-
run with bengal3
cd ~/DATA/Data_Denise_CalCov1 cp bacto-0.1.json ../Data_Denise_CalCov2 cp cluster.json ../Data_Denise_CalCov2 cp Snakefile ../Data_Denise_CalCov2 ln -s /home/jhuang/Tools/bacto/local . ln -s /home/jhuang/Tools/bacto/db . ln -s /home/jhuang/Tools/bacto/envs . mkdir raw_data; cd raw_data ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R1_001.fastq.gz HDM7_R1.fastq.gz ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R2_001.fastq.gz HDM7_R2.fastq.gz ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R1_001.fastq.gz HDM10_R1.fastq.gz ln -s ../Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R2_001.fastq.gz HDM10_R2.fastq.gz ln -s ../20240812_FS10003086_50_BSB09416-2831/Alignment_Imported_1/20240813_202730/Fastq/HDM1_S1_L001_R1_001.fastq.gz HDM1_R1.fastq.gz ln -s ../20240812_FS10003086_50_BSB09416-2831/Alignment_Imported_1/20240813_202730/Fastq/HDM1_S1_L001_R2_001.fastq.gz HDM1_R2.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R1_001.fastq.gz HDM7_R1.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM7_S1_L001_R2_001.fastq.gz HDM7_R2.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R1_001.fastq.gz HDM10_R1.fastq.gz ln -s ../20240913/Alignment_Imported_1/20240913_174420/Fastq/HDM10_S2_L001_R2_001.fastq.gz HDM10_R2.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM11-SF1_S1_L001_R1_001.fastq.gz HDM11-SF1_R1.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM11-SF1_S1_L001_R2_001.fastq.gz HDM11-SF1_R2.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM15-SF2_S2_L001_R1_001.fastq.gz HDM15-SF2_R1.fastq.gz ln -s ../20240919_FS10003086_61_BSB09416-2735/Alignment_Imported_1/20240920_173408/Fastq/HDM15-SF2_S2_L001_R2_001.fastq.gz HDM15-SF2_R2.fastq.gz ln -s ../20250203_FS10003086_95_BTR67811-0621/Alignment_Imported_1/20250204_154511/Fastq/HDM2-SF1_S1_L001_R1_001.fastq.gz HDM2-SF1_R1.fastq.gz ln -s ../20250203_FS10003086_95_BTR67811-0621/Alignment_Imported_1/20250204_154511/Fastq/HDM2-SF1_S1_L001_R2_001.fastq.gz HDM2-SF1_R2.fastq.gz ln -s ../20250203_FS10003086_95_BTR67811-0621/Alignment_Imported_1/20250204_154511/Fastq/HDM20-SF3_S2_L001_R1_001.fastq.gz HDM20-SF3_R1.fastq.gz ln -s ../20250203_FS10003086_95_BTR67811-0621/Alignment_Imported_1/20250204_154511/Fastq/HDM20-SF3_S2_L001_R2_001.fastq.gz HDM20-SF3_R2.fastq.gz # only activate the steps assembly and mlst in bacto-0.1.json. mamba activate /home/jhuang/miniconda3/envs/bengal3_ac3 cd ~/DATA/Data_Patricia_Sepi_7samples (bengal3_ac3) jhuang@WS-2290C:~/DATA/Data_Patricia_Sepi_7samples$ /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds -
MLST-results
cd mlst cat *.mlst.txt | sort > mlst_sorted shovill/HDM1/contigs.fa sepidermidis 5 1 1 1 2 2 1 1 shovill/HDM7/contigs.fa sepidermidis 59 2 1 1 1 2 1 1 shovill/HDM10/contigs.fa sepidermidis 59 2 1 1 1 2 1 1 shovill/HDM11-SF1/contigs.fa sepidermidis 224 19 16 19 6 3 19 10 shovill/HDM15-SF2/contigs.fa sepidermidis 87 7 1 1 2 2 1 1 shovill/HDM2-SF1/contigs.fa sepidermidis - 1 2 ~2 2 1 1 10 shovill/HDM20-SF3/contigs.fa sepidermidis - 7 1 ~2 2 4 1 1 -
(optional) mapping on assembly to calculate the coverage
#samtools depth input.bam > depth.txt #samtools depth input.bam | awk '{sum+=$3} END { print "Average coverage:",sum/NR}' #bedtools coverage -a regions.bed -b input.bam > coverage.txt #bedtools coverage -a regions.bed -b input.bam -d > coverage_per_base.txt bwa index ./shovill/HDM1/contigs.fa bwa mem ./shovill/HDM1/contigs.fa fastq/HDM1_1.fastq fastq/HDM1_2.fastq > aligned.sam samtools view -Sb aligned.sam > aligned.bam samtools sort aligned.bam -o sorted.bam samtools index sorted.bam samtools depth sorted.bam > depth.txt awk '{sum+=$3} END { print "Average coverage:",sum/NR}' depth.txt bedtools coverage -a regions.bed -b sorted.bam > coverage.txt bedtools genomecov -ibam sorted.bam -d > coverage_per_base.txt # Step 1: Calculate depth using samtools samtools depth sorted.bam > depth.txt # Step 2: Calculate average depth using awk awk '{sum+=$3; count++} END {print "Average Coverage:", sum/count}' depth.txt # Step 1: Calculate coverage with bedtools for a BED file #bedtools coverage -a regions.bed -b input.bam > coverage.txt # Step 2: Process the output with awk #awk '{ sum+=$7 } END { print "Average coverage depth:", sum/NR }' coverage.txt bwa index ./shovill/HDM7/contigs.fa bwa mem ./shovill/HDM7/contigs.fa fastq/HDM7_1.fastq fastq/HDM7_2.fastq > aligned_HDM7.sam samtools view -Sb aligned_HDM7.sam > aligned_HDM7.bam samtools sort aligned_HDM7.bam -o sorted_HDM7.bam samtools index sorted_HDM7.bam # Step 1: Calculate depth using samtools samtools depth sorted_HDM7.bam > depth_HDM7.txt # Step 2: Calculate average depth using awk awk '{sum+=$3; count++} END {print "Average Coverage:", sum/count}' depth_HDM7.txt #Average Coverage: 380.079 bwa index ./shovill/HDM10/contigs.fa bwa mem ./shovill/HDM10/contigs.fa fastq/HDM10_1.fastq fastq/HDM10_2.fastq > aligned_HDM10.sam samtools view -Sb aligned_HDM10.sam > aligned_HDM10.bam samtools sort aligned_HDM10.bam -o sorted_HDM10.bam samtools index sorted_HDM10.bam # Step 1: Calculate depth using samtools samtools depth sorted_HDM10.bam > depth_HDM10.txt # Step 2: Calculate average depth using awk awk '{sum+=$3; count++} END {print "Average Coverage:", sum/count}' depth_HDM10.txt #Average Coverage: 254.704 -
