RNA测序数据分析揭示了基因调控的复杂机制。
基因表达的转录组广泛应用于研究从 单细胞 到组织和复杂的微生物群落中的生物系统中的基因调控的研究。 RNA测序数据允许进行各种分析,以解决生物学和生物医学领域中无数的研究问题。
下面我们介绍了我们在RNA-seq数据上执行的一些最常见的分析。探索性、差异表达和通路分析大多也适用于其他高通量表达数据,如表达型芯片或蛋白质组学数据。
我们希望下面的示例能启发您欣赏RNA-测序的丰富多彩世界。
探索性基因表达分析
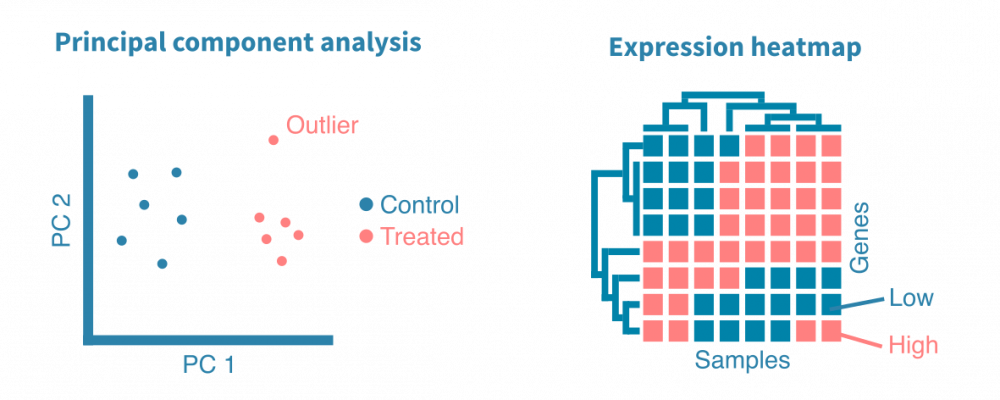
每个RNA-seq表达研究都包括探索性分析。在经过原始测序reads质量控制和基因计数之后,使用主成分分析(PCA)和表达热图来可视化数据集,以揭示其一般模式。这些可视化帮助我们回答以下问题:
- 生物学重复是否与其表达剖面相似?
- 不同样本组(例如不同组织、处理或时间点)是否形成单独的聚类?
- 是否存在异常样本?
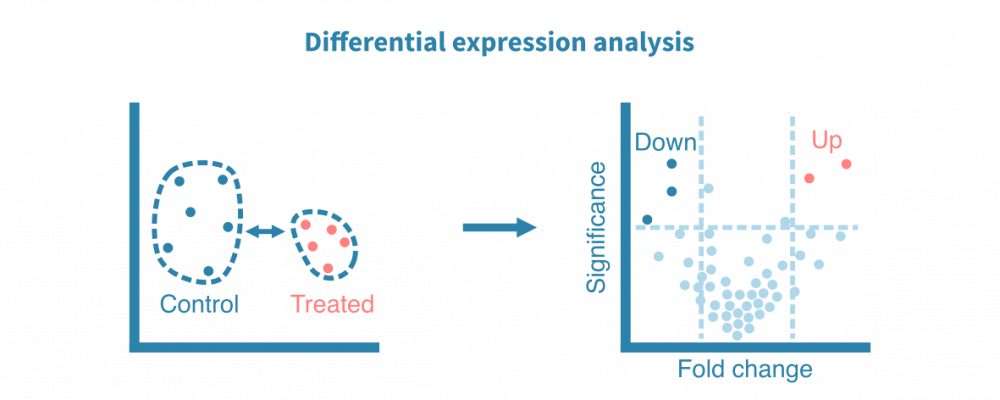
