-
Download the kmerfinder database: https://www.genomicepidemiology.org/services/ –> https://cge.food.dtu.dk/services/KmerFinder/ –> https://cge.food.dtu.dk/services/KmerFinder/etc/kmerfinder_db.tar.gz
# Download 20190108_kmerfinder_stable_dirs.tar.gz from https://zenodo.org/records/13447056 -
Run nextflow bacass
#–kmerfinderdb /path/to/kmerfinder/bacteria.tar.gz #–kmerfinderdb /mnt/nvme1n1p1/REFs/kmerfinder_db.tar.gz #–kmerfinderdb /mnt/nvme1n1p1/REFs/20190108_kmerfinder_stable_dirs.tar.gz nextflow run nf-core/bacass -r 2.5.0 -profile docker \ –input samplesheet.tsv \ –outdir bacass_out \ –assembly_type long \ –kraken2db /mnt/nvme1n1p1/REFs/k2_standard_08_GB_20251015.tar.gz \ –kmerfinderdb /mnt/nvme1n1p1/REFs/kmerfinder/bacteria/ \ -resume
-
KmerFinder summary
From the KmerFinder summary, the top hit is Gibbsiella quercinecans (strain FRB97; NZ_CP014136.1) with much higher score and coverage than the second hit (which is low coverage). So it’s fair to write:
“KmerFinder indicates the isolate is most consistent with Gibbsiella quercinecans.”
…but for a species call (especially for publication), you should confirm with ANI (or a genome taxonomy tool), because k-mer hits alone aren’t always definitive.
-
Using https://www.bv-brc.org/app/ComprehensiveGenomeAnalysis to annotate the genome using scaffolded results from bacass. ComprehensiveGenomeAnalysis provides comprehensive overview of the data.
-
Generate the Table 1 Summary of sequence data and genome features under the env gunc_env
activate the env that has openpyxl
mamba activate gunc_env mamba install -n gunc_env -c conda-forge openpyxl -y mamba deactivate
STEP_1
ENV_NAME=gunc_env AUTO_INSTALL=1 THREADS=32 ~/Scripts/make_table1_GE11174.sh
STEP_2
python export_table1_stats_to_excel_py36_compat.py \ –workdir table1_GE11174_work \ –out Comprehensive_GE11174.xlsx \ –max-rows 200000 \ –sample GE11174
STEP_1+2
ENV_NAME=gunc_env AUTO_INSTALL=1 THREADS=32 ~/Scripts/make_table1_with_excel.sh
#For the items “Total number of reads sequenced” and “Mean read length (bp)” pigz -dc GE11174.rawreads.fastq.gz | awk ‘END{print NR/4}’ seqkit stats GE11174.rawreads.fastq.gz
-
Antimicrobial resistance gene profiling and Resistome and Virulence Profiling with Abricate and RGI (Reisistance Gene Identifier)
Table 4. Specialty Genes
Source Genes
NDARO 1
Antibiotic Resistance CARD 15
Antibiotic Resistance PATRIC 55
Drug Target TTD 38
Metal Resistance BacMet 29
Transporter TCDB 250
Virulance factor VFDB 33
https://www.genomicepidemiology.org/services/
https://genepi.dk/
conda activate /home/jhuang/miniconda3/envs/bengal3_ac3 abricate –list #DATABASE SEQUENCES DBTYPE DATE #vfdb 2597 nucl 2025-Oct-22 #resfinder 3077 nucl 2025-Oct-22 #argannot 2223 nucl 2025-Oct-22 #ecoh 597 nucl 2025-Oct-22 #megares 6635 nucl 2025-Oct-22 #card 2631 nucl 2025-Oct-22 #ecoli_vf 2701 nucl 2025-Oct-22 #plasmidfinder 460 nucl 2025-Oct-22 #ncbi 5386 nucl 2025-Oct-22 abricate-get_db –list #Choices: argannot bacmet2 card ecoh ecoli_vf megares ncbi plasmidfinder resfinder vfdb victors (default ”).