SCCmec typing using Online-Service https://cge.food.dtu.dk/services/SCCmecFinder/ and drawing with clinker
#1. -- HDM1_contigs.fa -- One SCCmec element detected. Prediction based on genes: Predicted SCCmec element: SCCmec_type_IV(2B) Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: SCCmec_type_IV(2B) 79.92% Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig ccrA2:7:81108:AB096217 100.00 1350/1350 contig00032 3770..5119 ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00032 5120..6769 IS1272:3:AM292304 99.95 1844/1843 contig00032 8611..10454 dmecR1:1:AB033763 100.00 987/987 contig00032 10443..11429 mecA:12:AB505628 100.00 2010/2010 contig00032 11526..13535 samtools faidx shovill/HDM1_contigs.fa samtools faidx shovill/HDM1_contigs.fa contig00032:1-13635 > HDM1_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM1_sub.fna #2. -- HDM7_contigs.fa -- One SCCmec element detected. Prediction based on genes: Predicted SCCmec element: SCCmec_type_IVa(2B) Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: SCCmec_type_IVa(2B) 84.24% Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig mecA:12:AB505628 100.00 2010/2010 contig00014 2800..4809 dmecR1:1:AB033763 99.90 987/987 contig00014 4906..5892 IS1272:3:AM292304 100.00 1843/1843 contig00014 5881..7723 ccrB2:3:CA05:AB063172 100.00 1629/1629 contig00014 9565..11193 ccrA2:3:CA05:AB063172 100.00 1350/1350 contig00014 11215..12564 subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 16461..17951 #IS1272:2:AB033763 91.06 1577/1585 contig00001 369260..370836 samtools faidx shovill/HDM7_contigs.fa samtools faidx shovill/HDM7_contigs.fa contig00014:2700-18051 > HDM7_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM7_sub.fna mecA dmecR1 Type I restriction enzyme HindI endonuclease subunit-like C-terminal domain-containing protein IS1272 DUF1643 domain-containing protein Pyridoxal phosphate-dependent enzyme hypothetical protein ccrB2 ccrA2 DUF927 domain-containing protein hypothetical protein ACP synthase AAA family ATPase (= subtype-IVa(2B)) #3. -- HDM10_contigs.fa -- Prediction based on genes: Predicted SCCmec element: SCCmec_type_IV(2B&5) Prediction based on homology to whole cassette: Predicted whole cassette and % template coverage: SCCmec_type_IV(2B) 84.37% Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00020 4152..5642 ccrA2:3:CA05:AB063172 100.00 1350/1350 contig00020 9539..10888 ccrB2:3:CA05:AB063172 100.00 1629/1629 contig00020 10910..12538 IS1272:3:AM292304 100.00 1843/1843 contig00020 14380..16222 dmecR1:1:AB033763 100.00 987/987 contig00020 16211..17197 mecA:12:AB505628 100.00 2010/2010 contig00020 17294..19303 #IS1272:2:AB033763 90.75 1579/1585 contig00033 2..1580 #ccrC1-allele-2:1:AB512767 90.95 1680/1680 contig00022 9836..11515 samtools faidx shovill/HDM10_contigs.fa samtools faidx shovill/HDM10_contigs.fa contig00020:4052-19403 > HDM10_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM10_sub.fna #4. -- HDM11-SF1_contigs.fa -- No SCCmec element was detected Prediction based on genes: Predicted SCCmec element: none Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: none #5. -- HDM15-SF2_contigs.fa -- SCCmec_type_IV(2B) SCCmec_type_VI(4B) Following gene complexes based on prediction of genes was detected : ccr class 2 ccr class 4 mec class B Predicted genes: Fasta header % Identity Query/HSP Length Contig Position in contig ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3823..5172 ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5173..6822 IS1272:3:AM292304 99.95 1844/1843 contig00004 8664..10507 dmecR1:1:AB033763 100.00 987/987 contig00004 10496..11482 mecA:12:AB505628 100.00 2010/2010 contig00004 11579..13588 subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 20148..22082 ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 24570..25931 ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 25928..27562 subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 52228..53718 samtools faidx shovill/HDM7_contigs.fa samtools faidx shovill/HDM7_contigs.fa contig00014:2700-18051 > HDM7_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM7_sub.fna samtools faidx shovill/HDM10_contigs.fa samtools faidx shovill/HDM10_contigs.fa contig00020:4052-19403 > HDM10_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM10_sub.fna samtools faidx shovill/HDM15-SF2_contigs.fa samtools faidx shovill/HDM15-SF2_contigs.fa contig00004:1-27662 > HDM15-SF2_sub.fna samtools faidx shovill/HDM15-SF2_contigs.fa contig00015:52128-53818 >> HDM15-SF2_sub.fna bakta --db /mnt/nvme0n1p1/bakta_db HDM15-SF2_sub.fna #6. -- HDM2-SF1_contigs.fa -- The input organism was prediced as a MRSA isolate The mecA gene was detected One SCCmec element