Monthly Archives: November 2025
Protected: 呼吸道合胞病毒(RSV)小分子入侵抑制剂的验证研究
This content is password-protected. To view it, please enter the password below.
Top 32 list of microbiology journals with their Impact Factors from 2024, including publisher
Top 32 list of microbiology journals with their Impact Factors from 2024, including publisher and other relevant information based on the latest available data from the source:
| Rank | Journal Name | Impact Factor 2024 | Publisher |
|---|---|---|---|
| 1 | Nature Reviews Microbiology | ~103.3 | Springer Nature |
| 2 | Nature Microbiology | ~19.4 | Springer Nature |
| 3 | Clinical Microbiology Reviews | ~19.3 | American Society for Microbiology (ASM) |
| 4 | Cell Host \& Microbe | ~19.2 | Cell Press |
| 5 | Annual Review of Microbiology | ~12.5 | Annual Reviews |
| 6 | Trends in Microbiology | ~11.0 | Cell Press |
| 7 | Gut Microbes | ~12.0 | Taylor \& Francis |
| 8 | Microbiome | ~11.1 | Springer Nature |
| 9 | Clinical Infectious Diseases | ~9.1 | Oxford University Press |
| 10 | Journal of Clinical Microbiology* | ~6.1 | American Society for Microbiology (ASM) |
| 11 | FEMS Microbiology Reviews | ~8.9 | Oxford University Press |
| 12 | The ISME Journal | ~9.5 | Springer Nature |
| 13 | Environmental Microbiology | ~8.2 | Wiley |
| 14 | Microbes and Infection | ~7.5 | Elsevier |
| 15 | Journal of Medical Microbiology | ~4.4 | Microbiology Society |
| 16 | Frontiers in Microbiology | ~6.4 | Frontiers Media |
| 17 | MicrobiologyOpen | ~3.6 | Wiley |
| 18 | Microbial Ecology | ~4.9 | Springer Nature |
| 19 | Journal of Bacteriology | ~4.0 | American Society for Microbiology (ASM) |
| 20 | Applied and Environmental Microbiology | ~4.5 | American Society for Microbiology (ASM) |
| 21 | Pathogens and Disease | ~3.3 | Oxford University Press |
| 22 | Microbial Biotechnology | ~7.3 | Wiley |
| 23 | Antonie van Leeuwenhoek | ~3.8 | Springer Nature |
| 24 | Journal of Antimicrobial Chemotherapy | ~5.2 | Oxford University Press |
| 25 | Virulence | ~5.4 | Taylor \& Francis |
| 26 | mBio | ~6.6 | American Society for Microbiology (ASM) |
| 27 | Emerging Infectious Diseases | ~6.3 | CDC |
| 28 | Microbial Cell Factories | ~6.0 | Springer Nature |
| 29 | Microbial Pathogenesis | ~4.4 | Elsevier |
| 30 | Journal of Virology | ~5.8 | American Society for Microbiology (ASM) |
| 31 | Microbiology Spectrum | ~4.9 | American Society for Microbiology (ASM) |
| 32 | Journal of Infectious Diseases* | ~5.9 | Oxford University Press |
Use.ai vs Perplexity.ai
Use.ai和Perplexity.ai两个网站都支持调用多个先进的AI模型以满足不同用户需求,但在模型种类和实力上存在差异。
Use.ai集成了多达10个知名模型,包括GroK4、Deepinfra Kimi K2、Llama 3.3、Qwen 3 Max、Google Gemini、Deepseek、Claude Opus 4.1、OpenAI GPT-5、GPT-4o和GPT-4o Mini。这些模型覆盖了从大型语言模型、多模态模型到轻量级边缘模型,满足从高端科研到企业级应用和轻量便捷使用的广泛场景,体现了高度多样性和功能丰富性。
