full-length large T antigen (LT) followed by subsequent truncation of the LT antigen (part Da) or truncation of the LT antigen before viral genome integration (part Db).
-
Both of these two mutations lead to cellular transformation that drives tumour proliferation.
-
Although promising, many of these sequencing-based applications have yet to be incorporated into routine clinical practice.
-
-
Fig. 2 for chapter 4 | Targeted versus untargeted shotgun metagenomic next-generation sequencing approaches (USING: Amplicon sequencing vs Metagenomic sequencing).
A variety of patient samples, as well as cultured microbial colonies, can be analysed using targeted or untargeted metagenomic next-generation sequencing (mNGS) methods for pathogen identification, microbiome analyses and/or host transcriptome profiling. Universal PCR (left) is a targeted mNGS approach that uses primers designed from conserved regions such as the ribosomal RNA (rRNA) genes that are universally conserved among bacteria (16S or 23S rRNA) or fungi and parasites (18S rRNA, 28S rRNA or internal transcribed spacer (ITS)). Other sets of primers can be designed to target a defined set of pathogens and/or genes and used for multiplex reverse transcription PCR or PCR (multiplexed amplicon PCR). NGS library preparation and sequencing of the resultant amplicons enable pathogen identification down to the genus or species level. Metagenomic sequencing (right) entails unbiased shotgun sequencing of all microbial and host nucleic acids present in a clinical sample. Separate DNA and RNA libraries are constructed; the DNA library is used for identification of bacteria, fungi, DNA viruses and parasites, whereas the RNA library is used for identification of RNA viruses and RNA sequencing-based human host transcriptome profiling (heat map, bottom right). As no primers or probes are used in unbiased mNGS, the vast majority of reads corresponds to the human host and, thus, detection of pathogens from metagenomic libraries is a ‘needle-in-a-haystack’ endeavour. An optional capture probe enrichment step using magnetic beads enables targeted mNGS of pathogens and/or genes from metagenomic libraries. All these methods are compatible with sequencing on traditional benchtop instruments such as the Illumina HiSeq and portable nanopore sequencers such as the Oxford Nanopore Technologies MinION. -
Fig. 3 for chapter 5 [OPTIONAL, but the figure contains no content.
However, it is a good figure showing routine of future dignostics, they clinician like it] | Challenges to routine deployment of metagenomic sequencing in the clinical setting. At each step in the process, multiple factors (bullet points) must be taken into account when implementing a clinical metagenomic pipeline for diagnosis of infections to maximize accuracy and clinical relevance. In particular, it is often useful to interpret and discuss the results of metagenomic next-generation-sequencing (mNGS) testing in a clinical context as part of a clinical microbial sequencing board, akin to a tumour board in oncology. EMR, electronic medical record. -
Fig. 4 | A typical metagenomic next-generation sequencing [IGNORING] bioinformatics pipeline.
A next-generation sequencing (NGS) data set, generally in FASTQ or sequence alignment map (SAM) format, is analysed on a computational server, portable laptop or desktop computer or on the cloud. An initial preprocessing step consists of low- quality filtering, low-complexity filtering and adaptor trimming. Computational host subtraction is performed by mapping reads to the host (for example, human) genome and setting aside host reads for subsequent transcriptome (RNA) or genome (DNA) analysis. The remaining unmapped reads are directly aligned to large reference databases, such as the National Center for Biotechnology Information (NCBI) GenBank database or microbial reference sequence or genome collections, or are first assembled de novo into longer contiguous sequences (contigs) followed by alignment to reference databases. After taxonomic classification, in which individual reads or contigs are assigned into specific taxa (for example, species, genus and family), the data can be analysed and visualized in a number of different formats. These include coverage map and pairwise identity plots to determine how much of the microbial genome has been recovered and its similarity to reference genomes in the database; Krona plots to visualize taxonomic diversity in the metagenomic library ; phylogenetic analysis to compare assembled genes, gene regions or genomes to reference sequences; and heat maps to show microorganisms that were detected in the clinical samples. OTU, operational taxonomic unit.
SLAM-seq
SLAM-seq(thiol(SH)-linked Alkylation for the Metabolic sequencing of RNA)可以理解为“S4U烷基化RNA代谢测序技术”。
SLAM-seq技术原理
SLAM-seq技术是在培养的细胞中加入4-thiouridine (S4U)核酸类似物,转录时S4U可以与DNA序列上的A碱基互补配对,从而进入新合成的RNA链中。抽提总RNA,加入碘乙酰胺(IAA)使其发生化学修饰。在逆转录过程中发生修饰的S4U与G碱基发生错配生成cDNA单链,导致最终检测结果里原本的T变成了C。通过分析检测 T→C 的转变比例,即可得到新生RNA的信息。
实验流程简述如下:
- S4U标记:培养的细胞中加入S4U,得到带有S4U标记的新生RNA;
- 加化学修饰:抽提纯化RNA,并加入碘乙酰胺(IAA)诱导S4U发生化学修饰;
- 构建文库:逆转录时S4U与G发生错配,后续PCR中G又与C配对;
- 上机测序:最终测序结果中本来该为T的位点被识别为C;
- 生物信息学分析:检测 T→C 的转变比例,即可得到新生RNA。
结合 MYC 基因分析与 2 小时的 4-硫代尿苷(4sU)标记技术,在单细胞 SLAM 测序(scSLAM-seq)设置中,可以提供深入的见解,了解 MYC 基因的动态表达和调控机制。这种方法特别适合研究基因表达的瞬时动态和细胞间的异质性,以下是具体的解释和应用场景: 技术组合的优势
-
MYC 基因的角色:MYC 是一种重要的转录因子,涉及细胞周期调控、增殖、代谢和细胞死亡。在多种癌症中,MYC 基因经常发生过表达或异常激活。
-
4sU 标记:通过在细胞培养中加入 4sU,新合成的 RNA 分子会嵌入这种修饰的尿苷。这允许研究者区分在标记窗口期内新合成的 RNA(标记 RNA)与旧的 RNA(未标记 RNA),从而实现 RNA 合成和降解动态的精准测量。
-
单细胞 SLAM 测序:scSLAM-seq 技术使研究者能够在单细胞水平上对 RNA 进行精确分析,这对于揭示细胞内部复杂的分子机制和基因表达的细微差异至关重要。
应用场景
-
基因表达的时间分辨率:结合 4sU 标记与 scSLAM-seq,可以在单细胞水平上观察到 MYC 及其他基因的表达如何随时间变化,特别是如何响应外部信号或内部调控机制的变化。
-
转录动态研究:这种方法可以揭示 MYC 在不同细胞状态下(如细胞周期的不同阶段、癌症细胞与正常细胞之间)的表达和调控差异。
-
癌症研究:由于 MYC 在许多类型的癌症中都扮演着关键角色,通过分析其转录爆发和新旧 RNA 的动态,科研人员可以更好地理解其在疾病发展中的功能,为开发新的治疗策略提供依据。
结论
通过 MYC 基因与 2 小时 4sU 标记的结合使用,研究者可以获得关于 MYC 基因表达调控和功能的宝贵信息,特别是其在细胞增殖和癌症发展中的作用。这种方法为研究基因表达提供了一个强大的工具,尤其是在探索基因如何在单个细胞中瞬时和动态地响应生理和病理条件时。
成本效益考量的 TraDIS (转座子定向插入位点测序)
- Transposon directed insertion‑site sequencing (TraDIS), a variant of transposon insertion sequencing commonly known as Tn‑Seq, is a high‑throughput assay that defines essential bacterial genes across diverse growth conditions.