detected. Prediction based on genes: Predicted SCCmec element: SCCmec_type_IVa(2B) Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: SCCmec_type_IVa(2B) 87.68% #7. -- HDM20-SF3_contigs.fa -- The input organism was prediced as a MSSA isolate The mecA/mecC gene was not detected No SCCmec element was detected Prediction based on genes: Predicted SCCmec element: none Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: none #- The input organism was prediced as a MSSA isolate The mecA/mecC gene was not detected No SCCmec element was detected Prediction based on genes: Predicted SCCmec element: none Prediction based on homology to whole cassette: Predicted whole cassette and %template coverage: none #END #172.104.140.19 mkdir gbff_sub mv *_sub.gbff gbff_sub cd gbff_sub for f in *_sub.gbff; do mv "$f" "${f/_sub.gbff/.gbff}"; done #mv HDM1_sub.gbff HDM1.gbff #mv HDM7_sub.gbff HDM7.gbff #mv HDM10_sub.gbff HDM10.gbff #mv HDM15-SF2_sub.gbff HDM15-SF2.gbff rm *.json clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4 cp ./gbff_HDRNA_01/clinker.png HDRNA_01_clinker.png -
Agr typing
#under env (bakta) mamba activate /home/jhuang/miniconda3/envs/bakta for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2 HDM2-SF1 HDM20-SF3; do bakta --db /mnt/nvme0n1p1/bakta_db shovill/${sample}/contigs.fa --prefix ${sample} done mv HDM1* bakta_results mv HDM2* bakta_results mv HDM7* bakta_results cd bakta_results grep "agrD" *.gbff | sort HDM1.gbff: /gene="agrD" HDM1.gbff: /gene="agrD" HDM7.gbff: /gene="agrD" HDM7.gbff: /gene="agrD" HDM10.gbff: /gene="agrD" HDM10.gbff: /gene="agrD" HDM11-SF1.gbff: /gene="agrD" HDM11-SF1.gbff: /gene="agrD" HDM15-SF2.gbff: /gene="agrD" HDM15-SF2.gbff: /gene="agrD" HDM2-SF1.gbff: /gene="agrD" HDM2-SF1.gbff: /gene="agrD" HDM20-SF3.gbff: /gene="agrD" HDM20-SF3.gbff: /gene="agrD" MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE #* The agr typing is not defined, as I have compared the sequence with the amino acid sequences of AgrD described in the paper available at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187671/. It does not correspond to Type I, Type II, or Type III. (For more details, see below). -- AgrD I -- Query 1 MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE 46 M + L +K F+ + IG + + C Y DEP+VPEELTKL E Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -- AgrD II -- Query 1 MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE 46 MNLLGGLLLKIFSNFMAVIGNASKYNPC YLDEPQVPEELTKLDE Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -- AgrD III -- Query 1 MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE 46 MNLLGGLLLK+FSNFMAVIGNA+KYNPC YLDEPQVPEELTKLDE Sbjct 926825 MNLLGGLLLKIFSNFMAVIGNASKYNPCVMYLDEPQVPEELTKLDE 926688 -
(1st optional for mixed) Prepare the tree using roary from the mixed clinical (fastq.gz) and database (fasta) genomes —-roary—-> core_gene_alignement.aln —-iqtree2—-> core_gene_alignment.aln.treefile
python ~/Scripts/run_mlst.py complete_genome_1282_ncbi.fasta sepidermidis #73 python ~/Scripts/run_mlst.py complete_genome_1282_ena.fasta sepidermidis #73 python ~/Scripts/run_mlst.py complete_genome_1282_ena_taxon_descendants.fasta sepidermidis grep -P "\tsepidermidis\t2\t" all.mlst > ST2.mlst #73 grep -v "NZ_" ST2.mlst > ST2_.mlst cut -d'.' -f1 ST2_.mlst | sort > ST2.ids cut -d'|' -f2 ST2.mlst | sort > ST2.ids diff ST2.ids ../ena_descendants_all_ST2_mlst/ST2.ids for sample in AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do cp ../genomes_by_taxid/ncbi_all_ST2_mlst/${sample}.1.fasta ./ done cd typing_complete.csv grep -P "\tsepidermidis\t2\t" typing_complete.csv > typing_ST2.csv echo -e "AP029227\nCP013943\nCP040883\nCP045648\nCP052939\nCP052940\nCP052955\nCP052959\nCP052981\nCP052984\nCP052990\nCP052994\nCP053088\nCP064461\nCP064467\nCP064473\nCP064476\nCP064480\nCP064485\nCP064499\nCP064508\nCP064516\nCP064525\nCP064554\nCP064557\nCP064560\nCP064572\nCP064574\nCP064587\nCP064590\nCP064599\nCP064603\nCP064606\nCP064609\nCP064613\nCP064616\nCP064619\nCP064624\nCP064631\nCP064637\nCP064650\nCP073821\nCP073824\nCP073827\nCP073830\nCP073835\nCP073836\nCP073840\nCP073841\nCP073844\nCP073847\nCP073850\nCP073852\nCP073855\nCP073857\nCP073859\nCP073862\nCP073863\nCP073878\nCP073900\nCP073904\nCP084008\nCP093173\nCP093179\nCP093181\nCP093193\nCP093196\nCP093198\nCP093212\nCP097512\nCP097514\nCP120425\nCP133663" > ids.txt cat ids.txt | while read id; do efetch -db nuccore -id $id -format gff3 > "${id}.gff" efetch -db nuccore -id $id -format fasta > "${id}.fasta" # Append the FASTA sequence to the GFF3 file with ##FASTA header echo "##FASTA" >> "${id}.gff" cat "${id}.fasta" >> "${id}.gff" rm "${id}.fasta" # Optionally remove the separate FASTA file done for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319; do cp ../prokka/${sample}/${sample}.gff ./ done #run Roary: roary -f roary_ST2 -e --mafft -p 100 roary_input_ST2/*.gff #roary -p 15 -f ./roary -i 95 -cd 99 -s -e -n -v roary_.fa.gb.gff CP012351.gff3 CP003084.gff3 CP023676.gff3 #-i minimum percentage identity for blastp [95] #-cd FLOAT percentage of isolates a gene must be in to be core [99] #-f STR output directory [.] #-e create a multiFASTA alignment of core genes using PRANK #-n (==--mafft) fast core gene alignment with MAFFT, use with -e #-s dont split paralogs #-v verbose output to STDOUT #The file core_gene_alignment.aln can then be used for tree construction: iqtree2 -s core_gene_alignment.aln -m MFP -bb 1000 -nt AUTO -