CARD
abricate-get_db –db card
MEGARes (automatically install, if error try MANUAL install as below)
abricate-get_db –db megares
MANUAL install
wget -O megares_database_v3.00.fasta \ “https://www.meglab.org/downloads/megares_v3.00/megares_database_v3.00.fasta” #wget -O megares_drugs_database_v3.00.fasta \ “https://www.meglab.org/downloads/megares_v3.00/megares_drugs_database_v3.00.fasta“
1) Define dbdir (adjust to your env; from your logs it’s inside the conda env)
DBDIR=/home/jhuang/miniconda3/envs/bengal3_ac3/db
2) Create a custom db folder for MEGARes v3.0
mkdir -p ${DBDIR}/megares_v3.0
3) Copy the downloaded MEGARes v3.0 nucleotide FASTA to ‘sequences’
cp megares_database_v3.00.fasta ${DBDIR}/megares_v3.0/sequences
4) Build ABRicate indices
abricate –setupdb
#abricate-get_db –setupdb abricate –list | egrep ‘card|megares’ abricate –list | grep -i megares
chmod +x run_resistome_virulome.sh ASM=GE11174.fasta SAMPLE=GE11174 THREADS=32 ./run_resistome_virulome.sh
chmod +x run_resistome_virulome_dedup.sh ENV_NAME=/home/jhuang/miniconda3/envs/bengal3_ac3 ASM=GE11174.fasta SAMPLE=GE11174 THREADS=32 ./run_resistome_virulome_dedup.sh ENV_NAME=/home/jhuang/miniconda3/envs/bengal3_ac3 ASM=./vrap_HF/spades/scaffolds.fasta SAMPLE=HF THREADS=32 ~/Scripts/run_resistome_virulome_dedup.sh ENV_NAME=/home/jhuang/miniconda3/envs/bengal3_ac3 ASM=GE11174.fasta SAMPLE=GE11174 MINID=80 MINCOV=60 ./run_resistome_virulome_dedup.sh
grep -vc ‘^#’ resistome_virulence_GE11174/raw/GE11174.megares.tab grep -vc ‘^#’ resistome_virulence_GE11174/raw/GE11174.card.tab grep -vc ‘^#’ resistome_virulence_GE11174/raw/GE11174.resfinder.tab grep -vc ‘^#’ resistome_virulence_GE11174/raw/GE11174.vfdb.tab
grep -v ‘^#’ resistome_virulence_GE11174/raw/GE11174.megares.tab | grep -v ‘^[[:space:]]$’ | head -n 3 grep -v ‘^#’ resistome_virulence_GE11174/raw/GE11174.card.tab | grep -v ‘^[[:space:]]$’ | head -n 3 grep -v ‘^#’ resistome_virulence_GE11174/raw/GE11174.resfinder.tab | grep -v ‘^[[:space:]]$’ | head -n 3 grep -v ‘^#’ resistome_virulence_GE11174/raw/GE11174.vfdb.tab | grep -v ‘^[[:space:]]$’ | head -n 3
chmod +x make_dedup_tables_from_abricate.sh OUTDIR=resistome_virulence_GE11174 SAMPLE=GE11174 ./make_dedup_tables_from_abricate.sh
chmod +x run_abricate_resistome_virulome_one_per_gene.sh ENV_NAME=/home/jhuang/miniconda3/envs/bengal3_ac3 \ ASM=GE11174.fasta \ SAMPLE=GE11174 \ OUTDIR=resistome_virulence_GE11174 \ MINID=80 MINCOV=60 \ THREADS=32 \ ./run_abricate_resistome_virulome_one_per_gene.sh
#ABRicate thresholds: MINID=70 MINCOV=50 Database Hit_lines File MEGARes 35 resistome_virulence_GE11174/raw/GE11174.megares.tab CARD 28 resistome_virulence_GE11174/raw/GE11174.card.tab ResFinder 2 resistome_virulence_GE11174/raw/GE11174.resfinder.tab VFDB 18 resistome_virulence_GE11174/raw/GE11174.vfdb.tab
#ABRicate thresholds: MINID=80 MINCOV=60 Database Hit_lines File MEGARes 3 resistome_virulence_GE11174/raw/GE11174.megares.tab CARD 1 resistome_virulence_GE11174/raw/GE11174.card.tab ResFinder 0 resistome_virulence_GE11174/raw/GE11174.resfinder.tab VFDB 0 resistome_virulence_GE11174/raw/GE11174.vfdb.tab
python merge_amr_sources_by_gene.py python export_resistome_virulence_to_excel_py36.py \ –workdir resistome_virulence_GE11174 \ –sample GE11174 \ –out Resistome_Virulence_GE11174.xlsx
Methods sentence (AMR + virulence)
AMR genes were identified by screening the genome assembly with ABRicate against the MEGARes and ResFinder databases, using minimum identity and coverage thresholds of X% and Y%, respectively. CARD-based AMR determinants were additionally predicted using RGI (Resistance Gene Identifier) to leverage curated resistance models. Virulence factors were screened using ABRicate against VFDB under the same thresholds.
