-
Prepare input raw data
# -- Ringversuch -- ~/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20579/01_RV1_DNA_S1_R1_001.fastq.gz RV1_DNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20579/01_RV1_DNA_S1_R2_001.fastq.gz RV1_DNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20580/02_RV2_DNA_S2_R1_001.fastq.gz RV2_DNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20580/02_RV2_DNA_S2_R2_001.fastq.gz RV2_DNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20581/03_RV3_DNA_S3_R1_001.fastq.gz RV3_DNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20581/03_RV3_DNA_S3_R2_001.fastq.gz RV3_DNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20582/04_RV4_DNA_S4_R1_001.fastq.gz RV4_DNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20582/04_RV4_DNA_S4_R2_001.fastq.gz RV4_DNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20583/05_RV5_DNA_S5_R1_001.fastq.gz RV5_DNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20583/05_RV5_DNA_S5_R2_001.fastq.gz RV5_DNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20584/06_RV6_DNA_S6_R1_001.fastq.gz RV6_DNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20584/06_RV6_DNA_S6_R2_001.fastq.gz RV6_DNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20585/07_RV1_RNA_S7_R1_001.fastq.gz RV1_RNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20585/07_RV1_RNA_S7_R2_001.fastq.gz RV1_RNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20586/08_RV2_RNA_S8_R1_001.fastq.gz RV2_RNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20586/08_RV2_RNA_S8_R2_001.fastq.gz RV2_RNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20587/09_RV3_RNA_S9_R1_001.fastq.gz RV3_RNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20587/09_RV3_RNA_S9_R2_001.fastq.gz RV3_RNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20588/10_RV4_RNA_S10_R1_001.fastq.gz RV4_RNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20588/10_RV4_RNA_S10_R2_001.fastq.gz RV4_RNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20589/11_RV5_RNA_S11_R1_001.fastq.gz RV5_RNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20589/11_RV5_RNA_S11_R2_001.fastq.gz RV5_RNA_R2.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20590/12_RV6_RNA_S12_R1_001.fastq.gz RV6_RNA_R1.fastq.gz ln ../241213_VH00358_120_AAG523FM5_Ringversuch/p20590/12_RV6_RNA_S12_R2_001.fastq.gz RV6_RNA_R2.fastq.gz -
Prepare virus database and select 8 representatives for the eight given viruses from the database