(2nd tree-option for purely clinical samples) Prepare the tree under bengal3_ac3 generating phylogeny_fasttree or phylogeny_raxml using snakemake for all clinical samples
#http://xgenes.com/article/article-content/372/identify-all-occurrences-of-phages-mt880870-mt880871-and-mt880872-in-s-epidermidis-st2-genomes-from-public-and-clinical-isolates/ bacto-0.1.json "fastqc": false, "taxonomic_classifier": false, "assembly": true, "typing_ariba": false, "typing_mlst": true, "pangenome": true, "variants_calling": true, "phylogeny_fasttree": true, "phylogeny_raxml": true, "recombination": true, jhuang@WS-2290C:~/DATA/Data_Patricia_Sepi_7samples$ /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds -
Preparing gene seqences (The following steps for calulating the presence-absence-matrix for predefined gene list)
(Optional online search) https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=MicrobialGenomes #Title:Refseq prokaryote representative genomes (contains refseq assembly) #Molecule Type:mixed DNA #Update date:2024/10/16 #Number of sequences:1038672 # SCCmec,agr.typing, # gyrB (House keeper), # fumC,gltA,icd (Metabolic genes), # apsS,sigB,sarA,agrC,yycG (Virulence regulators), # psm.β (Toxins), #psm.β1 --> psm.β and delete hlb # atlE,sdrG,sdrH,ebh,ebpS,tagB (Biofilm formation), #ebp-->ebpS # pgsC,sepA,dltA,fmtC,lipA,sceD,SE0760 (Immune evasion & colonization), #capC-->pgsC # esp,ecpA (Serine protease) #under ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates #samtools faidx 20250204_Gene_sequences.fasta gltA > gltA_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta Psm > psm.β_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta ebpS > ebpS_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta tagB > tagB_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta PgsC > pgsC_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta sepA > sepA_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta fmtC > fmtC_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta sceD > sceD_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta esp > esp_gold.fasta #samtools faidx 20250204_Gene_sequences.fasta ecpA > ecpA_gold.fasta cd ~/DATA/Data_Patricia_Sepi_7samples/ mkdir presence_absence cut -d$'\t' -f1-12 typing.csv > typing_f12.csv cp typing_f12.csv presence_absence/typing.csv cd presence_absence cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/gyrB_revcomp.fasta gyrB.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/fumC_revcomp.fasta fumC.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/gltA_gold.fasta gltA.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/icd_revcomp.fasta icd.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/apsS.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sigB_revcomp.fasta sigB.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sarA_revcomp.fasta sarA.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/agrC.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/yycG.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/psm.β_gold.fasta psm.β.fasta #cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/hlb.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/atlE.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sdrG.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sdrH_revcomp.fasta sdrH.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ebh_revcomp.fasta ebh.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ebpS_gold.fasta ebpS.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/tagB_gold.fasta tagB.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/pgsC_gold.fasta pgsC.fasta #early is capC cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sepA_gold.fasta sepA.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/dltA.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/fmtC_gold.fasta fmtC.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/lipA.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sceD_gold.fasta sceD.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/SE0760.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/esp_gold.fasta esp.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ecpA_gold.fasta ecpA.fasta -
Perform blastn searching for short-sequencing