Replace X/Y with your actual values (e.g., 90/60) or state “default parameters” if you truly used defaults.
Table 2 caption (AMR)
Table 2. AMR gene profiling of the genome assembly. Hits were detected using ABRicate (MEGARes and ResFinder) and RGI (CARD). The presence of AMR-associated genes does not necessarily imply phenotypic resistance, which may depend on allele type, genomic context/expression, and/or SNP-mediated mechanisms; accordingly, phenotype predictions (e.g., ResFinder) should be interpreted cautiously.
Table 3 caption (virulence)
Table 3. Virulence factor profiling of the genome assembly based on ABRicate screening against VFDB, reporting loci with sequence identity and coverage above the specified thresholds.
-
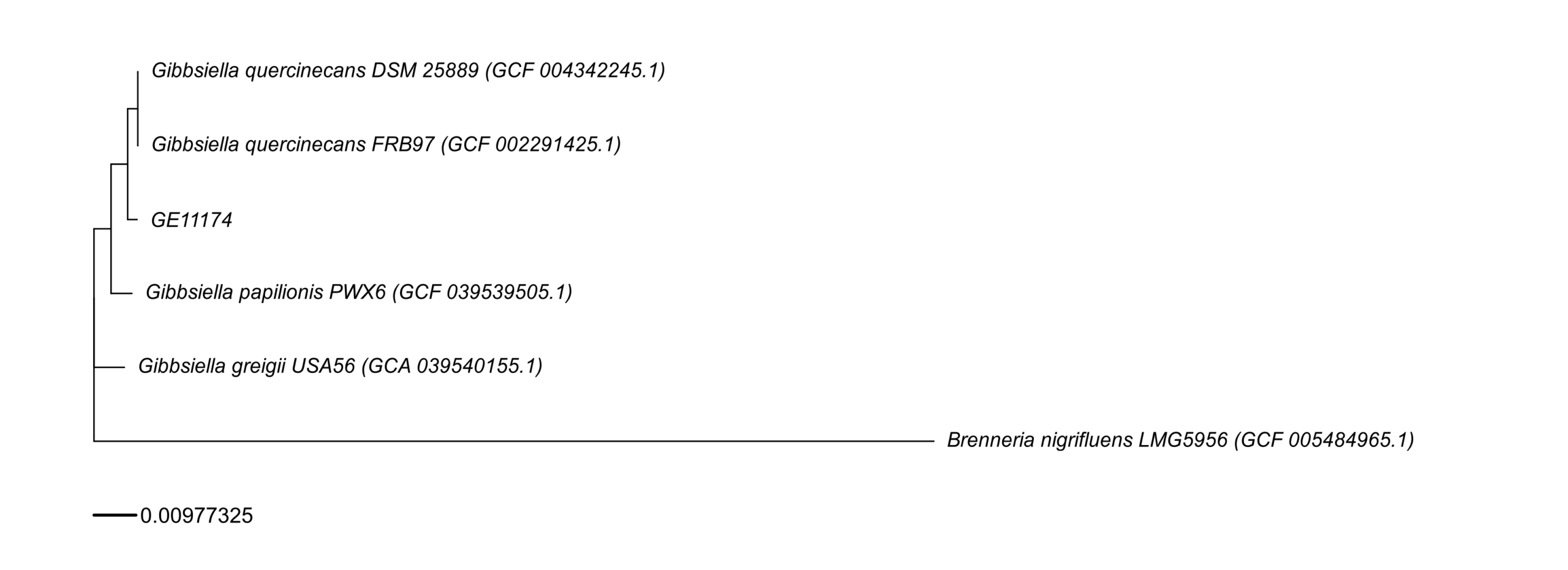
Generate phylogenetic tree
export NCBI_EMAIL=”j.huang@uke.de” ./resolve_best_assemblies_entrez.py targets.tsv resolved_accessions.tsv
Note the env bengal3_ac3 don’t have the following r-package, using r_env for the plot-step!
#mamba install -y -c conda-forge -c bioconda r-aplot bioconductor-ggtree r-ape r-ggplot2 r-dplyr r-readr
chmod +x build_wgs_tree_fig3B.sh export ENV_NAME=/home/jhuang/miniconda3/envs/bengal3_ac3 export NCBI_EMAIL=”j.huang@uke.de” ./build_wgs_tree_fig3B.sh
-
DEBUG (recommended): remove one genome and rerun Roary → RAxML; Example: drop GCF_047901425.1 (change to the other one if you prefer).