差异表达分析
差异表达分析是对两个样本组进行统计比较的过程。它会得到每个检测到的转录本的差异表达统计数据,例如折叠差异和统计显著性。这些统计数据通常使用火山图进行可视化。被发现上调或下调的基因可以进一步通过热图或箱线图进行可视化。
作为一种统计分析方法,表达研究中的这个阶段受益于生物复制品带来的统计功率。每个条件至少需要三个生物重复样本,但这仅适用于可靠检测具有相对较大表达差异的基因。通过谨慎的实验设计和足够的样本量,可以检测到更微妙的差异,并控制混杂因素。
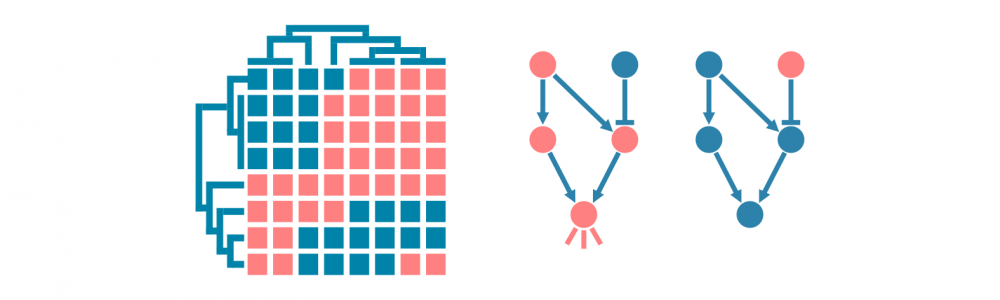
通路分析
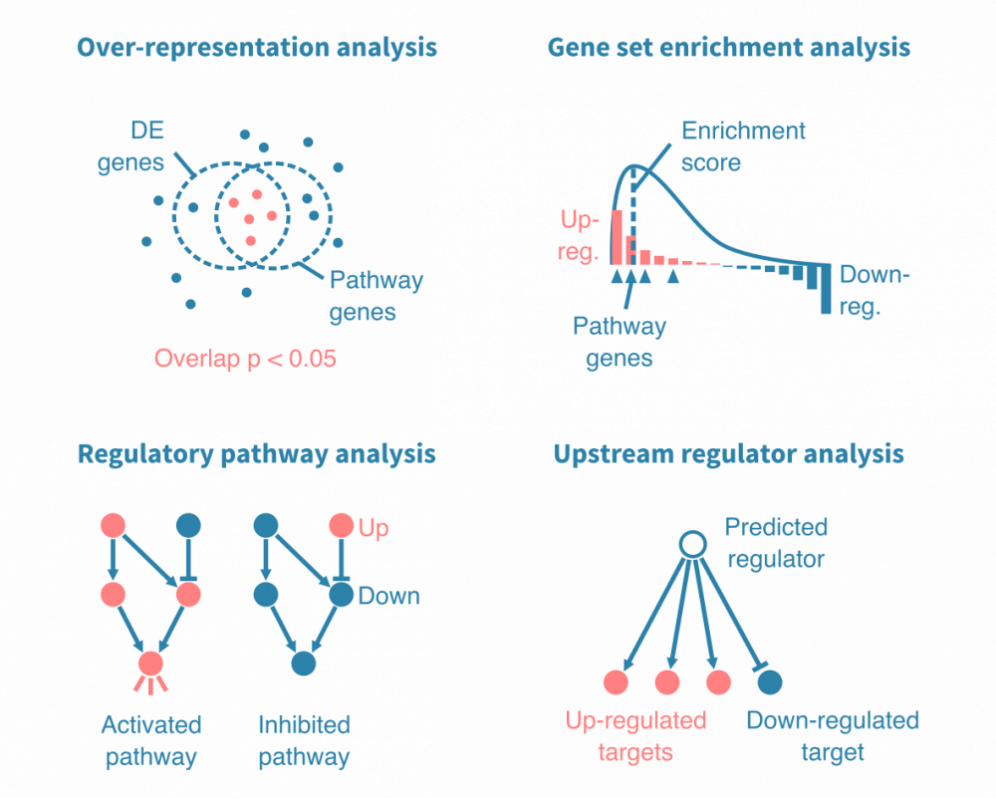
通路分析将差异表达分析中的基因放在更广泛的生物学背景中。简单的通路分析会将上调和下调基因与预定的基因列表进行统计学比较。这些列表被注释为生物学意义的术语,例如生物过程、信号通路或特定疾病。
这样的分析可能依靠过表达分析或基因集富集分析,两者都会得出具有相关统计学和注释的富集基因集列表。