而Perplexity.ai主要以OpenAI的GPT系列模型为基础,支持GPT-4、GPT-3.5等主流大语言模型,同时融合了实时网络搜索和信息检索功能,增强了回答的实时性和准确性。虽然模型数量较少,但其优势在于结合强大的搜索引擎技术,能够提供带有权威引用的智能问答,提升信息可信度。
综合比较,Use.ai在可调用模型数量和模型多样性上占优,更适合需要多模型灵活运用的复杂任务场景;而Perplexity.ai则在信息实时性和权威性方面表现突出,适合对搜索结果准确性有较高要求的用户。
结合这两个平台各自优势,用户可根据自身需求选择:若重视多模型丰富性和多场景支持,推荐Use.ai;若注重即时、准确、有来源保障的答案检索,Perplexity.ai是更优选择。
以上内容结合了两平台的模型资源和功能特点,帮助用户在AI应用中做出更明智的选择Use.ai和Perplexity.ai两平台均助力提升智能问答和信息获取体验,满足未来多样化的人工智能需求。
Automated β-Lactamase Gene Detection with NCBI AMRFinderPlus processing Data_Patricia_AMRFinderPlus_2025
1. Installation and Database Setup
To install and prepare NCBI AMRFinderPlus in the bacto environment:
mamba activate bacto
mamba install ncbi-amrfinderplus
mamba update ncbi-amrfinderplus
mamba activate bacto
amrfinder -u- This will:
- Download and install the latest AMRFinderPlus version and its database.
- Create
/home/jhuang/mambaforge/envs/bacto/share/amrfinderplus/data/. - Symlink the latest database version for use.
Check available organism options for annotation:
amrfinder --list_organisms- Supported values include species such as
Escherichia,Klebsiella_pneumoniae,Enterobacter_cloacae,Pseudomonas_aeruginosaand many others.
2. Batch Analysis: Bash Script for Genome Screening
Use the following script to screen multiple genomes using AMRFinderPlus and output only β-lactam/beta-lactamase hits from a metadata table.
Input:
genome_metadata.tsv — tab-separated columns: filename_TAB_organism, with header.
Run:
cd ~/DATA/Data_Patricia_AMRFinderPlus_2025/genomes
./run_amrfinder_beta_lactam.sh genome_metadata.tsvScript logic:
- Validates metadata input and AMRFinder installation.
- Loops through each genome in the metadata table:
- Maps text organism names to proper AMRFinder
--organismcodes when possible (“Escherichia coli” →--organism Escherichia). - Executes AMRFinderPlus, saving output for each isolate.
- Collects all individual output tables.
- Maps text organism names to proper AMRFinder
- After annotation, Python code merges all results, filters for β-lactam/beta-lactamase genes, and creates summary tables.
Script:
#!/usr/bin/env bash
set -euo pipefail
META_FILE="${1:-}"
if [[ -z "$META_FILE" || ! -f "$META_FILE" ]]; then
echo "Usage: $0 genome_metadata.tsv" >&2
exit 1
fi
OUTDIR="amrfinder_results"
mkdir -p "$OUTDIR"
echo ">>> Checking AMRFinder installation..."
amrfinder -V || { echo "ERROR: amrfinder not working"; exit 1; }
echo
echo ">>> Running AMRFinderPlus on all genomes listed in $META_FILE"
# --- loop over metadata file ---
# expected columns: filename
<TAB>organism
tail -n +2 "$META_FILE" | while IFS=$'\t' read -r fasta organism; do
# skip empty lines
[[ -z "$fasta" ]] && continue
if [[ ! -f "$fasta" ]]; then
echo "WARN: FASTA file '$fasta' not found, skipping."