1.1) remove from inputs so Roary cannot include it
rm -f work_wgs_tree/gffs/GCF_047901425.1.gff rm -f work_wgs_tree/fastas/GCF_047901425.1.fna rm -rf work_wgs_tree/prokka/GCF_047901425.1 rm -rf work_wgs_tree/genomes_ncbi/GCF_047901425.1 #optional
1.2) remove from accession list so it won’t come back
awk -F’\t’ ‘NR==1 || $2!=”GCF_047901425.1″‘ work_wgs_tree/meta/accessions.tsv > work_wgs_tree/meta/accessions.tsv.tmp \ && mv work_wgs_tree/meta/accessions.tsv.tmp work_wgs_tree/meta/accessions.tsv
2.1) remove from inputs so Roary cannot include it
rm -f work_wgs_tree/gffs/GCA_032062225.1.gff rm -f work_wgs_tree/fastas/GCA_032062225.1.fna rm -rf work_wgs_tree/prokka/GCA_032062225.1 rm -rf work_wgs_tree/genomes_ncbi/GCA_032062225.1 #optional
2.2) remove from accession list so it won’t come back
awk -F’\t’ ‘NR==1 || $2!=”GCA_032062225.1″‘ work_wgs_tree/meta/accessions.tsv > work_wgs_tree/meta/accessions.tsv.tmp \ && mv work_wgs_tree/meta/accessions.tsv.tmp work_wgs_tree/meta/accessions.tsv
3) delete old roary runs (so you don’t accidentally reuse old alignment)
rm -rf work_wgstree/roary*
4) rerun Roary (fresh output dir)
mkdir -p work_wgs_tree/logs ROARY_OUT=”work_wgstree/roary$(date +%s)” roary -e –mafft -p 8 -cd 95 -i 95 \ -f “$ROARY_OUT” \ work_wgs_tree/gffs/*.gff \
work_wgs_tree/logs/roary_rerun.stdout.txt \ 2> work_wgs_tree/logs/roary_rerun.stderr.txt
5) point meta file to new core alignment (absolute path)
echo “$(readlink -f “$ROARY_OUT/core_gene_alignment.aln”)” > work_wgs_tree/meta/core_alignment_path.txt
6) rerun RAxML-NG
rm -rf work_wgs_tree/raxmlng mkdir work_wgs_tree/raxmlng/ raxml-ng –all \ –msa “$(cat work_wgs_tree/meta/core_alignment_path.txt)” \ –model GTR+G \ –bs-trees 1000 \ –threads 8 \ –prefix work_wgs_tree/raxmlng/core
7) Run this to regenerate labels.tsv
bash regenerate_labels.sh
8) Manual correct the display name in vim work_wgs_tree/plot/labels.tsv
#Gibbsiella greigii USA56 #Gibbsiella papilionis PWX6 #Gibbsiella quercinecans strain FRB97 #Brenneria nigrifluens LMG 5956
9) Rerun only the plot step:
Rscript work_wgs_tree/plot/plot_tree.R \ work_wgs_tree/raxmlng/core.raxml.support \ work_wgs_tree/plot/labels.tsv \ 6 \ work_wgs_tree/plot/core_tree_like_fig3B.pdf \ work_wgs_tree/plot/core_tree_like_fig3B.png
-
fastaANI and busco explanations
find . -name “*.fna” #./work_wgs_tree/fastas/GCF_004342245.1.fna GCF_004342245.1 Gibbsiella quercinecans DSM 25889 (GCF_004342245.1) #./work_wgs_tree/fastas/GCF_039539505.1.fna GCF_039539505.1 Gibbsiella papilionis PWX6 (GCF_039539505.1) #./work_wgs_tree/fastas/GCF_005484965.1.fna GCF_005484965.1 Brenneria nigrifluens LMG5956 (GCF_005484965.1) #./work_wgs_tree/fastas/GCA_039540155.1.fna GCA_039540155.1 Gibbsiella greigii USA56 (GCA_039540155.1) #./work_wgs_tree/fastas/GE11174.fna #./work_wgs_tree/fastas/GCF_002291425.1.fna GCF_002291425.1 Gibbsiella quercinecans FRB97 (GCF_002291425.1)