更多机制通路分析依赖于基因之间实验验证的相互作用。它们不仅能够确定哪些通路由差异表达的基因表示,还能揭示通路是否被激活或抑制,以及由哪些基因激活或抑制。
更高级的通路分析我们使用Ingenuity Pathway Analysis (IPA, QIAGEN)。IPA能够进行深入分析已知和新颖的基因调控网络。
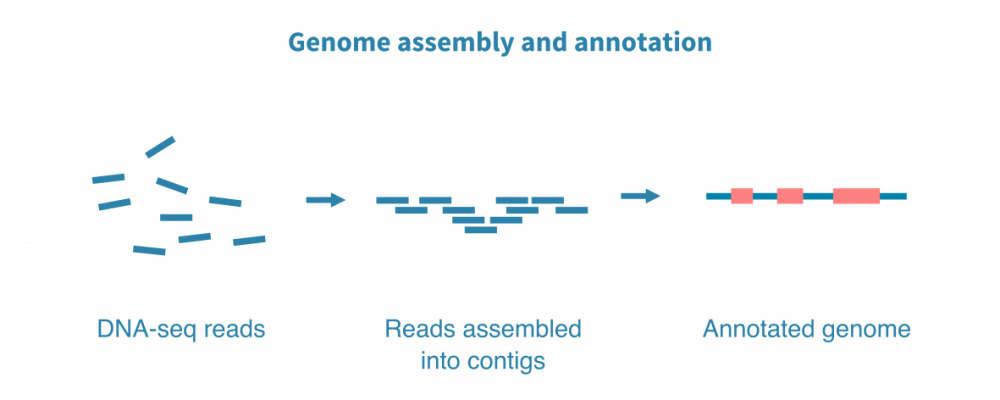
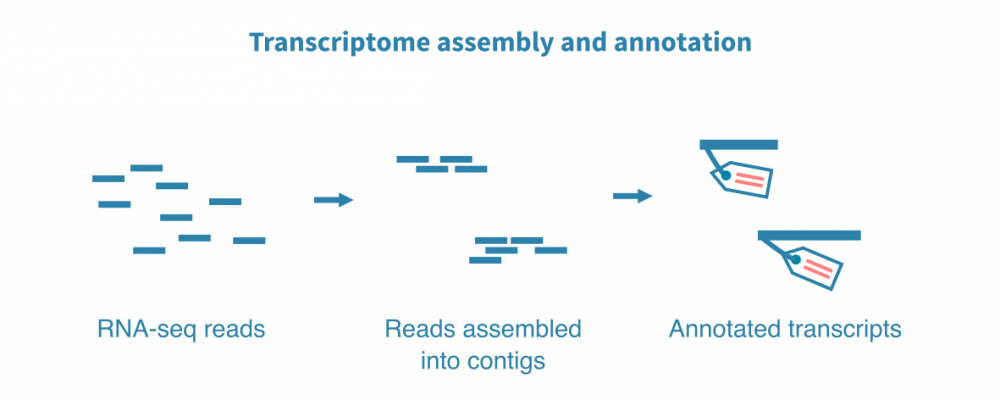
转录组组装
对于非模式生物以及具有非常动态的基因组,例如微生物,我们通常通过组装新的转录组来开始RNA测序数据分析,并使用相关物种的同源基因和计算基因预测来注释它。
一个新的参考转录组对您的进一步研究和整个研究社区的研究都是非常宝贵的资源。一旦建立了高质量的参考转录组,就可以打开大多数下游分析的大门,这些下游分析通常用于模型生物。
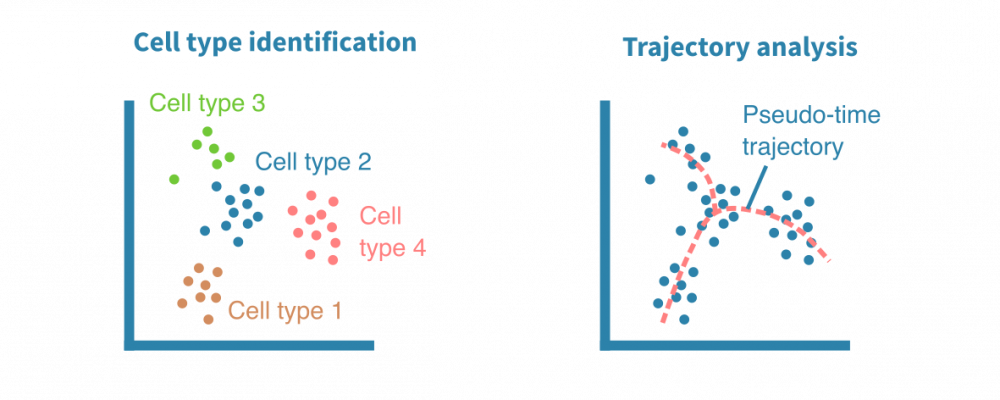
单细胞表达分析