# -- makeblastdb -- for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2 HDM2-SF1 HDM20-SF3; do makeblastdb -in ~/DATA/Data_Patricia_Sepi_7samples/shovill/${sample}/contigs.fa -dbtype nucl done # SCCmec,agr.typing, # gyrB(House keeper), # fumC,gltA,icd(Metabolic genes), # apsS,sigB,sarA,agrC,yycG(Virulence regulators), # psm.β (Toxins) #psm.β1 --> psm.β and delete hlb # atlE,sdrG,sdrH,ebh,ebpS,tagB(Biofilm formation), #ebp-->ebpS # pgsC,sepA,dltA,fmtC,lipA,sceD,SE0760(Immune evasion & colonization), #capC-->pgsC # esp,ecpA(Serine protease) #Note: The current -evalue is set to 1e-1; it might need to be made more stringent. for id in gyrB fumC gltA icd apsS sigB sarA agrC yycG psm.β atlE sdrG sdrH ebh ebpS tagB pgsC sepA dltA fmtC lipA sceD SE0760 esp ecpA; do echo "mkdir ${id}" echo "for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2 HDM2-SF1 HDM20-SF3; do" echo "blastn -db ~/DATA/Data_Patricia_Sepi_7samples/shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-1 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done python ~/Scripts/process_directory.py gyrB typing.csv typing_until_gyrB.csv python ~/Scripts/process_directory.py fumC typing_until_gyrB.csv typing_until_fumC.csv python ~/Scripts/process_directory.py gltA typing_until_fumC.csv typing_until_gltA.csv python ~/Scripts/process_directory.py icd typing_until_gltA.csv typing_until_icd.csv python ~/Scripts/process_directory.py apsS typing_until_icd.csv typing_until_apsS.csv python ~/Scripts/process_directory.py sigB typing_until_apsS.csv typing_until_sigB.csv python ~/Scripts/process_directory.py sarA typing_until_sigB.csv typing_until_sarA.csv python ~/Scripts/process_directory.py agrC typing_until_sarA.csv typing_until_agrC.csv python ~/Scripts/process_directory.py yycG typing_until_agrC.csv typing_until_yycG.csv python ~/Scripts/process_directory.py psm.β typing_until_yycG.csv typing_until_psm.β.csv #python ~/Scripts/process_directory.py hlb typing_until_psm.β.csv typing_until_hlb.csv python ~/Scripts/process_directory.py atlE typing_until_psm.β.csv typing_until_atlE.csv python ~/Scripts/process_directory.py sdrG typing_until_atlE.csv typing_until_sdrG.csv python ~/Scripts/process_directory.py sdrH typing_until_sdrG.csv typing_until_sdrH.csv python ~/Scripts/process_directory.py ebh typing_until_sdrH.csv typing_until_ebh.csv python ~/Scripts/process_directory.py ebpS typing_until_ebh.csv typing_until_ebpS.csv python ~/Scripts/process_directory.py tagB typing_until_ebpS.csv typing_until_tagB.csv python ~/Scripts/process_directory.py pgsC typing_until_tagB.csv typing_until_pgsC.csv python ~/Scripts/process_directory.py sepA typing_until_pgsC.csv typing_until_sepA.csv python ~/Scripts/process_directory.py dltA typing_until_sepA.csv typing_until_dltA.csv python ~/Scripts/process_directory.py fmtC typing_until_dltA.csv typing_until_fmtC.csv python ~/Scripts/process_directory.py lipA typing_until_fmtC.csv typing_until_lipA.csv python ~/Scripts/process_directory.py sceD typing_until_lipA.csv typing_until_sceD.csv python ~/Scripts/process_directory.py SE0760 typing_until_sceD.csv typing_until_SE0760.csv python ~/Scripts/process_directory.py esp typing_until_SE0760.csv typing_until_esp.csv python ~/Scripts/process_directory.py ecpA typing_until_esp.csv typing_until_ecpA.csv -
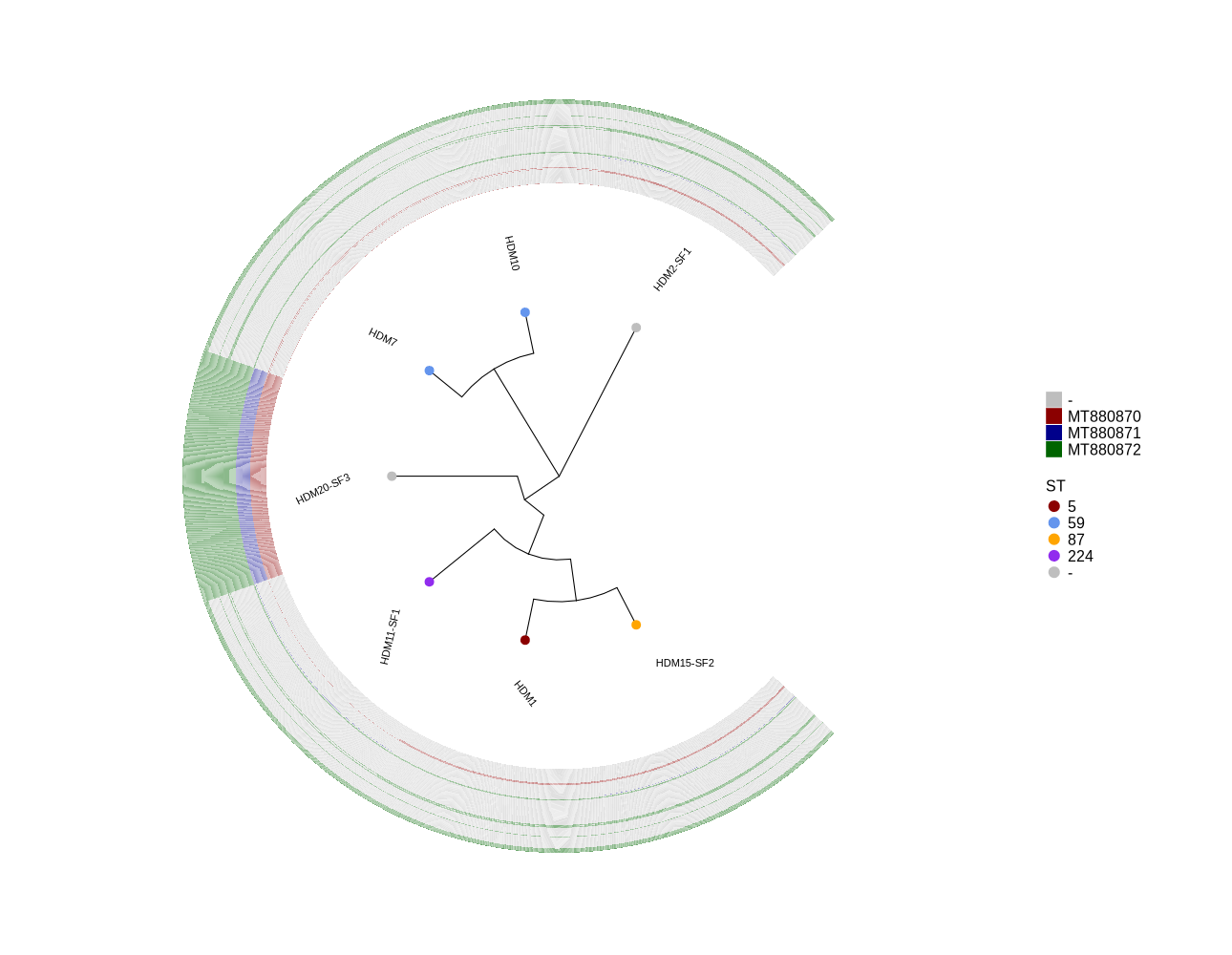
Identify all occurrences of Phages MT880870, MT880871 and MT880872 in S. epidermidis genomes
# http://xgenes.com/article/article-content/371/identify-all-occurrences-of-phage-hh1-mt880870-in-s-epidermidis-st2-genomes-from-public-and-clinical-isolates/ # http://xgenes.com/article/article-content/372/identify-all-occurrences-of-phages-mt880870-mt880871-and-mt880872-in-s-epidermidis-st2-genomes-from-public-and-clinical-isolates/ cd presence_absence cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/MT880870.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/MT880871.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/MT880872.fasta . samtools faidx MT880870.fasta samtools faidx MT880871.fasta samtools faidx MT880872.fasta for i in {1..51}; do num=$(printf "%03d" $((571 + i))) # Generates 572 to 622 id="QPB07${num}" samtools faidx MT880870.fasta "lcl|MT880870.1_cds_${id}.1_${i}" > ${id}.fasta done for i in {1..45}; do num=$((622 + i)) # Generates 623 to 667 id="QPB07${num}" samtools faidx MT880871.fasta "lcl|MT880871.1_cds_${id}.1_${i}" > ${id}.fasta done for i in {1..170}; do num=$((7667 + i)) # Generates 7668 to 7837 id="QPB0$num" samtools faidx MT880872.fasta "lcl|MT880872.1_cds_${id}.1_${i}" > ${id}.fasta done for i in {572..622}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2 HDM2-SF1 HDM20-SF3; do" echo "blastn -db ~/DATA/Data_Patricia_Sepi_7samples/shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {623..667}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2 HDM2-SF1 HDM20-SF3; do" echo "blastn -db ~/DATA/Data_Patricia_Sepi_7samples/shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {668..837}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in HDM1 HDM7 HDM10 HDM11-SF1 HDM15-SF2 HDM2-SF1 HDM20-SF3; do" echo "blastn -db ~/DATA/Data_Patricia_Sepi_7samples/shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done # -- under under the presence_absence DIR -- # - for generating a big Excel-table for showing all presence-absence info - for i in {572..837}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done #Adapted the first record as: python ~/Scripts/process_directory.py QPB07572 typing_until_ecpA.csv typing_until_QPB07572.csv #... #cp typing_until_QPB07837.csv typing_all.csv #Then save typing_all.csv as Excel table typing_all.xlsx! # - for drawing seperate plots for phages and selected genes - for i in {572..622}; do #echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done #Adapted the first record as: python ~/Scripts/process_directory.py QPB07572 typing.csv typing_until_QPB07572.csv #reprace all '+' with 'MT880870' in typing_until_QPB07622.csv sed -i 's/+/MT880870/g' typing_until_QPB07622.csv for i in {623..667}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done sed -i 's/+/MT880871/g' typing_until_QPB07667.csv for i in {668..837}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done sed -i 's/+/MT880872/g' typing_until_QPB07837.csv -
plotTreeHeatmap (draw plot for phages)
library(ggtree) library(ggplot2) library(dplyr) setwd("~/DATA/Data_Patricia_Sepi_7samples/presence_absence/") info <- read.csv("typing_until_QPB07837.csv", sep="\t") # Convert 'ST' column to factor with levels in the desired order info$ST <- factor(info$ST, levels = c("5", "59", "87", "224", "-")) info$name <- info$Isolate # Prepare the snippy.core.aln.raxml.tree by deleting the reference from snippy.core.aln.raxml.bestTree tree <- read.tree("~/DATA/Data_Patricia_Sepi_7samples/raxml-ng/snippy.core.aln.raxml.tree") #cols <- c("Public database"='purple2', "Clinical isolate"='darksalmon') #purple2 skyblue2 cols <- c("5"="darkred", "59"="cornflowerblue", "87"="orange", "224"="purple2", "-"="grey") heatmapData2 <- info %>% dplyr::select(Isolate, QPB07572, QPB07573, QPB07574, QPB07575, QPB07576, QPB07577, QPB07578, QPB07579, QPB07580, QPB07581, QPB07582, QPB07583, QPB07584, QPB07585, QPB07586, QPB07587, QPB07588, QPB07589, QPB07590, QPB07591, QPB07592, QPB07593, QPB07594, QPB07595, QPB07596, QPB07597, QPB07598, QPB07599, QPB07600, QPB07601, QPB07602, QPB07603, QPB07604, QPB07605, QPB07606, QPB07607, QPB07608, QPB07609, QPB07610, QPB07611, QPB07612, QPB07613, QPB07614, QPB07615, QPB07616, QPB07617, QPB07618, QPB07619, QPB07620, QPB07621, QPB07622, QPB07623, QPB07624, QPB07625, QPB07626, QPB07627, QPB07628, QPB07629, QPB07630, QPB07631, QPB07632, QPB07633, QPB07634, QPB07635, QPB07636, QPB07637, QPB07638, QPB07639, QPB07640, QPB07641, QPB07642, QPB07643, QPB07644, QPB07645, QPB07646, QPB07647, QPB07648, QPB07649, QPB07650, QPB07651, QPB07652, QPB07653, QPB07654, QPB07655, QPB07656, QPB07657, QPB07658, QPB07659, QPB07660, QPB07661, QPB07662, QPB07663, QPB07664, QPB07665, QPB07666, QPB07667, QPB07668, QPB07669, QPB07670, QPB07671, QPB07672, QPB07673, QPB07674, QPB07675, QPB07676, QPB07677, QPB07678, QPB07679, QPB07680, QPB07681, QPB07682, QPB07683, QPB07684, QPB07685, QPB07686, QPB07687, QPB07688, QPB07689, QPB07690, QPB07691, QPB07692, QPB07693, QPB07694, QPB07695, QPB07696, QPB07697, QPB07698, QPB07699, QPB07700, QPB07701, QPB07702, QPB07703, QPB07704, QPB07705, QPB07706, QPB07707, QPB07708, QPB07709, QPB07710, QPB07711, QPB07712, QPB07713, QPB07714, QPB07715, QPB07716, QPB07717, QPB07718, QPB07719, QPB07720, QPB07721, QPB07722, QPB07723, QPB07724, QPB07725, QPB07726, QPB07727, QPB07728, QPB07729, QPB07730, QPB07731, QPB07732, QPB07733, QPB07734, QPB07735, QPB07736, QPB07737, QPB07738, QPB07739, QPB07740, QPB07741, QPB07742, QPB07743, QPB07744, QPB07745, QPB07746, QPB07747, QPB07748, QPB07749, QPB07750, QPB07751, QPB07752, QPB07753, QPB07754, QPB07755, QPB07756, QPB07757, QPB07758, QPB07759, QPB07760, QPB07761, QPB07762, QPB07763, QPB07764, QPB07765, QPB07766, QPB07767, QPB07768, QPB07769, QPB07770, QPB07771, QPB07772, QPB07773, QPB07774, QPB07775, QPB07776, QPB07777, QPB07778, QPB07779, QPB07780, QPB07781, QPB07782, QPB07783, QPB07784, QPB07785, QPB07786, QPB07787, QPB07788, QPB07789, QPB07790, QPB07791, QPB07792, QPB07793, QPB07794, QPB07795, QPB07796, QPB07797, QPB07798, QPB07799, QPB07800, QPB07801, QPB07802, QPB07803, QPB07804, QPB07805, QPB07806, QPB07807, QPB07808, QPB07809, QPB07810, QPB07811, QPB07812, QPB07813, QPB07814, QPB07815, QPB07816, QPB07817, QPB07818, QPB07819, QPB07820, QPB07821, QPB07822, QPB07823, QPB07824, QPB07825, QPB07826, QPB07827, QPB07828, QPB07829, QPB07830, QPB07831, QPB07832, QPB07833, QPB07834, QPB07835, QPB07836, QPB07837) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn heatmap.colours <- c("darkred", "darkblue", "darkgreen", "grey") names(heatmap.colours) <- c("MT880870","MT880871","MT880872","-") #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular #geom_tippoint(aes(color=Source)) + scale_color_manual(values=cols) p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=ST), size=4) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree_phages.png", width=1260, height=1260) #svg("ggtree_phages.svg", width=1260, height=1260) p dev.off() #png("ggtree_and_gheatmap_phages.png", width=1290, height=1000) #svg("ggtree_and_gheatmap_phages.svg", width=17, height=15) #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) png("ggtree_and_gheatmap_phages.png", width=1290, height=1000) gheatmap(p, heatmapData2, width=0.5, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.0, hjust=0.5, font.size=0, offset = 3) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() -