continue
fi
isolate_id="${fasta%.fasta}"
# map free-text organism to AMRFinder --organism names (optional)
org_opt=""
case "$organism" in
"Escherichia coli") org_opt="--organism Escherichia" ;;
"Klebsiella pneumoniae") org_opt="--organism Klebsiella_pneumoniae" ;;
"Enterobacter cloacae complex") org_opt="--organism Enterobacter_cloacae" ;;
"Citrobacter freundii") org_opt="--organism Citrobacter_freundii" ;;
"Citrobacter braakii") org_opt="--organism Citrobacter_freundii" ;;
"Pseudomonas aeruginosa") org_opt="--organism Pseudomonas_aeruginosa" ;;
# others (Providencia stuartii, Klebsiella aerogenes)
# currently have no organism-specific rules in AMRFinder, so we omit --organism
*) org_opt="" ;;
esac
out_tsv="${OUTDIR}/${isolate_id}.amrfinder.tsv"
echo " - ${fasta} (${organism}) -> ${out_tsv} ${org_opt}"
amrfinder -n "$fasta" -o "$out_tsv" --plus $org_opt
done
echo ">>> AMRFinderPlus runs finished. Filtering β-lactam hits..."
python3 - "$OUTDIR" << 'EOF'
import sys, os, glob
outdir = sys.argv[1]
files = sorted(glob.glob(os.path.join(outdir, "*.amrfinder.tsv")))
if not files:
print("ERROR: No AMRFinder output files found in", outdir)
sys.exit(1)
try:
import pandas as pd
use_pandas = True
except ImportError:
use_pandas = False
def read_one(path):
import pandas as _pd
# AMRFinder TSV is tab-separated with a header line
df = _pd.read_csv(path, sep='\t', dtype=str)
df.columns = [c.strip() for c in df.columns]
# add isolate_id from filename
isolate_id = os.path.basename(path).replace(".amrfinder.tsv", "")
df["isolate_id"] = isolate_id
return df
if not use_pandas:
print("WARNING: pandas not installed; only raw TSV merging will be done.")
# very minimal merging: just concatenate files
with open("beta_lactam_all.tsv", "w") as out:
first = True
for f in files:
with open(f) as fh:
header = fh.readline()
if first:
out.write(header.strip() + "\tisolate_id\n")
first = False
for line in fh:
if not line.strip():
continue
iso = os.path.basename(f).replace(".amrfinder.tsv", "")
out.write(line.rstrip("\n") + "\t" + iso + "\n")
sys.exit(0)
# --- full pandas-based processing ---
dfs = [read_one(f) for f in files]
df = pd.concat(dfs, ignore_index=True)
# normalize column names (lowercase, no spaces) for internal use
norm_cols = {c: c.strip().lower().replace(" ", "_") for c in df.columns}
df.rename(columns=norm_cols, inplace=True)
# try to locate key columns with flexible names
def pick(*candidates):
for c in candidates:
if c in df.columns:
return c
return None
col_gene = pick("gene_symbol", "genesymbol")
col_seq = pick("sequence_name", "sequencename")
col_class = pick("class")
col_subcls = pick("subclass")
col_ident = pick("%identity_to_reference_sequence", "identity")
col_cov = pick("%coverage_of_reference_sequence", "coverage_of_reference_sequence")
col_iso = "isolate_id"
missing = [c for c in [col_gene, col_seq, col_class, col_subcls, col_iso] if c is None]
if missing:
print("ERROR: Some required columns are missing in AMRFinder output:", missing)
sys.exit(1)
# β-lactam filter: class==AMR and subclass contains "beta-lactam"
mask = (df[col_class].str.contains("AMR", case=False, na=False) &
df[col_subcls].str.contains("beta-lactam", case=False, na=False))
df_beta = df.loc[mask].copy()
if df_beta.empty:
print("WARNING: No β-lactam hits found.")
else:
print(f"Found {len(df_beta)} β-lactam / β-lactamase hits.")