mamba activate /home/jhuang/miniconda3/envs/bengal3_ac3 fastANI \ -q GE11174.fasta \ -r ./work_wgs_tree/fastas/GCF_004342245.1.fna \ -o fastANI_out_Gibbsiella_quercinecans_DSM_25889.txt fastANI \ -q GE11174.fasta \ -r ./work_wgs_tree/fastas/GCF_039539505.1.fna \ -o fastANI_out_Gibbsiella_papilionis_PWX6.txt fastANI \ -q GE11174.fasta \ -r ./work_wgs_tree/fastas/GCF_005484965.1.fna \ -o fastANI_out_Brenneria_nigrifluens_LMG5956.txt fastANI \ -q GE11174.fasta \ -r ./work_wgs_tree/fastas/GCA_039540155.1.fna \ -o fastANI_out_Gibbsiella_greigii_USA56.txt fastANI \ -q GE11174.fasta \ -r ./work_wgs_tree/fastas/GCF_002291425.1.fna \ -o fastANI_out_Gibbsiella_quercinecans_FRB97.txt cat fastANIout*.txt > fastANI_out.txt
GE11174.fasta ./work_wgs_tree/fastas/GCF_005484965.1.fna 79.1194 597 1890 GE11174.fasta ./work_wgs_tree/fastas/GCA_039540155.1.fna 95.9589 1547 1890 GE11174.fasta ./work_wgs_tree/fastas/GCF_039539505.1.fna 97.2172 1588 1890 GE11174.fasta work_wgs_tree/fastas/GCF_004342245.1.fna 98.0889 1599 1890 GE11174.fasta ./work_wgs_tree/fastas/GCF_002291425.1.fna 98.1285 1622 1890 #在细菌基因组比较里,一个常用经验阈值是:
- ANI ≥ 95–96%:通常认为属于同一物种(species)的范围
- 你这里 97.09% → 很大概率表示 An6 与 HITLi7 属于同一物种,但可能不是同一株(strain),因为还存在一定差异。 是否“同一菌株”通常还要结合:
- 核心基因 SNP 距离、cgMLST
- 组装质量/污染
- 覆盖率是否足够高
#BUSCO 结果的快速解读(顺便一句). The results have been already packaged in the Table 1.
- Complete 99.2%,Missing 0.0%:说明你的组装非常完整(对细菌来说很优秀)
- Duplicated 0.0%:重复拷贝不高,污染/混样风险更低
- Scaffolds 80、N50 ~169 kb:碎片化还可以,但总体质量足以做 ANI/物种鉴定
-
fastANI explanation
From your tree and the fastANI table, GE11174 is clearly inside the Gibbsiella quercinecans clade, and far from the outgroup (Brenneria nigrifluens). The ANI values quantify that same pattern.
1) Outgroup check (sanity)
-
GE11174 vs Brenneria nigrifluens (GCF_005484965.1): ANI 79.12% (597/1890 fragments)
- 79% ANI is way below any species boundary → not the same genus/species.
- On the tree, Brenneria sits on a long branch as the outgroup, consistent with this deep divergence.
- The relatively low matched fragments (597/1890) also fits “distant genomes” (fewer orthologous regions pass the ANI mapping filters).
2) Species-level placement of GE11174
A common rule of thumb you quoted is correct: ANI ≥ 95–96% ⇒ same species.
Compare GE11174 to the Gibbsiella references:
-
vs GCA_039540155.1 (Gibbsiella greigii USA56): 95.96% (1547/1890)
- Right at the boundary. This suggests “close but could be different species” or “taxonomy/labels may not reflect true species boundaries” depending on how those genomes are annotated.
- On the tree, G. greigii is outside the quercinecans group but not hugely far, which matches “borderline ANI”.
-
vs GCF_039539505.1 (Gibbsiella papilionis PWX6): 97.22% (1588/1890)
-
vs GCF_004342245.1 (G. quercinecans DSM 25889): 98.09% (1599/1890)
-
vs GCF_002291425.1 (G. quercinecans FRB97): 98.13% (1622/1890)
These are all comfortably above 96%, especially the two quercinecans genomes (~98.1%). That strongly supports:
GE11174 belongs to the same species as Gibbsiella quercinecans (and is closer to quercinecans references than to greigii).
This is exactly what your tree shows: GE11174 clusters in the quercinecans group, not with the outgroup.