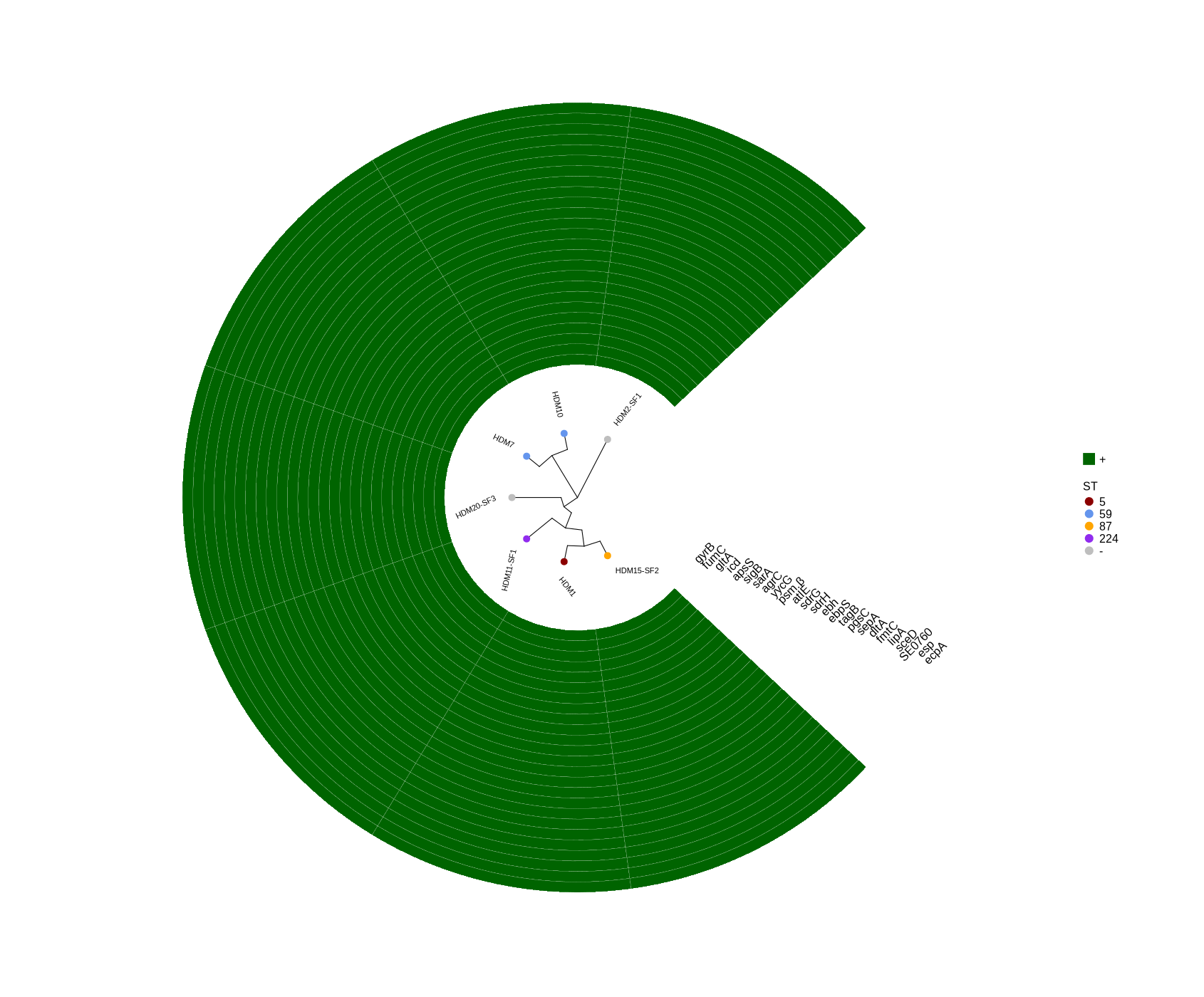
plotTreeHeatmap (draw plot for selected genes)
#FastTree -gtr -nt variants/snippy.core_without_reference.aln > plotTreeHeatmap/snippy.core.tree #cp ../Data_Holger_S.epidermidis_short/plotTreeHeatmap/typing_104.csv . #cp ../Data_Holger_S.epidermidis_short/results_HDRNA_01-20/variants/snippy.core.aln_104.tree . #Run step 4 library(ggtree) library(ggplot2) library(dplyr) setwd("~/DATA/Data_Patricia_Sepi_7samples/presence_absence/") # -- edit tree -- info <- read.csv("typing_until_ecpA.csv", sep="\t") # Convert 'ST' column to factor with levels in the desired order info$ST <- factor(info$ST, levels = c("5", "59", "87", "224", "-")) info$name <- info$Isolate tree <- read.tree("~/DATA/Data_Patricia_Sepi_7samples/raxml-ng/snippy.core.aln.raxml.tree") cols <- c("5"="darkred", "59"="cornflowerblue", "87"="orange", "224"="purple2", "-"="grey") heatmapData2 <- info %>% dplyr::select(Isolate, gyrB, fumC, gltA, icd, apsS, sigB, sarA, agrC, yycG, psm.β, atlE, sdrG, sdrH, ebh, ebpS, tagB, pgsC, sepA, dltA, fmtC, lipA, sceD, SE0760, esp, ecpA) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn heatmap.colours <- c("darkgreen", "grey") names(heatmap.colours) <- c("+","-") #heatmap.colours <- c("darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("MT880870","MT880871","MT880872","-") #heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "gold", "green","orange","pink","purple","magenta","brown", "darksalmon","chocolate4","darkkhaki", "azure3", "maroon","lightgreen", "blue","cyan", "skyblue2", "blueviolet", "darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("2","5","7","9","14", "17","23", "35","59","73", "81","86","87","89","130","190","290", "297","325", "454","487","558","766", "MT880870","MT880871","MT880872","-") #TEMP_DEACTIVATED! heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "purple", "green","cyan", "darkred", "darkblue", "darkgreen", "grey", "darkgreen", "grey") #TEMP_DEACTIVATED! names(heatmap.colours) <- c("SCCmec_type_II(2A)", "SCCmec_type_III(3A)", "SCCmec_type_III(3A) and SCCmec_type_VIII(4A)", "SCCmec_type_IV(2B)", "SCCmec_type_IV(2B&5)", "SCCmec_type_IV(2B) and SCCmec_type_VI(4B)", "SCCmec_type_IVa(2B)", "SCCmec_type_IVb(2B)", "SCCmec_type_IVg(2B)", "I", "II", "III", "none", "+","-") ## The heatmap colours should correspond to the factor levels #heatmap.colours <- #setNames(c("cornflowerblue","darkgreen","seagreen3","tan","red","green","orange","pink","brown","magenta","cyan", #"magenta","navyblue","cornflowerblue","gold","orange","darkgreen", "green", "seagreen3", #"chocolate4","brown","purple","pink", "tan","cyan","red"), #c("5", "23", "35", "69", "86", "87", "130", "224", "487", "640", "none", "P01", "P02", "P03", "P04", "P05", "P06", "P07", #"P08", "P10", "P11", "P12", "P16", "P17", "P19", "P20")) #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular #p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=ST)) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=ST), size=4) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree_selected_genes.png", width=1260, height=1260) #svg("ggtree_selected_genes.svg", width=1260, height=1260) p dev.off() #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) #svg("ggtree_and_gheatmap_mibi_selected_genes.svg", width=17, height=15) png("ggtree_and_gheatmap_selected_genes.png", width=1690, height=1400) gheatmap(p, heatmapData2, width=4, colnames_position="top", colnames_angle=45, colnames_offset_y = 0.0, hjust=0.5, font.size=5.8, offset = 3.8) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() -
(optional) Prepare the long fasta if we use only the long sequencing
ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_01_K01_conservative.fasta HDRNA_01_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_03_K01_bold_bandage.fasta HDRNA_03_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_06_K01_conservative.fasta HDRNA_06_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_07_K01_conservative.fasta HDRNA_07_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_08_K01_conservative.fasta HDRNA_08_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_12_K01_bold.fasta HDRNA_12_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_16_K01_conservative.fasta HDRNA_16_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_17_K01_conservative.fasta HDRNA_17_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_19_K01_bold.fasta HDRNA_19_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_20_K01_conservative.fasta HDRNA_20_K01.fasta -