单细胞RNA测序(scRNA-seq)实验可以以比批量RNA测序更高的规模和分辨率对细胞类型进行编目和揭示分化轨迹。
特别是用于研究复杂组织的组成和发展,scRNA-seq数据集通常包含数千个单个细胞。大多数用于分析批量RNA-seq数据的方法也可以为单细胞RNA-seq数据量身定制。
了解更多
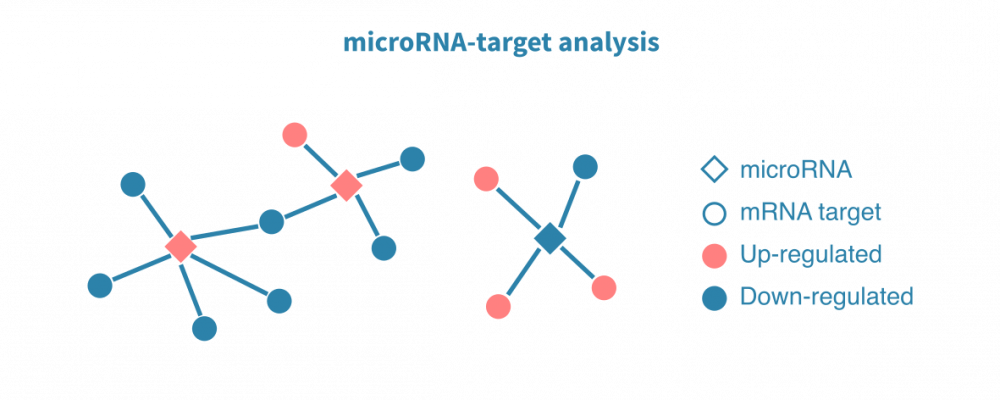
MicroRNA数据分析
小RNA测序可用于研究各种短RNA物种,尤其是microRNAs。MicroRNA-seq分析与mRNAs的分析主要类似,但路径和调节分析利用预测和/或先前验证过的microRNA靶基因。
从匹配样品中同时测序mRNA和小RNA可估计microRNAs与其靶标之间的调节关系。为了确定在给定条件下受microRNA调节的基因,可以使用argo naute CLIP-测序(和相关协议)。
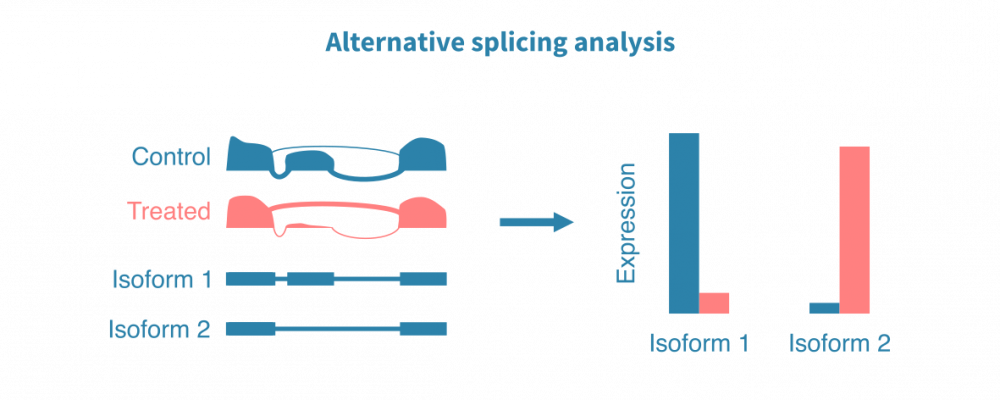
可变剪接 分析
除了在基因水平上研究表达外,RNA测序还允许进行更详细的视图:剪接变异水平的表达。可靠地鉴定可变剪接事件需要比典型的基因水平表达分析更深的测序。