- However, the variability between laboratory environments often requires laborious, time‑consuming modifications to its protocol.
- In this technical study, we aimed to refine the protocol by identifying key parameters that can impact the complexity of mutant libraries.
- Firstly, we discovered that adjusting electroporation parameters including transposome concentration, transposome assembly conditions, and cell densities can significantly improve the recovery of viable mutants for different Escherichia coli strains.
- Secondly, we found that post‑electroporation conditions, such as recovery time and the use of different mediums for selecting mutants may also impact the complexity of viable mutants in the library.
- Finally, we developed a simplified sequencing library preparation workflow based on a Nextera‑TruSeq hybrid design where ~ 80% of sequenced reads correspond to transposon‑DNA junctions.
- The technical improvements presented in our study aim to streamline TraDIS protocols, making this powerful technique more accessible for a wider scientific audience.
Introduction
-
Transposon directed insertion-site sequencing (TraDIS) is a powerful high-throughput assay used to identify essential bacterial genes in various growth conditions 1 .
-
This technique operates by randomly integrating Tn5 mini-transposons into the bacterial genome through the action of the transposase enzyme, a process that disrupts or alters gene function.
-
While widely applied, TraDIS protocols are not standardized and often require custom modifications to suit the unique circumstances of different laboratories, such as variations in available equipment and bacterial strains.
-
These adjustments can be particularly time-consuming and labor-intensive for those new to the technique.
-
Our current study aims to streamline the construction of complex mutant libraries by identifying key parameters that enhance labor efficiency and practicality.
-
We employed Escherichia coli strains from diverse sources, including freshwater environments, the human gut, and laboratory settings, to demonstrate the versatility and applicability of our methods.
-
Our investigation included optimizing electroporation conditions to improve transposon insertion and mutant recovery.
-
Specifically, we fine-tuned the concentration of the transposome, evaluated the effect of varying transposon DNA quantities during transposome assembly, and explored how cell density affects the outcomes of electroporation reactions.
-
Following the optimization of electroporation conditions, our study investigated the impact of post-electroporation parameters on the diversity of mutants with unique insertion sites.
-
We assessed the length of recovery time after electroporation and compared the efficacy of different mediums for selecting mutants, namely agar plates versus liquid broth.
-
While using liquid broth is a simpler approach, it potentially compromises the complexity of the recovered mutant library, as mutants compete for shared resources before the culture can be collected.
-
These comparisons aimed to determine the most effective method for constructing complex mutant libraries while considering factors such as labor efficiency and practicality.
-
Another important aspect of TraDIS assay is a robust PCR protocol to enrich transposon-DNA junctions, while minimizing amplification of non-specific fragments without such junctions.
-
Prior strategies include the use of splinkerette adaptors followed by a sequencing run with custom primers and operation protocols2,3.
-
However, this strategy requires a dedicated run for TraDIS libraries, which can limit the use of a higher capacity, more cost-effective sequencer like NovaSeq.
-
Another strategy is a nested PCR approach using ‘all-in-one’ primers that combine both the Illumina flow cell and transposon-specific sequences4,5.
-
Although this method is effective, it involves the use of long primers (exceeding 60 base pairs), which often necessitates more expensive synthesis.
-
Also, if one were to use a different transposon for the assay, this requires the synthesis of a new set of ‘all-in-one’ primers.
-
To this end, we present a PCR strategy that simplifies the library preparation workflow.
-
Our library construction scheme is compatible with standard TruSeq/Nextera indexing primers used for other Illumina assays, thereby reducing the overall cost.
Methods
Bacterial strains (Supplemental Table 1)
- We utilized three strains of Escherichia coli, each selected for its unique origin and characteristics.
- AB_00116 was isolated from a freshwater environment and provided by Aaron Best’s laboratory at Hope College, Holland, Michigan, USA.
- MS 69-1 was obtained from BEI Resources (HM347) and originally isolated from the colon of a control patient with abnormal histology in New York, New York, USA.
- D43HC1 was derived from a laboratory E. coli strain ATCC25922, previously characterized as multi-drug resistant after being exposed to a gut relevant- concentration of the antipsychotic drug quetiapine in vitro for 6 weeks6.
- These strains were chosen to represent a diverse range of environmental and physiological conditions, thereby ensuring the robustness and applicability of our experimental findings across different bacterial backgrounds.
Synthesis of transposon encoding the kanamycin resistance gene (KAN2)
- Using the Twist Bioscience Gene Fragments service, we ordered a custom DNA fragment corresponding to the KAN2 transposon (1221 bp), whose full sequence is available on the product manual for the EZ-Tn5 Tnp Transposome Kit (Lucigen #TSM99K2).
- We PCR amplified 1 ng of the fragment with Q5® High-Fidelity 2X Master Mix (NEB #M0492) using a PCR primer set: Fwd 5′—/5′Phos/CTGTCTCTTATACAC ATCTCAACCATCATCGA—3′ and Rev 5′—/5′Phos/CTGTCTCTTATACACATCTCAACCCTGAAGCT—3′ and the following PCR cycle parameters: 98 °C for 30 s, 18 cycles of (98 °C for 10 s, 69 °C for 30 s, 72 °C for 45 s), and 72 °C for 2 min (50 uL reactions in triplicate).
- After confirming the amplicon size by gel electrophoresis, we purified the amplicon using the DNA Clean & Concentrator-5 kit (Zymo Research #D4013).
- To increase the DNA concentration in the final eluate, we used three purification columns (i.e. one column for each PCR reaction).
- At the elution step, we eluted the first column with 25 uL of the fresh elution buffer for the DNA Clean & Concentrator-5 kit (10 mM Tris-HCol, ph 8.5, 0.1 mM EDTA).
- For the second and third columns, we used the first and second eluates as the elution medium.