# -- Download all genomes -- # enterovirus D68 # HSV-1 # HSV-2 # Influenza A H1N1 # Cytomegalovirus AD169 (The genome size of Human herpesvirus 5 (HHV-5) — more commonly known as Cytomegalovirus (CMV)) # Influenza A H3N2 # Monkeypox # HIV-1 esearch -db nucleotide -query "txid42789[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_42789_ncbi.fasta python ~/Scripts/filter_fasta.py genome_42789_ncbi.fasta complete_42789_ncbi.fasta #899 esearch -db nucleotide -query "txid10298[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_10298_ncbi.fasta python ~/Scripts/filter_fasta.py genome_10298_ncbi.fasta complete_10298_ncbi.fasta #162 esearch -db nucleotide -query "txid10310[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_10310_ncbi.fasta python ~/Scripts/filter_fasta.py genome_10310_ncbi.fasta complete_10310_ncbi.fasta #33 esearch -db nucleotide -query "txid1323429[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_1323429_ncbi.fasta python ~/Scripts/filter_fasta2.py genome_1323429_ncbi.fasta complete_1323429_ncbi.fasta #465 esearch -db nucleotide -query "txid10360[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_10360_ncbi.fasta python ~/Scripts/filter_fasta2.py genome_10360_ncbi.fasta complete_10360_ncbi.fasta #1 esearch -db nucleotide -query "txid41857[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_41857_ncbi.fasta python ~/Scripts/filter_fasta2.py genome_41857_ncbi.fasta complete_41857_ncbi.fasta #120 esearch -db nucleotide -query "txid10244[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_10244_ncbi.fasta python ~/Scripts/filter_fasta.py genome_10244_ncbi.fasta complete_10244_ncbi.fasta #2525 esearch -db nucleotide -query "txid11676[Organism:exp]" | efetch -format fasta -email j.huang@uke.de > genome_11676_ncbi.fasta python ~/Scripts/filter_fasta.py genome_11676_ncbi.fasta complete_11676_ncbi.fasta #485995-->7416 # ---- Alternatively, using ENA instead to download the genomes ---- # https://www.ebi.ac.uk/ena/browser/view/11676 (1138065 records) # #Click "Sequence" and download "Counts" (1132648) and "Taxon descendants count" (1138065) if there is enough time! Downloading time points is 09.04.2025. # python ~/Scripts/filter_fasta.py ena_11676_sequence.fasta complete_11676_ena.fasta #1138065-->???? # Virus Name NCBI TaxID # ------------------------ # Enterovirus D68 42789 >PQ895337.1 Enterovirus D68 isolate SH2024-25870 # HSV-1 (Herpes Simplex Virus 1) 10298 >PQ569920.1 Human alphaherpesvirus 1 isolate MacIntyre, complete genome # HSV-2 (Herpes Simplex Virus 2) 10310 >OM370995.1 Human alphaherpesvirus 2 strain G, complete genome samtools faidx complete_42789_ncbi.fasta PQ895337.1 > Enterovirus_D68_isolate_SH2024-25870.fasta samtools faidx complete_10298_ncbi.fasta PQ569920.1 > HSV-1_isolate_MacIntyre.fasta samtools faidx complete_10310_ncbi.fasta OM370995.1 > HSV-2_strain_G.fasta # Influenza A virus (H1N1) 1323429 # The Influenza A virus (H1N1) genome is composed of eight single-stranded negative-sense RNA segments, and the total genome