3) Closest reference and “same strain?” question
GE11174’s closest by ANI in your list is:
- FRB97 (GCF_002291425.1): 98.1285%
- DSM 25889 (GCF_004342245.1): 98.0889%
- Next: PWX6 97.2172%
These differences are small, but 98.1% ANI is not “same strain” evidence by itself. Within a species, different strains commonly sit anywhere from ~96–99.9% ANI depending on diversity. To claim “same strain / very recent transmission,” people usually look for much tighter genome-wide similarity:
- core-genome SNP distance (often single digits to tens, depending on organism and context)
- cgMLST allele differences
- recombination filtering (if relevant)
- assembly QC/contamination checks
- and confirming that alignment/ANI coverage is high and not biased by missing regions
Your fragment matches (e.g., 1622/1890 for FRB97) are reasonably high, supporting that the ANI is meaningful, but it still doesn’t equate to “same strain.”
4) How to phrase the combined interpretation (tree + ANI)
A clear summary you can use:
- The phylogenetic tree places GE11174 within the Gibbsiella quercinecans lineage, with Brenneria nigrifluens as a distant outgroup.
-
fastANI supports this:
- ~98.1% ANI to G. quercinecans FRB97 and DSM 25889 → strong same-species support.
- 97.2% to G. papilionis → still same-species range, but more distant than the quercinecans references.
- 95.96% to G. greigii → borderline; consistent with being a close neighboring lineage but not the best species match for GE11174.
- 79.1% to Brenneria → confirms it is an appropriate outgroup and far outside the species/genus boundary.
- Therefore, GE11174 is very likely Gibbsiella quercinecans (species-level), and appears most similar to FRB97/DSM 25889, but additional high-resolution analyses are required to assess “same strain.”
If you want, paste the fastANI line for GE11174 vs GE11174? (or run fastANI against a broader set), and/or your core SNP distances from the Roary alignment, and I can help you write a tighter “strain-level” conclusion.
#TODO_NEXT_MONDAY: * phylogenetic tree + fastaANI + nf-core/pairgenomealign (compare to the closest isoalte https://nf-co.re/pairgenomealign/2.2.1/)
* #summarize all results with a mail to send them back, mentioned that we can submit the genome to NCBI to obtain a high-quality annotation. What strain name would you like to assign to this isolate?
* If they agree, I can submit the two new isolates to the NCBI-database!-
submit both sequences in a batch to NCBI-server!
-
Find the more “closest isolate” from GenBank (robust approach) for STEP_7
4. Find the more “closest isolate” from GenBank (robust approach)
# download all available genomes for the genus Gibbsiella (includes assemblies + metadata) #--assembly-level" flag: must be 'chromosome', 'complete', 'contig', 'scaffold' datasets download genome taxon Gibbsiella --include genome,gff3,gbff --assembly-level complete,chromosome,scaffold --filename gibbsiella.zip unzip -q gibbsiella.zip -d gibbsiella_ncbi mamba activate /home/jhuang/miniconda3/envs/bengal3_ac3 # make a Mash sketch of your isolate mash sketch -o isolate bacass_out/Unicycler/GE11174.scaffolds.fa # sketch all reference genomes (example path—adjust) find gibbsiella_ncbi -name "*.fna" -o -name "*.fasta" > refs.txt mash sketch -o refs -l refs.txt # get closest genomes mash dist isolate.msh refs.msh | sort -gk3 | head -n 20 > top20_mash.txt ## What your Mash results mean * The **best hits** to your assembly (`GE11174.scaffolds.fa`) are: * **GCA/GCF_002291425.1** (shows up twice: GenBank **GCA** and RefSeq **GCF** copies of the *same assembly*) * **GCA/GCF_004342245.1** (same duplication pattern) * **GCA/GCF_047901425.1** (FRB97; also duplicated) * Mash distances around **0.018–0.020** are **very close** (typically same species; often same genus and usually within-species). * The `0` in your output is just Mash’s p-value being printed as 0 due to underflow (i.e., extremely significant). So yes: your isolate looks **very close to those Gibbsiella genomes**, and FRB97 being in that set is consistent with your earlier KmerFinder result.
5. — Remove duplicates (GCA vs GCF)
Right now you’re seeing the same genome twice (GenBank + RefSeq). For downstream work, keep one. Example: keep only **GCF** if available, else GCA: ```bash # Take your top hits, prefer GCF over GCA cat top20_mash.txt \ | awk '{print $2}' \ | sed 's|/GCA_.*||; s|/GCF_.*||' \ | sort -u ``` But easiest: just manually keep one of each pair: * keep `GCF_002291425.1` (drop `GCA_002291425.1`) * keep `GCF_004342245.1` * keep `GCF_047901425.1` (and maybe keep `GCA_032062225.1` if it’s truly different and you want a more distant ingroup point)