Transposome assembly
- We purchased the unloaded Tagmentase (Tn5 transposase) from Diagenode (#C01070010-10; 2 mg/mL = 37.52 uM), and adjusted the protein concentration to 1 uM by diluting it in the storage buffer described in the manual for the EZ-Tn5 Transposase (LGC Biosearch Technologies) (50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 0.1% Triton® X-100 (Rohm & Haas) and 1 mM DTT).
- To assemble the transposome, we mixed 1 part of the KAN2 transposon DNA solution, 2 parts of the diluted Tn5 at 1 uM, and 1 part of 100% glycerol, and incubated in a thermocycler at 23 °C for 60 min before use (or stored at − 20 °C if not used immediately).
Electroporation experimental setup (Basic workflow)
- To make E. coli electrocompetent, we first inoculated a colony of bacteria in 3 mL of LB medium and shake-incubated at 37 °C overnight.
- Next day, we transferred the overnight culture in fresh LB medium (1/100 dilution) and shake-incubated again at 37 °C until the optical density at 600 nm ( OD600) of the culture reached ~ 0.4.
- Cells were harvested by centrifugation at 4000g for 10 min at 4 °C, washed twice in 2/3 culture volume of sterile water (4 °C) (centrifugation after each wash), and then resuspended in 1/100 culture volume of sterile water (4 °C) (e.g. for 45 mL of culture, 30 mL water was used for each wash, and 450 uL water was used for final resuspension).
- Cells and electroporation cuvettes (1 mm gap) were kept on ice until electroporation.
- We used BioRad Gene Pulser to electroporate the bacteria using the following parameters: 2000 V, 25 uF and 200 Ω.
- Immediately after electroporation, we added 1 mL of SOC medium, pipette up and down several times, and transferred to a sterile 2 mL screw-cap tube for recovery in a shaker incubator at 37 °C (with tubes placed in a horizontal position for better aeration).
- We recorded the time constant for each electroporation reaction.
抗卡那霉素基因 (KAN2) 转座子的合成
- 我们使用 Twist Bioscience Gene Fragments 服务订购了一个自定义 DNA 片段,该片段对应于 KAN2 转座子(1221 bp),其完整序列可以在 EZ-Tn5 Tnp Transposome Kit(Lucigen #TSM99K2)的产品手册中找到。
- 我们使用 Q5® 高保真 2X Master Mix(NEB #M0492)和一组 PCR 引物:Fwd 5′—/5′Phos/CTGTCTCTTATACACATCTCAACCATCATCGA—3′ 和 Rev 5′—/5′Phos/CTGTCTCTTATACACATCTCAACCCTGAAGCT—3′,对片段进行了 PCR 扩增,使用的 PCR 循环参数为:98 °C 30秒,18个循环(98 °C 10秒,69 °C 30秒,72 °C 45秒),以及 72 °C 2分钟(50 uL 反应,三次重复)。
- 通过凝胶电泳确认了扩增产物的大小后,我们使用 DNA Clean & Concentrator-5 kit(Zymo Research #D4013)纯化了扩增产物。
- 为了增加最终洗脱液中的 DNA 浓度,我们使用了三个纯化柱(即每个 PCR 反应一个柱)。
- 在洗脱步骤中,我们用 25 uL 新鲜的洗脱缓冲液为第一个柱洗脱,该洗脱缓冲液来自于 DNA Clean & Concentrator-5 kit(10 mM Tris-HCl,pH 8.5,0.1 mM EDTA)。
- 对于第二和第三个柱,我们使用了第一和第二次洗脱液作为洗脱介质。
Transposome 组装: Tn5 transposase [转座酶] (1. protein) + KAN2 transposon DNA (2. DNA) = Tn5 transposome [转座体] (3. protein-DNA complex)
- 我们从 Diagenode 购买了未加载的 Tagmentase (Tn5 转座酶)(#C01070010-10; 2 mg/mL = 37.52 uM),并按照 EZ-Tn5 转座酶(LGC Biosearch Technologies)手册中描述的储存缓冲液(50 mM Tris–HCl (pH 7.5),100 mM NaCl,0.1 mM EDTA,0.1% Triton® X-100 (Rohm & Haas) 和 1 mM DTT)调整蛋白浓度至 1 uM。
- 为了组装 transposome,我们将 1份 KAN2 转座子 DNA 溶液、2份稀释至 1 uM 的 Tn5 以及 1份 100% 甘油混合,并在热循环仪中于 23 °C 孵育 60分钟,使用前取出(或若暂时不使用则储存于 −20 °C)。
- 转座体组装: 在这个步骤中,未装载的Tagmentase(Tn5转座酶)与转座子DNA(如KAN2转座子)按照一定比例混合,并加入甘油进行保护,然后在特定温度下孵育一段时间。这个过程的目的是使转座酶和DNA片段结合,形成可活性的转座体复合物。这种复合物可以将DNA片段插入到目标细胞的基因组中。
- 转座酶和DNA片段结合后形成的转座体(transposome)的大小主要取决于使用的DNA片段的长度。在实验中通常使用的转座子DNA可以是几百到几千碱基对长。因此,转座体的大小会略大于所使用的DNA片段的长度,加上转座酶本身的尺寸。例如,如果使用的转座子DNA片段长度为2000碱基对(大约等于2千个核苷酸),转座酶(如Tn5转座酶)则具有一定的三维结构和体积,可能将整个复合物的长度增加数十到数百纳米。转座酶一般会与DNA的特定位点结合,形成稳定的蛋白-DNA复合体。这种复合物的实际尺寸和结构取决于具体的转座酶和DNA的结合方式,以及转座酶自身的空间结构。
电转化实验设置(基础工作流程)+ Cells [4]
- 为了使大肠杆菌电转化成分子,我们首先在 3 mL LB 培养基中接种一个细菌菌落,并在 37 °C 下振荡培养过夜。
- 第二天,我们将过夜培养物转移到新鲜的 LB 培养基中(1/100 稀释)并再次在 37 °C 下振荡培养,直到培养液的光密度在 600 nm(OD600)达到约 0.4。
- 细胞通过在 4 °C 下 4000g 离心 10 分钟收集,洗涤两次,每次使用 2/3 培养体积的无菌水(4 °C)(每次洗涤后离心),然后用 1/100 培养体积的无菌水(4 °C)重新悬浮(例如,对于 45 mL 的培养物,每次洗涤使用 30 mL 水,最终重悬时使用 450 μL 水)。
- 在进行电转化前,将细胞和电转化池(1 mm 间隙)保持在冰上。
- 我们使用 BioRad Gene Pulser 进行电转化,参数为 2000 V、25 μF 和 200 Ω。
- 电转化后立即加入 1 mL SOC 培养基,上下吸吮数次,然后转移到一个无菌的 2 mL 螺旋盖试管中,在 37 °C 的摇床孵育器中恢复,试管置于水平位置以便更好地通气。
- 我们记录了每次电转化反应的时间常数。
- 尽管转座体已经组装好,但它们仍需被有效地送入宿主细胞(如大肠杆菌)内才能发挥作用。电转化是一个高效的细胞转化技术,通过对细胞暂时施加一个高电压电场,造成细胞膜上微小的孔隙,使得转座体可以通过这些孔隙进入细胞内部。在电转化后,这些孔隙会迅速自我修复,从而将外源DNA锁定在细胞内部。
Analyzing the effect of selection medium (agar plate vs. liquid culture) on mutant library complexity
- After 1 h of recovery from electroporation, we selected mutants as follows: Agar plates: We spread one portion of the culture on LB agar plates containing 40 μg/mL kanamycin (each plate received an appropriate volume of culture to generate 1000–2000 CFUs), and incubated at 37 °C overnight.