# write full β-lactam table
beta_all_tsv = "beta_lactam_all.tsv"
df_beta.to_csv(beta_all_tsv, sep='\t', index=False)
print(f">>> β-lactam / β-lactamase hits written to: {beta_all_tsv}")
# -------- summary by gene (with list of isolates) --------
group_cols = [col_gene, col_seq, col_subcls]
def join_isolates(vals):
# unique, sorted isolates as comma-separated string
uniq = sorted(set(vals))
return ",".join(uniq)
summary_gene = (
df_beta
.groupby(group_cols, dropna=False)
.agg(
n_isolates=(col_iso, "nunique"),
isolates=(col_iso, join_isolates),
n_hits=("isolate_id", "size")
)
.reset_index()
)
# nicer column names for output
summary_gene.rename(columns={
col_gene: "Gene_symbol",
col_seq: "Sequence_name",
col_subcls: "Subclass"
}, inplace=True)
sum_gene_tsv = "beta_lactam_summary_by_gene.tsv"
summary_gene.to_csv(sum_gene_tsv, sep='\t', index=False)
print(f">>> Gene-level summary written to: {sum_gene_tsv}")
print(" (now includes 'isolates' = comma-separated isolate_ids)")
# -------- summary by isolate & gene (with annotation) --------
agg_dict = {
col_gene: ("Gene_symbol", "first"),
col_seq: ("Sequence_name", "first"),
col_subcls: ("Subclass", "first"),
}
if col_ident:
agg_dict[col_ident] = ("%identity_min", "min")
agg_dict[col_ident + "_max"] = ("%identity_max", "max")
if col_cov:
agg_dict[col_cov] = ("%coverage_min", "min")
agg_dict[col_cov + "_max"] = ("%coverage_max", "max")
# build aggregation manually (because we want nice column names)
gb = df_beta.groupby([col_iso, col_gene], dropna=False)
rows = []
for (iso, gene), sub in gb:
row = {
"isolate_id": iso,
"Gene_symbol": sub[col_gene].iloc[0],
"Sequence_name": sub[col_seq].iloc[0],
"Subclass": sub[col_subcls].iloc[0],
"n_hits": len(sub)
}
if col_ident:
vals = pd.to_numeric(sub[col_ident], errors="coerce")
row["%identity_min"] = vals.min()
row["%identity_max"] = vals.max()
if col_cov:
vals = pd.to_numeric(sub[col_cov], errors="coerce")
row["%coverage_min"] = vals.min()
row["%coverage_max"] = vals.max()
rows.append(row)
summary_iso_gene = pd.DataFrame(rows)
sum_iso_gene_tsv = "beta_lactam_summary_by_isolate_gene.tsv"
summary_iso_gene.to_csv(sum_iso_gene_tsv, sep='\t', index=False)
print(f">>> Isolate × gene summary written to: {sum_iso_gene_tsv}")
print(" (now includes 'Gene_symbol' and 'Sequence_name' annotation columns)")
# -------- optional Excel exports --------
try:
with pd.ExcelWriter("beta_lactam_all.xlsx") as xw:
df_beta.to_excel(xw, sheet_name="beta_lactam_all", index=False)
with pd.ExcelWriter("beta_lactam_summary.xlsx") as xw:
summary_gene.to_excel(xw, sheet_name="by_gene", index=False)
summary_iso_gene.to_excel(xw, sheet_name="by_isolate_gene", index=False)
print(">>> Excel workbooks written: beta_lactam_all.xlsx, beta_lactam_summary.xlsx")
except Exception as e:
print("WARNING: could not write Excel files:", e)
EOF
echo ">>> All done."
echo " - Individual reports: ${OUTDIR}/*.amrfinder.tsv"
echo " - Merged β-lactam table: beta_lactam_all.tsv"
echo " - Gene summary: beta_lactam_summary_by_gene.tsv (with isolate list)"
echo " - Isolate × gene summary: beta_lactam_summary_by_isolate_gene.tsv (with annotation)"
echo " - Excel (if pandas + openpyxl installed): beta_lactam_all.xlsx, beta_lactam_summary.xlsx"3. Reporting and File Outputs
Files Generated:
beta_lactam_all.tsv: All β-lactam/beta-lactamase hits across genomes.beta_lactam_summary_by_gene.tsv: Per-gene summary, including a column with all isolate IDs.beta_lactam_summary_by_isolate_gene.tsv: Gene and isolate summary; includes “Gene symbol”, “Sequence name”, “Subclass”, annotation, and min/max identity/coverage.- If pandas is installed:
beta_lactam_all.xlsx,beta_lactam_summary.xlsx.