size is approximately 13,500 nucleotides (13.5 kb). # Segment Gene Protein Product(s) Approx. Length (nt) # 1 PB2 Polymerase basic 2 ~2,341 # 2 PB1 Polymerase basic 1, PB1-F2 ~2,341 # 3 PA Polymerase acidic ~2,233 # 4 HA Hemagglutinin ~1,778 # 5 NP Nucleoprotein ~1,565 # 6 NA Neuraminidase ~1,413 # 7 M Matrix proteins (M1, M2) ~1,027 # 8 NS Nonstructural (NS1, NS2) ~890 # >LC662544.1 Influenza A virus (H1N1) A/PR/8/34 NEP, NS1 genes for nonstructural protein 2, nonstructural protein 1, complete cds # >LC662543.1 Influenza A virus (H1N1) A/PR/8/34 M2, M1 genes for matrix protein 2, matrix protein 1, complete cds # >LC662542.1 Influenza A virus (H1N1) A/PR/8/34 NA gene for neuraminidase, complete cds # >LC662541.1 Influenza A virus (H1N1) A/PR/8/34 NP gene for nucleoprotein, complete cds # >LC662540.1 Influenza A virus (H1N1) A/PR/8/34 HA gene for haemagglutinin, complete cds # >LC662539.1 Influenza A virus (H1N1) A/PR/8/34 PA, PA-X genes for polymerase PA, PA-X protein, complete cds # >LC662538.1 Influenza A virus (H1N1) A/PR/8/34 PB1, PB1-F2 genes for polymerase PB1, PB1-F2 protein, complete cds # >LC662537.1 Influenza A virus (H1N1) A/PR/8/34 PB2 gene for polymerase PB2, complete cds samtools faidx complete_1323429_ncbi.fasta LC662537.1 > H1N1_A-PR-8-34_PB2.fasta samtools faidx complete_1323429_ncbi.fasta LC662538.1 > H1N1_A-PR-8-34_PB1.fasta samtools faidx complete_1323429_ncbi.fasta LC662539.1 > H1N1_A-PR-8-34_PA.fasta samtools faidx complete_1323429_ncbi.fasta LC662540.1 > H1N1_A-PR-8-34_HA.fasta samtools faidx complete_1323429_ncbi.fasta LC662541.1 > H1N1_A-PR-8-34_NP.fasta samtools faidx complete_1323429_ncbi.fasta LC662542.1 > H1N1_A-PR-8-34_NA.fasta samtools faidx complete_1323429_ncbi.fasta LC662543.1 > H1N1_A-PR-8-34_M.fasta samtools faidx complete_1323429_ncbi.fasta LC662544.1 > H1N1_A-PR-8-34_NS.fasta # Human cytomegalovirus AD169 10360 # Influenza A virus (H3N2) 41857 # >LC817411.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 8, complete sequence # >LC817410.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 7, complete sequence # >LC817409.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 6, complete sequence # >LC817408.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 5, complete sequence # >LC817407.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 4, complete sequence # >LC817406.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 3, complete sequence # >LC817405.