- On the next day, we first measured CFUs using a ProtoCOL3 automatic colony counter (Synbiosis), and then scraping off colonies from agar plates using a sterile cell spreader and 1 mL of LB.
- We recovered an equal number of mutants for each strain under investigation.
- We extracted DNA from a dilute sample of the pooled culture using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s instructions. Liquid culture: We transferred the equal volume of the
Analyzing the effect of recovery time on mutant library complexity
- After electroporation and addition of SOC medium for each strain, we combined multiple reactions into one to normalize the potential difference in electroporation efficiency of each reaction, and then immediately split them into equal portions among 2 mL screw-cap tubes.
- All portions were shake-incubated in parallel at 37 °C, and they were spread onto LB-kanamycin plates at designated time points.
- All plates were incubated at 37 °C overnight.
- Each experiment was performed in duplicate.
- On the next day, we first measured CFUs using the ProtoCOL3 automatic colony counter, and then scraped off colonies from agar plates using a sterile cell spreader and 1 mL of LB.
- We extracted DNA from a dilute solution of the pooled culture using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s instructions.
Statistics
- Statistical analysis involved applying a two-way analysis of variance (ANOVA) to assess significance, followed by Tukey’s Honest Significant Difference test for post hoc comparisons.
Construction of DNA libraries and sequencing
- We sonicated extracted DNA samples using the Covaris S2 sonicator (parameters: Intensity – 5; Duty Factor – 10; time – 90 s) to fragment the genome into 300–400 bp pieces.
- Proper fragmentation was verified via gel electrophoresis.
- We used the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB #E7103) to constructed DNA library (input = 500 ng) according to the manufacturer’s protocol (version 7.0_9/22) with the following adjustments:
- (1) During fragment size selection, we used 22.5 μL (Step 3A.2) and 10 μL (Step 3A.6) of the NEBNext Sample Purification Beads;
- (2) At “Step 4.1 PCR Amplification” we enriched the transposon-DNA junction using the following PCR parameters: 1 cycle of 98°C for 30 s, 22 cycles of [98 °C for 10 s, 65 °C for 75 s], and 1 cycle of 65 °C for 5 min (primer sequences are listed in Supplemental Table 2; and
- (3) We eluted the resulting PCR products using 52.5 uL of 10 mM Tris (pH 8.0) (Step 5.9) and collected 50 uL of the eluate in Step 5.11.
- We measured the concentration of each eluate using Qubit reagent (Invitrogen #Q32851) and adjusted concentrations of each sample to 4 ng/uL with 10 mM Tris (pH 8.0).
- Using 20 ng of 1st PCR product as input material, we conducted index PCR (50 uL reaction) with 2X KAPA HiFi HotStart ReadyMix (Roche Diagnostics #07958927001), Illumina Nextera/TruSeq P5 and TruSeq P7 index primers, and the following PCR parameters: 1 cycle of 95 °C for 3 min, 8 cycles of [95 °C for 10 s, 55 °C for 30 s, 72 °C for 30 s], and 1 cycle of 72 °C for 5 min.
- We purified the resulting product with the HighPrep PCR beads (MagBio Genomics #AC-60005) according to the manufacturer’s protocol (version v2.0) using the beads-to-sample ratio of 1:1.12.
- We pooled libraries in equimolar concentration and sequenced on the Illumina NovaSeq 6000 platform (150 bp PE).
Sequencing data analysis (TODO: NEXT_WEEK_REPERT_THE_PIPELINE_ON_THE_PAPER_DATA_and_ON_OWN_DATA!)
- We first subjected fastq reads to two rounds of filtering using the cutadapt software (ver. 4.1) to remove reads that did not have expected fragment layout.
- For the first round of filtering, we trimmed a total of 29 bp sequences from Read 1 (5′—GCA TGCAAGCTTCAGGGTTGAGATGTGTA—3′), which corresponded to the PCR anchor (20 bp) and the 5′ portion of the 19-bp Tn5 mosaic end (ME) sequence (9 bp), with a maximum of 10% mismatches allowed.
- For the second round, we trimmed the remaining 10-bp ME sequence (5′—TAAGAGACAG—3′), with no mismatch allowed.
- In both rounds of filtering, we discarded reads that did not pass the filtering criteria (‘–discard-untrimmed’), and corresponding read mates were discarded from Read 2 fastq files.
- We mapped the filtered reads (Read 1 only) to respective reference FASTA in both forward and reverse complement orientations using the bwa software (version 0.7.17-r1188) with default options.
- We defined the transposon-DNA junctions, which is equivalent to the number of unique mutants in the library, as the leftmost coordinate of the mapped reads, and we used a custom bash script to determine this.
- We first sorted the mapped reads based on their genomic coordinates using the samtools software (version 1.14) and filtered for reads that align to forward orientation (FLAG = 0 in the second column).
- We then de-duplicated reads with identical coordinates (chromosome and the leftmost position of the mapped reads) and counted the number of unique reads as ‘putative junctions’.
- We filtered out putative junctions with low number of reads (< 1 count per million reads mapped), and then classified the remaining junctions as ‘true junctions’, which are reported in the Results section.
-
The code used for this analysis can be found here: https://github.com/StephanieAFlowers/TraDIS_techical_paper.git.
#### Calculate the number of putative junctions grep -v "@PG" ${SORTED.SAM} |grep -v "@SQ" |grep -v "@HD" |awk '{if ($2 == 0) print $3"_"$4}' |uniq |wc -l #### Calculate the number of putative junctions with low read counts (CPM < 1) to be subtracted #### grep -v "@PG" ${SORTED.SAM} |grep -v "@SQ" |grep -v "@HD" |awk '{if ($2 == 0) print $3"_"$4}' |uniq -c |awk '{print $1}' |sort -k1,1n |uniq -c |head -${LOW_CPM_THRESHOLD} |awk 'BEGIN {OFS="\t"} {print $2,$1}' |awk '{sum += $2} END {print sum}'
Results
Diluting transposome [3] improves electroporation efficiency (稀释转座酶复合体可以提高电穿孔效率, https://zhuanlan.zhihu.com/p/664630787)
- We examined how electroporation efficiency is affected by the amount of transposome in electroporation reaction (Fig. 1A).