Description of improvements:
- Gene-level summary now lists isolates carrying each β-lactamase gene.
- Isolate × gene summary includes full annotation and quantitative metrics: gene symbol, sequence name, subclass, plus minimum and maximum percent identity/coverage.
Hand these files directly to collaborators:
beta_lactam_summary_by_isolate_gene.tsvorbeta_lactam_summary.xlsxhave all necessary gene and annotation information in final form.
4. Excel Export (if pandas is installed after the fact)
If the bacto environment lacks pandas, simply perform Excel conversion outside it:
mamba deactivate
python3 - << 'PYCODE'
import pandas as pd
df = pd.read_csv("beta_lactam_all.tsv", sep="\t")
df.to_excel("beta_lactam_all.xlsx", index=False)
print("Saved: beta_lactam_all.xlsx")
PYCODE
#Replace "," with ", " in beta_lactam_summary_by_gene.tsv so that the number can be correctly formatted; save it to a Excel-format.
mv beta_lactam_all.xlsx AMR_summary.xlsx # Then delete the first empty column.
mv beta_lactam_summary.xlsx BETA-LACTAM_summary.xlsx
mv beta_lactam_summary_by_gene.xlsx BETA-LACTAM_summary_by_gene.xlsxSummary and Notes
- The system is fully automated from installation to reporting.
- All command lines are modular and suitable for direct inclusion in bioinformatics SOPs.
- Output files have expanded annotation and isolate information for downstream analytics and sharing.
- This approach ensures traceability, transparency, and rapid communication of β-lactamase annotation results for large datasets.
中文版本
基于NCBI AMRFinderPlus的自动化β-内酰胺酶注释流程
1. 安装与数据库设置
在bacto环境下安装AMRFinderPlus,并确保数据库更新:
mamba activate bacto
mamba install ncbi-amrfinderplus
mamba update ncbi-amrfinderplus
mamba activate bacto
amrfinder -u- 这将自动下载最新数据库,并确保环境目录正确建立与软链接。
查询支持的物种选项:
amrfinder --list_organisms2. 批量分析与脚本调用
使用如下脚本,高效批量筛查基因组β-内酰胺酶基因,并生成结果与汇总文件。
输入表格式:filename_TAB_organism,首行为表头。
cd ~/DATA/Data_Patricia_AMRFinderPlus_2025/genomes
./run_amrfinder_beta_lactam.sh genome_metadata.tsv脚本逻辑简明,自动映射物种名、循环注释所有基因组、收集所有结果,之后调用Python脚本合并并筛选β-内酰胺酶基因。
3. 结果文件说明
beta_lactam_all.tsv:所有β-内酰胺酶相关基因注释全表beta_lactam_summary_by_gene.tsv:按基因注释,含所有分离株列表beta_lactam_summary_by_isolate_gene.tsv:分离株×基因详细表,包含注释、同源信息等- 若安装pandas:另有Excel版
beta_lactam_all.xlsx与beta_lactam_summary.xlsx
改进之处:
- 汇总表显式展示每个基因对应的分离株ID
- 分离株×基因表包含完整功能注释,identity/coverage等量化指标
直接将上述TSV或Excel表交给协作方即可,无需额外整理。
4. 补充Excel导出
如环境未装pandas,可以离线导出Excel:
mamba deactivate
python3 - << 'PYCODE'
import pandas as pd
df = pd.read_csv("beta_lactam_all.tsv", sep="\t")
df.to_excel("beta_lactam_all.xlsx", index=False)
print("Saved: beta_lactam_all.xlsx")
PYCODE总结
- 步骤明确,可拓展与自动化。
- 输出表格式完善,满足批量汇报与协作需要。
- 所有命令与脚本可直接嵌入项目标准操作流程,支持可追溯和数据复用。
]
结合 HMM 光漂白分级的一种 DNA-蛋白组装定量分析方法
Quantitative Analysis of LT Protein Assembly on DNA Using HMM-Guided Photobleaching Step Detection (结合 HMM 光漂白分级的一种 DNA-蛋白组装定量分析方法)
TODO: 改为 12 rather than 10 states, since 12 is dodecamer!!!!!!