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 2, complete sequence # >LC817404.1 Influenza A virus H3N2 A_Fukushima_OR808_2023 RNA, seqment 1, complete sequence samtools faidx complete_41857_ncbi.fasta LC817404.1 > H3N2_A-Fukushima-OR808-2023_PB2.fasta samtools faidx complete_41857_ncbi.fasta LC817405.1 > H3N2_A-Fukushima-OR808-2023_PB1.fasta samtools faidx complete_41857_ncbi.fasta LC817406.1 > H3N2_A-Fukushima-OR808-2023_PA.fasta samtools faidx complete_41857_ncbi.fasta LC817407.1 > H3N2_A-Fukushima-OR808-2023_HA.fasta samtools faidx complete_41857_ncbi.fasta LC817408.1 > H3N2_A-Fukushima-OR808-2023_NP.fasta samtools faidx complete_41857_ncbi.fasta LC817409.1 > H3N2_A-Fukushima-OR808-2023_NA.fasta samtools faidx complete_41857_ncbi.fasta LC817410.1 > H3N2_A-Fukushima-OR808-2023_M.fasta samtools faidx complete_41857_ncbi.fasta LC817411.1 > H3N2_A-Fukushima-OR808-2023_NS.fasta # Monkeypox virus 10244: >OP689666.1 Monkeypox virus isolate MPXV/Germany/2022/RKI513, complete genome samtools faidx complete_10244_ncbi.fasta OP689666.1 > Monkeypox_isolate_MPXV-Germany-2022-RKI513.fasta # Human immunodeficiency virus 1 11676: >AJ866558.1 Human immunodeficiency virus 1 complete genome, isolate 01IC-PCI127 samtools faidx complete_11676_ncbi.fasta AJ866558.1 > HIV-1_isolate_01IC-PCI127.fasta # -- Selected genomes saved in the fasta-files -- # Enterovirus_D68_isolate_SH2024-25870.fasta (7391 nt) # HSV-1_isolate_MacIntyre.fasta (151817 nt) # HSV-2_strain_G.fasta (155498 nt) # H1N1_A-PR-8-34_PB2.fasta (2341 nt) # H1N1_A-PR-8-34_PB1.fasta (2341 nt) # H1N1_A-PR-8-34_PA.fasta (2233 nt) # H1N1_A-PR-8-34_HA.fasta (1775 nt) # H1N1_A-PR-8-34_NP.fasta (1565 nt) # H1N1_A-PR-8-34_NA.fasta (1413 nt) # H1N1_A-PR-8-34_M.fasta (1027 nt) # H1N1_A-PR-8-34_NS.fasta (890 nt) # Human_cytomegalovirus_strain_AD169.fasta (229354 nt) # H3N2_A-Fukushima-OR808-2023_PB2.fasta (2301 nt) # H3N2_A-Fukushima-OR808-2023_PB1.fasta (2316 nt) # H3N2_A-Fukushima-OR808-2023_PA.fasta (2208 nt) # H3N2_A-Fukushima-OR808-2023_HA.fasta (1722 nt) # H3N2_A-Fukushima-OR808-2023_NP.fasta (1536 nt) # H3N2_A-Fukushima-OR808-2023_NA.fasta (1440 nt) # H3N2_A-Fukushima-OR808-2023_M.fasta (1002 nt) # H3N2_A-Fukushima-OR808-2023_NS.fasta (865 nt) # Monkeypox_isolate_MPXV-Germany-2022-RKI513.fasta (197140 nt) # HIV-1_isolate_01IC-PCI127.fasta (9752 nt) -
(Optional) Run the first round of vrap (–virus==viruses_selected.fasta)
ln -s ~/Tools/vrap/ . mamba activate /home/jhuang/miniconda3/envs/vrap cd ~/DATA/Data_Damian/vrap_Ringversuch cat complete_10244_ncbi.fasta complete_10298_ncbi.fasta complete_10310_ncbi.fasta complete_1323429_ncbi.fasta complete_10360_ncbi.fasta complete_41857_ncbi.fasta complete_10244_ncbi.fasta complete_11676_ncbi.fasta > viruses_selected.fasta #Run vrap (first round): replace --virus to the specific taxonomy (e.g. viruses_selected.fasta) --> change virus_user_db --> specific_bacteria_user_db (vrap) for sample in RV1_DNA RV2_DNA RV3_DNA RV4_DNA RV5_DNA RV6_DNA RV1_RNA RV2_RNA RV3_RNA RV4_RNA RV5_RNA RV6_RNA; do vrap/vrap.py -1 ${sample}_R1.fastq.gz -2 ${sample}_R2.fastq.gz -o vrap_${sample} --bt2idx=/home/jhuang/REFs/genome --host=/home/jhuang/REFs/genome.fa --virus=/home/jhuang/DATA/Data_Damian/vrap_Ringversuch/viruses_selected.fasta --nt=/mnt/nvme1n1p1/blast/nt --nr=/mnt/nvme1n1p1/blast/nr -t 100 -l 200 -g done -