- To analyze this, we serially diluted the most concentrated transposome (1 U) to 0.5, 0.25, 0.1, 0.05, 0.025, and 0.01 U, mixed into electrocompetent cells, and assessed the electroporation efficiency by measuring CFUs the next day.
- All conditions for each strain were performed in duplicate. We found that CFUs increased as we used more diluted solutions of transposome, peaked around 0.05 U for AB_00116 strain or 0.1 U for MS 69–1 strain, and then declined.
- The effect of transposome concentration on electroporation efficiency, assessed via two-way ANOVA, was significant (p = 3.95e–08).
- The time constant (τ) is measured by the product of resistance and capacitance and often correlates with electroporation efficiency.
- We found that the time constants for reactions containing 1, 0.5, and 0.25 U transposome were 8.7%, 4.3%, and 2.2% lower compared to those containing less than 0.25 U, which correlated with lower CFUs (Fig. 1B).
Higher quantity of transposon DNA [2] during transposome assembly improves electroporation efficiency
- We also examined how electroporation efficiency is affected by the quantity of transposon DNA during transposome assembly (Fig. 2). To analyze this, we assembled transposomes by mixing 1 U of Tn5 with 400, 200, or 100 ng of KAN2 transposons.
- We mixed 0.05 U of the assembled transposomes with electrocompetent cells, and assessed the electroporation efficiency by counting the resulting CFUs the next day.
- All conditions were performed in duplicate.
- The CFUs were the highest for those electroporated with the transposome with 400 ng KAN2 transposon, followed by 200 ng (18.0% CFU on average relative to 400 ng), and then 100 ng, which yielded virtually no colonies like the mock-electroporated negative control.
- We assessed the effect of transposon DNA quantity on electroporation efficiency by two-way ANOVA (p = 7.95e–12).
- The time constant (τ) was not affected by the transposon quantity.
Cell density [4] in electroporation reaction affects its efficiency
- We determined how electroporation efficiency is affected by the cell density in the reaction mixture (Fig. 3).
- We mixed 0.05 U of the transposomes with cell suspension of different densities (50X, 100X or 200X of the starter culture volume), and assessed the efficiency by counting the resulting CFUs the next day.
- All conditions were performed in duplicate.
- We found that overall, 100X concentrate of the starter culture volume had the highest electroporation efficiency, followed by 50X (21.4% less efficient on average) and 200X (56.8% less efficient on average; p = 0.0014).
- The density producing the highest electroporation efficiency was somewhat strain specific.
- The time constant (τ) was not affected by cell density.
Increasing recovery time after electroporation does not significantly affect the number of unique insertion events
- Electroporation of transposome into bacterial cells is immediately followed by a recovery period in rich growth medium such as SOC.
- This recovery period provides time for bacterial cells that successfully took up the transposome to express selection marker genes.
- However, the length of recovery time is not standardized in the TraDIS assay and varies among previous studies (from 1–2 h 4,5,7,8).
- This is an important parameter to consider when creating the mutant strain library, as the number of unique mutants is often estimated from CFUs (Colony-forming unit).
- Therefore, we sought to examine the relationship between the number of CFUs and unique mutants, as a function of recovery time length.
- We hypothesized that longer recovery time would result in a lower proportion of unique mutants because longer recovery time would allow expansion of clones in culture.
- To test the hypothesis, we electroporated our E. coli cohort with the Tn5 transposome carrying the KAN2 gene, and let them recover for 0.5, 1, and 2 h before plating on kanamycin selection plates.
- After an overnight incubation (Digital bacterial incubator), we harvested the same number of colonies for each time point per strain, and determined the number of unique mutants via sequencing.
- Not surprisingly, a longer recovery time yielded higher CFUs, doubling roughly every 0.5 h of recovery time (Fig. 4A).
- Contrary to our expectation, however, we did not observe a corresponding decrease in the number of unique insertion events (p = 0.4), and the number of unique mutants recovered was somewhat strain dependent (Fig. 4B).
- For two of the three test strains (MS 69-1 and D43HC1), we found that the number of unique mutants detected were comparable across time points; whereas, for strain AB_00116, we observed a reduced number of mutants as the recovery time lengthens.
Based on the information you provided, your understanding that each colony contains an isolate with multiple insertion events is not necessarily correct. Here’s a more precise breakdown:
- Transposome and Insertion Events: When transposomes are electroporated into bacterial cells, they can integrate into the genome at multiple locations. However, each individual transposome typically only carries one or a few copies of a DNA fragment, which could insert into different sites within the bacterial genome. The number of insertion events per cell depends on how many transposomes enter the cell and successfully integrate into the genome.
- Colony Formation: After electroporation, cells are allowed to recover and then plated on a selective medium that only allows the growth of cells where the transposome has successfully integrated (because it carries an antibiotic resistance gene like the KAN2 gene). Each colony that forms on the plate theoretically originates from a single cell that took up the transposome. Therefore, each colony represents a unique isolate, which could potentially have multiple insertion events if multiple transposomes entered that particular cell and integrated at different sites.
- Unique Insertion Events: The research you described was testing whether the length of recovery time after electroporation affects the number of unique insertion events within these colonies. The results indicated that the number of unique insertion events did not significantly change with different recovery times, although the number of colonies (CFUs) increased with longer recovery times.
- Conclusion: Each colony on the selective plate indeed originates from a single cell that has taken up the transposome. While it’s possible for multiple insertion events to occur within a single cell (leading to possibly multiple insertion sites within the genome of the cell that founded the colony), each colony primarily represents a unique transformation event. The total number of unique insertion events across the population might not necessarily increase with longer recovery times, indicating that the dynamics of how transposomes integrate are complex and may be influenced by factors other than just the length of recovery time.
- Therefore, each colony generally represents a unique transformant, potentially with multiple insertion events.
- Combination of Sites: The diversity also involves the combination of sites where insertions occur within a single cell or across a population of cells. For example, in experiments involving transposons, some cells may have insertions at unique single sites, while others may have multiple insertions at different sites. The range and pattern of these combinations add to the diversity of insertion sites.
If it means that after long time, more colonies have the same combination of sites, or means that after long time, more different combinitions of sites occours based on the infomation: after long time, we have more colonies, but total unique insertion sites are not changed?
Based on the information provided in your earlier message about the experimental findings, it seems that extending the recovery time after electroporation results in a higher number of colonies (CFUs), but does not significantly alter the total number of unique insertion sites across all colonies. This indicates a couple of key points regarding the diversity and combination of insertion sites:
* Increase in Colonies but Steady Number of Unique Insertion Sites: As recovery time increases, more cells survive and form colonies. However, the overall diversity of insertion sites—the different combinations and locations where the transposome integrates into the genome—does not significantly change. This suggests that while there are more colonies, they may not necessarily exhibit a greater variety of insertion site combinations.