(optional) Perform blastn searching for long-sequencing
#under ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/plotTreeHeatmap_long cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/gltA_gold.fasta gltA.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/psm.β_gold.fasta psm.β.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ebpS_gold.fasta ebpS.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/tagB_gold.fasta tagB.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/pgsC_gold.fasta pgsC.fasta #early is capC cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sepA_gold.fasta sepA.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/fmtC_gold.fasta fmtC.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sceD_gold.fasta sceD.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/esp_gold.fasta esp.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ecpA_gold.fasta ecpA.fasta # -- makeblastdb -- for sample in HDRNA_01_K01 HDRNA_03_K01 HDRNA_06_K01 HDRNA_07_K01 HDRNA_08_K01 HDRNA_12_K01 HDRNA_16_K01 HDRNA_17_K01 HDRNA_19_K01 HDRNA_20_K01; do makeblastdb -in ../assembly/${sample}.fasta -dbtype nucl done for id in gyrB fumC gltA icd apsS sigB sarA agrC yycG psm.β hlb atlE sdrG sdrH ebh ebpS tagB pgsC sepA dltA fmtC lipA sceD SE0760 esp ecpA; do #for id in gltA psm.β ebpS tagB pgsC sepA fmtC sceD esp ecpA; do echo "mkdir ${id}" echo "for sample in in HDRNA_01_K01 HDRNA_03_K01 HDRNA_06_K01 HDRNA_07_K01 HDRNA_08_K01 HDRNA_12_K01 HDRNA_16_K01 HDRNA_17_K01 HDRNA_19_K01 HDRNA_20_K01; do" echo "blastn -db ../assembly/${sample}.fasta -query ${id}.fasta -evalue 1e-40 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done python ~/Scripts/process_directory.py gyrB typing.csv typing_until_gyrB.csv python ~/Scripts/process_directory.py fumC typing_until_gyrB.csv typing_until_fumC.csv python ~/Scripts/process_directory.py gltA typing_until_fumC.csv typing_until_gltA.csv python ~/Scripts/process_directory.py icd typing_until_gltA.csv typing_until_icd.csv python ~/Scripts/process_directory.py apsS typing_until_icd.csv typing_until_apsS.csv python ~/Scripts/process_directory.py sigB typing_until_apsS.csv typing_until_sigB.csv python ~/Scripts/process_directory.py sarA typing_until_sigB.csv typing_until_sarA.csv python ~/Scripts/process_directory.py agrC typing_until_sarA.csv typing_until_agrC.csv python ~/Scripts/process_directory.py yycG typing_until_agrC.csv typing_until_yycG.csv python ~/Scripts/process_directory.py psm.β typing_until_yycG.csv typing_until_psm.β.csv python ~/Scripts/process_directory.py hlb typing_until_psm.β1.csv typing_until_hlb.csv python ~/Scripts/process_directory.py atlE typing_until_hlb.csv typing_until_atlE.csv python ~/Scripts/process_directory.py sdrG typing_until_atlE.csv typing_until_sdrG.csv python ~/Scripts/process_directory.py sdrH typing_until_sdrG.csv typing_until_sdrH.csv python ~/Scripts/process_directory.py ebh typing_until_sdrH.csv typing_until_ebh.csv python ~/Scripts/process_directory.py ebpS typing_until_ebh.csv typing_until_ebpS.csv python ~/Scripts/process_directory.py tagB typing_until_ebp.csv typing_until_tagB.csv python ~/Scripts/process_directory.py pgsC typing_until_tagB.csv typing_until_pgsC.csv python ~/Scripts/process_directory.py sepA typing_until_capC.csv typing_until_sepA.csv python ~/Scripts/process_directory.py dltA typing_until_sepA.csv typing_until_dltA.csv python ~/Scripts/process_directory.py fmtC typing_until_dltA.csv typing_until_fmtC.csv python ~/Scripts/process_directory.py lipA typing_until_fmtC.csv typing_until_lipA.csv python ~/Scripts/process_directory.py sceD typing_until_lipA.csv typing_until_sceD.csv python ~/Scripts/process_directory.py SE0760 typing_until_sceD.csv typing_until_SE0760.csv python ~/Scripts/process_directory.py esp typing_until_SE0760.csv typing_until_esp.csv python ~/Scripts/process_directory.py ecpA typing_until_esp.csv typing_until_ecpA.csv -
Report
Attached is the presence-absence table (typing_all.xlsx), which includes the presence and absence status of all selected genes (listed below), as well as all genes from the three phages MT880870, MT880871, and MT880872.
You may notice some differences in the results for the selected genes compared to the ones I sent earlier. This is because Holger provided updated sequences, correcting those I previously used for the selected genes.
Additionally, I’ve included two graphics visualizing the presence-absence table:
* Selected genes: ggtree_and_gheatmap_selected_genes.png * Phage genes (MT880870, MT880871, and MT880872): ggtree_and_gheatmap_phages.pngHere’s the list of selected genes:
* Housekeeping gene: gyrB * Metabolic genes: fumC, gltA, icd * Virulence regulators: apsS, sigB, sarA, agrC, yycG * Toxins: psmβ * Biofilm formation: atlE, sdrG, sdrH, ebh, ebpS, tagB * Immune evasion & colonization: pgsC, sepA, dltA, fmtC, lipA, sceD, SE0760 * Serine protease: esp, ecpA
Presence-Absence Table and Graphics for Selected Genes in Data_Patricia_Sepi_7samples
Leave a reply