Run the second round of vrap (–host==${virus}.fasta)
cat Enterovirus_D68_isolate_SH2024-25870.fasta HSV-1_isolate_MacIntyre.fasta HSV-2_strain_G.fasta H1N1_A-PR-8-34_PB2.fasta H1N1_A-PR-8-34_PB1.fasta H1N1_A-PR-8-34_PA.fasta H1N1_A-PR-8-34_HA.fasta H1N1_A-PR-8-34_NP.fasta H1N1_A-PR-8-34_NA.fasta H1N1_A-PR-8-34_M.fasta H1N1_A-PR-8-34_NS.fasta Human_cytomegalovirus_strain_AD169.fasta H3N2_A-Fukushima-OR808-2023_PB2.fasta H3N2_A-Fukushima-OR808-2023_PB1.fasta H3N2_A-Fukushima-OR808-2023_PA.fasta H3N2_A-Fukushima-OR808-2023_HA.fasta H3N2_A-Fukushima-OR808-2023_NP.fasta H3N2_A-Fukushima-OR808-2023_NA.fasta H3N2_A-Fukushima-OR808-2023_M.fasta H3N2_A-Fukushima-OR808-2023_NS.fasta Monkeypox_isolate_MPXV-Germany-2022-RKI513.fasta HIV-1_isolate_01IC-PCI127.fasta > viruses_representative.fasta # Run vrap (second round): selecte some representative viruses from the generated Excel-files generated by the last step as --host (vrap) for virus in Enterovirus_D68_isolate_SH2024-25870 HSV-1_isolate_MacIntyre HSV-2_strain_G H1N1_A-PR-8-34_PB2 H1N1_A-PR-8-34_PB1 H1N1_A-PR-8-34_PA H1N1_A-PR-8-34_HA H1N1_A-PR-8-34_NP H1N1_A-PR-8-34_NA H1N1_A-PR-8-34_M H1N1_A-PR-8-34_NS Human_cytomegalovirus_strain_AD169 H3N2_A-Fukushima-OR808-2023_PB2 H3N2_A-Fukushima-OR808-2023_PB1 H3N2_A-Fukushima-OR808-2023_PA H3N2_A-Fukushima-OR808-2023_HA H3N2_A-Fukushima-OR808-2023_NP H3N2_A-Fukushima-OR808-2023_NA H3N2_A-Fukushima-OR808-2023_M H3N2_A-Fukushima-OR808-2023_NS Monkeypox_isolate_MPXV-Germany-2022-RKI513 HIV-1_isolate_01IC-PCI127; do for sample in RV1_DNA RV2_DNA RV3_DNA RV4_DNA RV5_DNA RV6_DNA RV1_RNA RV2_RNA RV3_RNA RV4_RNA RV5_RNA RV6_RNA; do vrap/vrap_until_bowtie2.py -1 ${sample}_R1.fastq.gz -2 ${sample}_R2.fastq.gz -o vrap_${sample}_on_${virus} --host /home/jhuang/DATA/Data_Damian/vrap_Ringversuch/${virus}.fasta -t 100 -l 200 --gbt2 --noblast done done -
Generate the mapping statistics for the sam-files generated from last step
“) sys.exit(1) # Parse arguments coverage_file = sys.argv[1] genome_length = int(sys.argv[2]) # Extract accession from file name (e.g., “PQ895337” from “PQ895337_coverage.txt”) file_name = os.path.basename(coverage_file) accession_match = re.match(r”([A-Z0-9]+)_coverage\.txt”, file_name) accession = accession_match.group(1) if accession_match else “” # Extract sample name from the grandparent directory of the file sample_dir = os.path.basename(os.path.dirname(os.path.dirname(coverage_file))) sample_name = re.sub(r’^vrap_’, ”, sample_dir).replace(‘_’, ‘ ‘) # Create title and filename plot_title = f”{sample_name} ({accession})” output_filename = plot_title.replace(” “, “_”) + “.png” # Load coverage data df = pd.read_csv( coverage_file, sep=”\t”, header=None, names=[“chr”, “pos”, “coverage”] ) # Create a full genome position index full_index = pd.DataFrame({‘pos’: range(1, genome_length + 1)}) # Merge coverage data with full index df_full = pd.merge(full_index, df[[‘pos’, ‘coverage’]], on=’pos’, how=’left’) df_full[‘coverage’].fillna(0, inplace=True) # Plot plt.figure(figsize=(10, 4)) plt.plot(df_full[“pos”], df_full[“coverage”], color=”blue”, linewidth=0.5) plt.xlabel(“Genomic Position”) plt.ylabel(“Coverage Depth”) plt.title(plot_title) plt.tight_layout() # Save plot to file plt.savefig(output_filename, dpi=150) print(f”Plot saved to {output_filename}”) # Optionally show the plot # plt.show()#Enterovirus_D68_isolate_SH2024-25870 #for virus in HSV-1_isolate_MacIntyre HSV-2_strain_G H1N1_A-PR-8-34_PB2 H1N1_A-PR-8-34_PB1 H1N1_A-PR-8-34_PA H1N1_A-PR-8-34_HA H1N1_A-PR-8-34_NP H1N1_A-PR-8-34_NA H1N1_A-PR-8-34_M H1N1_A-PR-8-34_NS Human_cytomegalovirus_strain_AD169; do for virus in H3N2_A-Fukushima-OR808-2023_PB2 H3N2_A-Fukushima-OR808-2023_PB1 H3N2_A-Fukushima-OR808-2023_PA H3N2_A-Fukushima-OR808-2023_HA H3N2_A-Fukushima-OR808-2023_NP H3N2_A-Fukushima-OR808-2023_NA H3N2_A-Fukushima-OR808-2023_M H3N2_A-Fukushima-OR808-2023_NS Monkeypox_isolate_MPXV-Germany-2022-RKI513 HIV-1_isolate_01IC-PCI127; do for sample in RV1_DNA RV2_DNA RV3_DNA RV4_DNA RV5_DNA RV6_DNA RV1_RNA RV2_RNA RV3_RNA RV4_RNA RV5_RNA RV6_RNA; do echo "-----${sample}_on_${virus}------" >> LOG_mapping cd vrap_${sample}_on_${virus}/bowtie # Rename and convert SAM to BAM mv mapped mapped.sam 2>> ../../LOG_mapping samtools view -S -b mapped.sam > mapped.bam 2>> ../../LOG_mapping samtools sort mapped.bam -o mapped_sorted.bam 2>> ../../LOG_mapping samtools index mapped_sorted.bam 2>> ../../LOG_mapping # Write flagstat output to log (go up two levels to write correctly) samtools flagstat mapped_sorted.bam >> ../../LOG_mapping 2>&1 cd ../.. done done #draw some plots for some representative isolates which found in the first round (see Excel-file). samtools depth -m 0 -a mapped_sorted.bam > coverage.txt grep "PQ895337.1" coverage.txt > PQ895337_coverage.txt import pandas as pd import matplotlib.pyplot as plt import sys import os import re # Check for required arguments if len(sys.argv) != 3: print("Usage: python script.py -
Report
Subject: Mapping Results and Selected Reference Genomes
Dear XXXX,
Please find below the results of the mapping analysis. For each virus you provided, I have selected a representative reference isolate, listed as follows:
Selected Reference Isolates
Enterovirus D68
• PQ895337.1 – Enterovirus D68 isolate SH2024-25870
HSV-1 (Herpes Simplex Virus 1)
• PQ569920.1 – Human alphaherpesvirus 1 isolate MacIntyre, complete genome
HSV-2 (Herpes Simplex Virus 2)
• OM370995.1 – Human alphaherpesvirus 2 strain G, complete genome
Influenza A Virus (H1N1)
• LC662537.1 – PB2 gene, complete CDS
• LC662538.1 – PB1 and PB1-F2 genes, complete CDS
• LC662539.1 – PA and PA-X genes, complete CDS
• LC662540.1 – HA gene, complete CDS
• LC662541.1 – NP gene, complete CDS
• LC662542.1 – NA gene, complete CDS
• LC662543.1 – M1 and M2 genes, complete CDS
• LC662544.1 – NS1 and NEP genes, complete CDS
Cytomegalovirus (strain AD169)
• X17403.1 – Human cytomegalovirus strain AD169, complete genome
Influenza A Virus (H3N2)
• LC817404.1 – PB2 gene
• LC817405.1 – PB1 genes
• LC817406.1 – PA genes
• LC817407.1 – HA gene
• LC817408.1 – NP gene
• LC817409.1 – NA gene
• LC817410.1 – M genes
• LC817411.1 – NS genes
Monkeypox Virus
• OP689666.1 – Isolate MPXV/Germany/2022/RKI513, complete genome
Human Immunodeficiency Virus 1 (HIV-1)
• AJ866558.1 – Isolate 01IC-PCI127, complete genomeMapping Results
We mapped paired-end reads from 12 Ringversuch project samples against the selected reference genomes.
Below are the mapping statistics for Enterovirus D68, HSV-1, HSV-2, and H1N1. Coverage plots are attached for all cases where the percentage of reads mapping to the reference genome is greater than 0.00%. Results for the remaining viruses will follow next week.
(* An asterisk indicates cases with non-zero mapping percentages.)Mapping Statistics
Enterovirus D68 (SH2024-25870):
RV1_DNA: 0 (0.00%)
RV2_DNA: 0 (0.00%)
RV3_DNA: 0 (0.00%)
RV4_DNA: 0 (0.00%)
RV5_DNA: 0 (0.00%)
RV6_DNA: 0 (0.00%)
RV1_RNA: 66 (0.00%)
RV2_RNA: 55 (0.00%)
RV3_RNA: 15 (0.00%)
RV4_RNA: 1701 (0.02%) *
RV5_RNA: 26 (0.00%)
RV6_RNA: 35 (0.00%)
HSV-1 (isolate MacIntyre):
RV1_DNA: 387 (0.02%) *
RV2_DNA: 6232 (0.26%) *
RV3_DNA: 0 (0.00%)
RV4_DNA: 1443 (0.03%) *
RV5_DNA: 2 (0.00%)
RV6_DNA: 0 (0.00%)
RV1_RNA: 6 (0.00%)
RV2_RNA: 32 (0.00%)
RV3_RNA: 4 (0.00%)
RV4_RNA: 13 (0.00%)
RV5_RNA: 4 (0.00%)
RV6_RNA: 10 (0.00%)
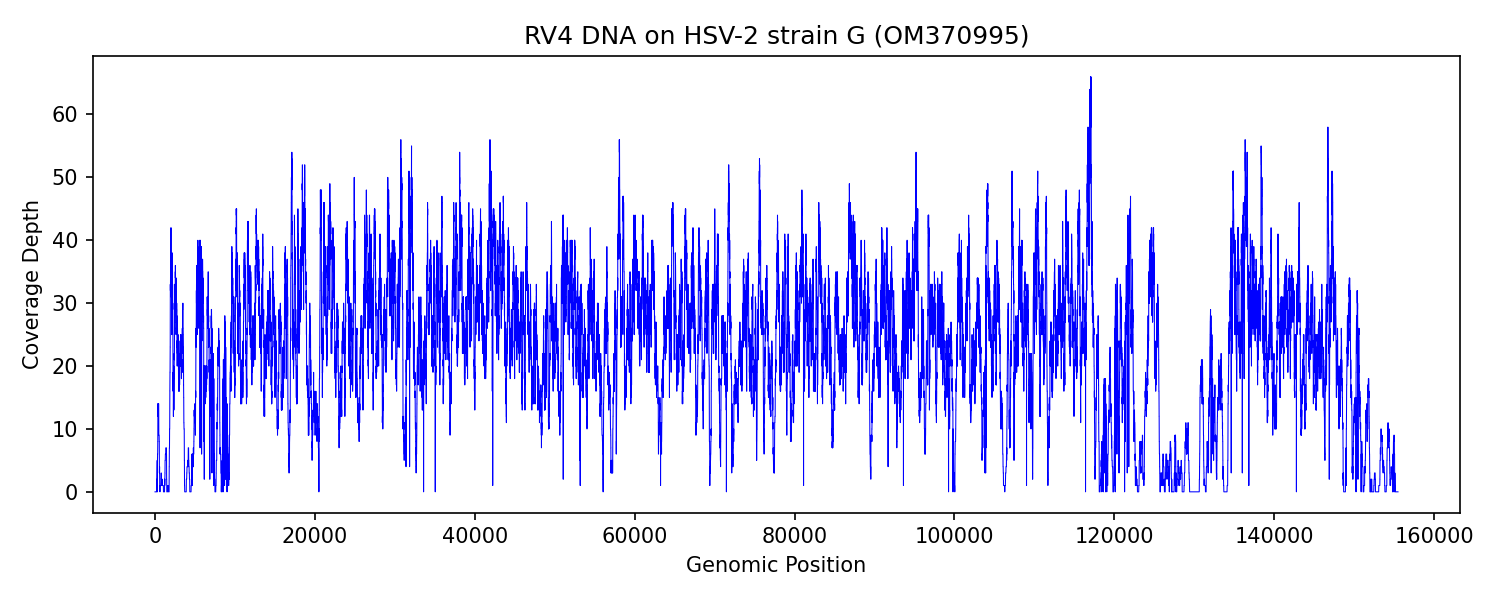
HSV-2 (strain G):
RV1_DNA: 201 (0.01%) *
RV2_DNA: 376 (0.02%) *
RV3_DNA: 0 (0.00%)
RV4_DNA: 19670 (0.46%) *
RV5_DNA: 0 (0.00%)
RV6_DNA: 0 (0.00%)
RV1_RNA: 0 (0.00%)
RV2_RNA: 3 (0.00%)
RV3_RNA: 0 (0.00%)
RV4_RNA: 25 (0.00%)
RV5_RNA: 5 (0.00%)
RV6_RNA: 24 (0.00%)
Influenza A Virus (H1N1, A/PR/8/34):
RV1_DNA: 0 (0.00%)
RV2_DNA: 0 (0.00%)
RV3_DNA: 0 (0.00%)
RV4_DNA: 0 (0.00%)
RV5_DNA: 0 (0.00%)
RV6_DNA: 0 (0.00%)
RV1_RNA: 0 (0.00%)
RV2_RNA: 0 (0.00%)
RV3_RNA: 0 (0.00%)
RV4_RNA: 13 + 354 (0.00%)
RV5_RNA: 0 (0.00%)
RV6_RNA: 0 (0.00%)