* Implications for Insertion Site Combinations: The fact that the total number of unique insertion sites remains roughly constant despite more colonies forming can suggest two things:
- Redundancy in Insertion Events: Longer recovery times might allow for more cells to replicate before plating, leading to a higher number of colonies that may share similar insertion profiles. This would mean that some of the additional colonies formed under longer recovery times are clones or near-clones of each other, representing the same or very similar insertion events.
- Saturation of Potential Sites: It's also possible that there is a kind of saturation effect where, despite more cells becoming transformants, the transposome tends to integrate into a somewhat fixed repertoire of favored genomic sites. Thus, while more cells are transformed, the variety of unique genomic integration events does not proportionally increase.
* Conclusion Regarding Diversity: In summary, more colonies after a longer recovery time do not equate to a higher diversity of insertion site combinations. Instead, the diversity of insertion sites in terms of unique integration events remains consistent, indicating that more time allows for greater cell survival and colony formation, but not necessarily a greater diversity in the genetic outcomes of those colonies.
This conclusion can be valuable in experimental design and interpretation in genetics, particularly when using transposon mutagenesis to explore gene function, as it highlights the importance of considering both the number of transformants and the actual diversity of genetic alterations achieved through the process.Selecting mutants in liquid broth may be a viable alternative to construct a complex mutant library
- Constructing the TraDIS mutant library is a laborious task.
- To achieve the desired complexity of the library, it is necessary to prepare, plate, and scrape (/skreɪp/) hundreds of agar plates to collect hundreds of thousands of colonies.
- A potential alternative to this method is to select the bacteria in a pooled fashion using liquid broth with a selection agent.
- Previously, Fels and colleagues developed a variant of the Tn-seq approach called Transposon Liquid Enrichment Sequencing (TnLE-seq) and demonstrated that a liquid broth can be successfully used to build a complex mutant strain library of Desulfovibrio vulgaris Hildenborough9.
- However, only a handful of studies have adopted the strategy10,11, despite its potential to save time, labor and cost.
- We sought to examine whether the liquid broth approach could be adopted for constructing complex mutant libraries after electroporation of Tn5 mini-transposons.
- We electroporated our E. coli cohort with the Tn5 transposome carrying the KAN2 selection gene, and after 1 h of recovery, selected the resulting transformants either: (1) by plating them onto LB-kanamycin plates and collecting colonies after overnight incubation; or (2) by incubating them in a liquid broth with kanamycin and later collecting as the culture reached early-, mid-, or late log growth phase.
- All experiments were done in duplicate.
- We isolated DNA from each condition and determined the number of unique mutants via sequencing after subsampling reads to 5 million for each strain and condition (Table 1).
- For two of the three test strains (AB_00116 and MS 69-1), we found that the number of unique mutants detected were comparable between the two selection methods; whereas, for one of the test strains (D43HC1), we observed on average 15% reduction in the number of mutants when liquid broth was used for selection. For all strains, the growth phase of the liquid cultures at the time of harvest did not affect the number of unique mutants (0 = 0.44).
- NOTE that the Tn5 mini-transposon corresponds to the “KAN2 transposon DNA” part in the information you provided. In the assembly of the Tn5 transposome, the Tn5 mini-transposon would be the DNA component that pairs with the Tn5 transposase (the protein) to form the Tn5 transposome, which is a protein-DNA complex.
- NOTE that in the context provided, “unique mutants” refers to individual bacterial cells in which the Tn5 mini-transposons have inserted into different genomic locations, resulting in genetic variations that are distinct from each other. Each unique mutant represents a different insertion site of the transposon within the bacterial genome. This variation is crucial for constructing a complex mutant library because it allows for a wide exploration of gene functions and interactions within the organism.
Nextera‐TruSeq hybrid library design enables a simple PCR approach to enrich transposon‐DNA junctions
- Our simplified workflow to prepare the TraDIS libraries is shown in Fig. 5.
- Following DNA fragmentation, we used a standard protocol to end repair, dA-tail, and ligate the TruSeq adaptors (see “Methods”).
- Adaptor-ligated DNA samples were then subjected to the first round of PCR using a transposon-specific primer (< 60 bp) that had three components: (1) a partial Nextera i5 adapter (34 bp); (2) a ‘balancer’ (3–6 bp) to increase nucleotide diversity during a sequencing run as previously done4,5; and (3) the anchor sequences that target the KAN2 transposon (20 bp). The reverse primer was targeted to the TruSeq i7 adapter.
- During indexing PCR, we added necessary components for Illumina sequencing such as i5/i7 indexes for sample multiplexing and P5/P7 flow cell adaptors (primer sequences are available in Supplemental Table 2).
- We sequenced the resulting libraries on the Illumina NovaSeq platform without custom sequencing primers or machine operation protocol, and then calculated the fraction of usable reads that contained transposon-DNA junctions (see “Methods” for filtering criteria and analysis workflow).
- Our simplified workflow generated a high fraction of usable reads (on average 79.3 ± 1.1% among 9 samples with at least 6 million reads each).
- Using the Nextera i5 adapter in the transposon-specific primer was crucial, as the fraction of usable reads plummeted to on average 0.34 ± 0.01% when we replaced the Nextera i5 adapter with the TruSeq i5 adapter.
-
Poor performance of the TruSeq adaptor primer is likely due to non-specific amplification, as > 95% of non-usable reads were properly-paired and mappable to appropriate genomes (see “Discussion” for a potential cause).