根据数据的数量和质量,可变剪接分析可以集中于量化已知的、先前注释的剪接亚型的表达水平,或检测新的剪接事件。
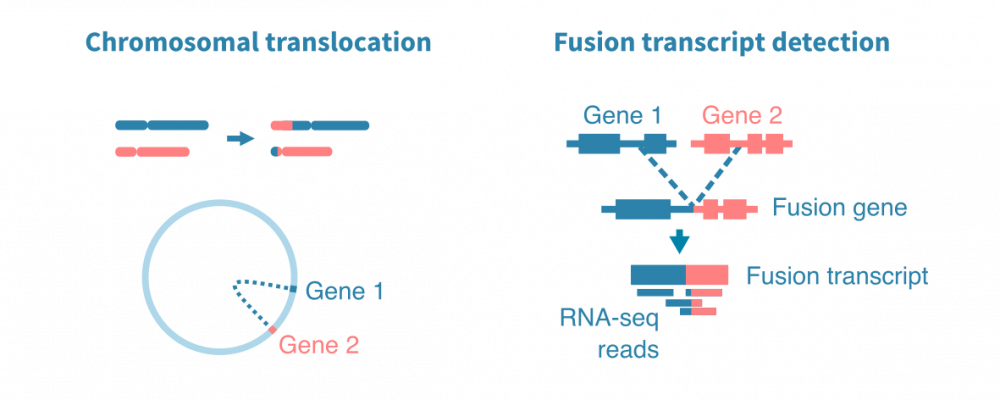
融合基因检测
在癌症中,某些结构变异已知会导致融合基因。DNA中两个分开的基因融合在一起可能导致融合转录本。反过来,融合转录本可能导致融合蛋白质具有新的、潜在的癌症驱动调控和功能组合。
可以使用识别和分析discordantly mapping RNA-seq读数或读取对的工具从RNA-seq数据中检测融合基因。
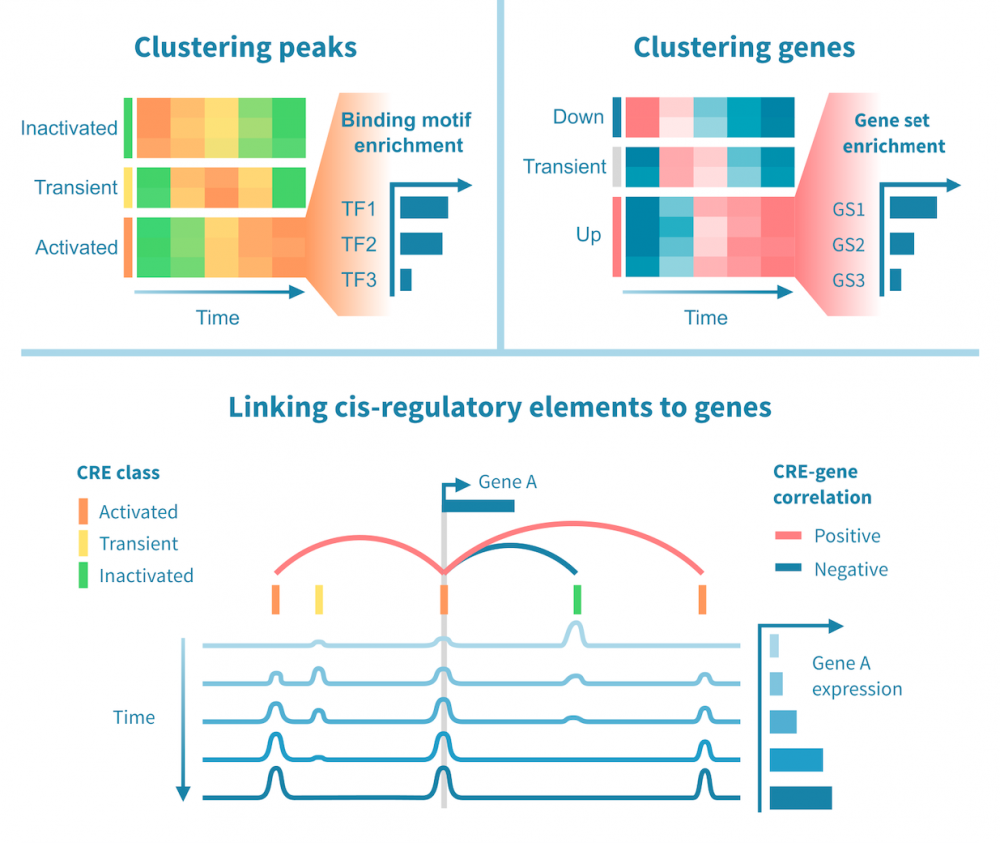
整合RNA-seq和表观基因组数据
在同一样本上进行RNA-seq和表观基因组测序(例如ChIP或ATAC-seq)可以进行整合分析,研究基因调控程序的全基因组范围。
可以在基因表达和调控元素的表观基因组状态的证据基础上,确定增强子与其靶基因以及转录因子与其靶基因之间的调控联系。
了解更多