#Purpose Sequence (5' to 3') #Amplify KAN2 transposon (5' phosphorylated) /5'Phos/CTGTCTCTTATACACATCTCAACCATCATCGA #Amplify KAN2 transposon (5' phosphorylated) /5'Phos/CTGTCTCTTATACACATCTCAACCCTGAAGCT Fwd primer for 1st PCR (Nextera i5 adaptor + 3N balancer) tcgtcggcagcgtcAGATGTGTATAAGAGACAGNNNGCATGCAAGCTTCAGGGTTGA Fwd primer for 1st PCR (Nextera i5 adaptor + 3N+C balancer) tcgtcggcagcgtcAGATGTGTATAAGAGACAGNNNcGCATGCAAGCTTCAGGGTTGA Fwd primer for 1st PCR (Nextera i5 adaptor + 3N+TA balancer) tcgtcggcagcgtcAGATGTGTATAAGAGACAGNNNtaGCATGCAAGCTTCAGGGTTGA Fwd primer for 1st PCR (Nextera i5 adaptor + 3N+ATT balancer) tcgtcggcagcgtcAGATGTGTATAAGAGACAGNNNattGCATGCAAGCTTCAGGGTTGA #Rev primer for 1st PCR (rev. comp of NEBNext adaptor read 1) gtgactggagttcagACGTG #Fwd primer for 1st PCR (TruSeq i5 adaptor + 3N balancer) acactctttccctacACGACGCTCTTCCGATCTNNNGCATGCAAGCTTCAGGGTTGA #Fwd primer for 1st PCR (TruSeq i5 adaptor + 3N+C balancer) acactctttccctacACGACGCTCTTCCGATCTNNNcGCATGCAAGCTTCAGGGTTGA #Fwd primer for 1st PCR (TruSeq i5 adaptor + 3N+TA balancer) acactctttccctacACGACGCTCTTCCGATCTNNNtaGCATGCAAGCTTCAGGGTTGA #Fwd primer for 1st PCR (TruSeq i5 adaptor + 3N+ATT balancer) acactctttccctacACGACGCTCTTCCGATCTNNNattGCATGCAAGCTTCAGGGTTGA Index primer for Nextera P5 end (X = 8 bp i5 index) AATGATACGGCGACCACCGAGATCTACACXXXXXXXXtcgtcggcagcgtc #Index primer for TruSeq P5 end (X = 8 bp i5 index) AATGATACGGCGACCACCGAGATCTACACXXXXXXXXacactctttccctac Index primer for TruSeq P7 end (X = 8 bp i7 index) CAAGCAGAAGACGGCATACGAGATXXXXXXXXgtgactggagttcag
Discussion
-
Transposon insertion sequencing is a powerful high-throughput method of identifying essential bacterial genes in various growth conditions.
-
Yet, there are several key technical hurdles that one has to overcome to successfully conduct the assay: an efficient construction of a complex mutant library, and a robust detection of the resulting transposon-DNA junctions using deep sequencing.
-
In this study, we have identified a number of electroporation parameters that are readily adjustable that can enhance the number of transformants, and also described a simpler sequencing library workflow that is more cost-effective and flexible than previous approaches.
-
We believe that these technical improvements can make this powerful technique more accessible for a wider audience.
-
We found that more dilute concentration of the Tn5 transposome was more effective in generating higher CFUs, at least among our cohort of three E. coli strains we tested, as the optimal concentrations ranged between 10 and 20 times lower than the concentration frequently used in prior TraDIS studies (1 U).
-
Poorer performance at higher transposome concentrations is likely due to the carryover of the Tn5 storage buffer into electroporation reactions, whose salt content may interfere with its efficiency.
-
In support of this, reactions with higher transposome quantity had parallel decrease in the time constant, which is often a good indicator of electroporation efficiency.
-
On the other hand, overly diluted transposome solutions result in a low overall yield of viable mutants, although these dilutions do not necessarily compromise the electroporation process itself as indicated by time constant.
-
The ability to use more diluted transposomes has several advantages.
-
First, since the commercially available Tn5 transposase is an expensive reagent of the TraDIS assay, this reduces the overall cost of each electroporation reaction by 10–20 fold.
-
Second, because less amount of the enzyme is required for each round of electroporation, this allows researchers to optimize electroporation conditions directly using Tn5 transposomes, instead of using other molecules like plasmids as a proxy, whose optimized parameters may not be necessarily the same as those for Tn5 transposomes.
-
Optimization of optical density (OD) and/or growth phase prior to electroporation is another well-documented factor influencing mutant yields (PMID: 35804085, PMCID: PMC3939052) that may vary between bacteria.
-
As our experiments were specific to E. coli, we acknowledge that researchers applying these methods to other bacterial species might need to consider the OD/growth phase as a critical parameter.
-
More experiments are required to see if our findings are generalizable to other microbial species.
-
We also observed that the transposon concentration during transposome assembly has a striking impact on CFU recovery.
-
A standard protocol (EZ-Tn5 Transposase manual) suggests the pairing of 200 ng of a transposon with 0.5 μM of Tn5 transposase (53.3 kDa) during assembly.
-
We found that doubling the quantity of transposon to 400 ng enhanced CFUs by roughly fourfold, while halving the quantity to 100 ng reduced CFUs virtually to none.
-
Our findings clearly show that using an insufficient amount of transposon during transposome assembly is detrimental to obtaining viable mutants, and therefore, a proper calculation of the molar ratio between the Tn5 transposase and the transposon is crucial.
-
In our most successful condition with 400 ng of the KAN2 transposon (1221 bp), the molar ratio during assembly was roughly 15:1 Tn5 transposase to the transposon.
-
Because a complete transposome requires 2 molecules of Tn5 for every molecule of transposon, 400 ng of the KAN2 transposon would allow us to form at maximum ~ 15% of complete transposomes in the assembly mixture.
-
Therefore, it is theoretically possible that adding more quantities of transposons can yield higher CFUs; however, we did not test this in the current study.
-
Similar to the Tn5-to-transposon ratio during assembly, our analysis also found that the cell density during electroporation reactions is another factor that can impact CFUs.
-
We found that concentrating a greater number of cells does not necessarily yield a higher number of viable mutants, and that there were strain-specific variations in the optimal densities.
-
When constructing the mutant libraries, CFUs are often used as a proxy for the number of unique mutants until sequencing is complete.
-
While this is often a good proxy, existing TraDIS studies report a wide range of the CFU-to-unique mutant ratio (10–90%), and it is hard to draw conclusions about what parameters might be affecting this ratio.
-
To address this, we investigated one potential factor influencing this relationship by varying the length of recovery time after electroporation.
-
We reasoned that longer recovery time would result in higher CFUs but would also result in a lower proportion of unique mutants as longer incubation might facilitate expansion of clones.
-
As expected, longer recovery time generated higher CFUs with an estimated doubling time of 0.5 h for our E. coli cohort.
-
However, contrary to what we expected, our results showed that the number of unique mutants did not decline in proportion; therefore, a longer recovery time may increase the overall yield of unique mutants perhaps by allowing more time for transposomes to mutate the genome.
-
Constructing the TraDIS mutant library is a laborious and time-consuming task.
-
Inspired by the success of TnLE-seq, a variant of Tn-seq method that uses liquid broth as a selection medium to construct a complex mutant library, we explored the possibility of adopting this approach to TraDIS.
-
Based on the side-by-side comparison of the number of unique mutants obtained from agar plates or liquid broth, our results suggest that liquid broth may be a viable option for selecting mutants for the TraDIS assay.
-
Because of the pilot nature of our study, we intentionally kept the complexity of our mutant libraries low (~ 10,000 unique mutants per condition).
-
Future studies should address whether more complex libraries can be built using the liquid broth method.
In a typical transposon mutagenesis experiment, the terms "unique insertion sites" and "unique mutants" are often used interchangeably because each unique mutant is defined by a unique insertion of the transposon into the genome. Thus, if you have identified 10,000 unique mutants, this usually implies that you have also identified 10,000 unique insertion sites. Each mutant corresponds to a single, distinct insertion site where the transposon has integrated into the genome. Here's how the terms align: * Unique Mutants: Each represents a different bacterial cell or clone that carries the transposon inserted at a distinct location in the genome. * The uniqueness of each mutant is determined by this unique insertion site. * Unique Insertion Sites: These are the specific genomic coordinates where the transposon has landed. Each site is unique to a particular mutant. In other words, the number of unique insertion sites should typically match the number of unique mutants in a well-controlled experiment, as each unique insertion site gives rise to a unique mutant. However, if there are methodological errors, such as cloning or sampling biases, or if multiple clones of the same insertion are picked up, the numbers might not perfectly align. But under ideal experimental and sequencing conditions, these two numbers should be equivalent. -
Successful TraDIS assay also relies on efficient detection of transposon-DNA junctions via deep sequencing, and in this study, we introduced a simpler library design that does not involve the ligation of custom adaptors, custom sequencing primers during a sequencing run, or custom operation protocol of the sequencer such as the need for dark cycles.
-
Despite its simplicity, our Nextera-TruSeq layout design generated a high proportion (~ 80%) of reads that correspond to transposon-DNA junctions. This is likely due to the ‘hybrid’ nature of the fragment layout because the same library preparation workflow failed if both ends of the fragments had TruSeq adaptors, as less than 0.5% of sequenced fragments corresponded to transposon-DNA junctions.
-
The suboptimal performance of the TruSeq-TruSeq fragment layout can be attributed to the mechanics of the two-stage PCR process.
-
Like previous approaches, the first round of PCR is designed to specifically amplify DNA fragments that contains transposon-DNA junctions; however, in the 2nd round of PCR, indexing primers are targeted to partial adaptor sequences that would also be present on all DNA fragments even if they do not contain junctions (Fig. 5).
-
Consequently, depending on the relative proportion of target DNA and non-specific DNA fragments at the end of the first PCR, non-specific DNA can dominate the sequencing outputs.
-
The Nextera-TruSeq hybrid layout minimizes this type of non-specific amplification during index PCR because only target junction fragments would have the partial Nextera adapter on their 5’ end.
-
Our library design is also simpler in terms of indexing, as it uses the standard TruSeq and Nextera indexing primers that are compatible with other Illumina sequencing assays.
-
Therefore, adapting the assay to a different transposon simply involves replacing the primers in the first PCR to target transposon-specific sequences, which is more cost-effective than synthesizing long all-in-one primers.
-
Although we did not directly demonstrate this versatility in the current study, our hybrid library design should be compatible using other DNA library preparation kits, such as Illumina DNA Prep kit that integrates Nextera adaptors during tagmentation.
-
In this instance, one should set up the first round of PCR using a transposon-specific forward primer with the TruSeq i5 adaptor, and a reverse primer that targets the Nextera i7 adaptor to generate TruSeq-Nextera library fragments to minimize non-specific amplification.
-
TraDIS protocols still face several challenges that require improvement.
-
Firstly, the efficiency and uniformity of transposon insertion across the genome needs enhancement to ensure comprehensive mutagenesis coverage.
-
Even with strains that are amenable to genetic manipulation, the transformation efficiency using the transposome complex or plasmid vector, along with specific growth parameters and timing, can significantly limit mutant production and the accuracy of downstream analysis in TraDIS protocols.
-
Secondly, data analysis pipelines must be optimized to identify and quantify insertional mutants accurately.
-
Lastly, the development of more scalable library construction methods is necessary to facilitate the study of diverse organisms, enabling broader applications of TraDIS in functional genomics.
Top Agricultural Biological Companies in Germany by Revenue (2021)
-
BayWa AG
- Revenue: €19.84 billion (2021)
- Description: The largest agricultural company in Germany, involved in trading agricultural products, building materials, and energy, with comprehensive agricultural services including biological products.
- Source: HitHorizons
-
AGRAVIS Raiffeisen AG
- Revenue: €7.28 billion (2021)
- Description: A significant player in the agricultural market, offering crop production, animal nutrition, and agricultural technology services, including biological solutions.
- Source: HitHorizons
-
DMK Deutsches Milchkontor GmbH
- Revenue: €5.47 billion (2021)
- Description: Germany’s largest dairy cooperative, focusing on milk production and processing, with agricultural services including biological products.
- Source: HitHorizons
-
Hauptgenossenschaft Nord AG
- Revenue: €2.47 billion (2021)
- Description: Involved in agricultural trade, offering fertilizers and crop protection products, including biologicals.
- Source: HitHorizons
-
Hochwald Milch eG
- Revenue: €1.59 billion (2021)
- Description: A dairy cooperative that deals in agricultural products and services, including biological solutions for sustainable farming.
- Source: HitHorizons
-
KWS SAAT SE & Co. KGaA
- Revenue: €1.31 billion (2021)
- Description: A leading seed company providing a wide range of agricultural seeds and biological solutions for crop protection and productivity enhancement.
- Source: HitHorizons
-
Arla Foods Deutschland GmbH
- Revenue: €1.30 billion (2021)
- Description: Focuses on dairy products but also supplies agricultural inputs, including biologicals, to its cooperative members.
- Source: HitHorizons
-
Molkerei Ammerland eG
- Revenue: €1.12 billion (2021)
- Description: Known for dairy products, Molkerei Ammerland supplies various agricultural inputs, including biological solutions.
- Source: HitHorizons
-
Ekosem-Agrar AG
- Revenue: €657.3 million (2021)
- Description: Focuses on large-scale agricultural production, mainly in the dairy sector, but also offers agricultural products including biologicals.
- Source: HitHorizons
-
Müller Fleisch GmbH
- Revenue: €574.1 million (2021)
- Description: Primarily a meat-processing company, Müller Fleisch also engages in related agricultural services.
- Source: HitHorizons
-
Bayer CropScience
- Revenue: Part of Bayer AG, which had total revenues of €44.08 billion in 2021. CropScience contributes significantly to this.
- Description: A global leader in agricultural science, Bayer CropScience offers a wide range of crop protection products, including biologicals.
- Source: Bayer AG Annual Report
-
BASF SE
- Revenue: Part of BASF Group, which had total revenues of €78.6 billion in 2021. The Agricultural Solutions segment is a significant contributor.
- Description: BASF provides a wide range of agricultural products including fungicides, insecticides, herbicides, seed treatments, and biologicals.
- Source: BASF Annual Report
These companies are leaders in the agricultural biologicals market in Germany, providing a range of products to enhance sustainable and eco-friendly farming practices.