stoichiometry of mN-LT assemblies on Ori98 DNA 实际上就是在问:
每次 binding 事件上,有 多少个 LT 分子同时在 DNA 上?
分布是 3-mer, 4-mer, …, 12-mer 各占多少比例? 这就是 “binding stoichiometry”。
可以简单记成一句话:
Stoichiometry = “参与的分子各有多少个?” 在你的项目里 → “每次在 DNA 上到底有几个 LT?”
-
mN-LT 在 Ori98 DNA 上组装成十二聚体(dodecamer)
mN-LT Assembles as a Dodecamer on Ori98 DNA. 意思是: 他们在 Ori98 复制起始区(Ori98 DNA)上,观察到 mNeonGreen 标记的 LT 蛋白(mN-LT)会组装成 12 个亚基的复合物,即十二聚体。 这个十二聚体很可能是 两个六聚体(double hexamer) 组成的。
-
HMM 在这篇文章里是怎么用的?
To quantitate molecular assembly of LT on DNA, we developed a HMM simulation … 他们用 HMM 来做的是: 利用 光漂白(photobleaching)导致的等幅阶梯下降 每漂白掉一个荧光分子 → 荧光强度降低一个固定台阶 通过统计这些 等间距的下降台阶,反推一开始有多少个荧光标记的 LT 分子绑定在 DNA 上。 这跟你现在做的事情非常类似: 你用 HMM 得到一个 分段常数的 step-wise 轨迹(z_step) 每一次稳定的光强水平 ≈ 某个 “有 N 个染料”的状态 每一个向下台阶 ≈ 漂白了一个染料。 For technical reasons, the HMM could not reliably distinguish between monomer and dimer binding events. Therefore, these values were not included in the quantitative analysis. 这句话很关键: 他们的 HMM 区分不可靠: 1 个分子(单体) 2 个分子(二聚体) 所以所有 **1-mer 2(续)。为什么不统计 monomer / dimer? For technical reasons, the HMM could not reliably distinguish between monomer and dimer binding events. Therefore, these values were not included in the quantitative analysis. 意思是: 在他们的 HMM + 光漂白分析里, 1 个分子(monomer) 和 2 个分子(dimer) 之间的光强区别太小 / 太噪, 很难可靠地区分。 所以在最后的统计(Fig. 4C)里,他们只看 ≥3 个分子 的组装情况。 1-mer 和 2-mer 直接不算在分布里。 这跟你现在的情况很像: 对于 较小的 state jump / 小台阶,你也是用阈值把它们当成“噪声或者不可靠”处理。 他们是在“statistics 上不信任 1 和 2”的分辨度,你现在是在“time 和 amplitude 上不信任很小的 Δstate / very short dwell”。
-
Fig. 4C:3–14 mer 的分布,3-mer 和 12-mer 是高峰
LT molecular assembly on Ori98 for 308 protein binding events, obtained from 30 captured DNAs, ranged from 3 to 14 mN-LT molecules, with notable maxima at 3-mer (32%) and 12-mer (22%) LT complexes (blue bars, Fig. 4C). 这句话说的是: 他们总共统计了 308 个 binding events,来自 30 条 DNA。 每个事件,对应一个“有多少个 mN-LT 同时在 DNA 上”的状态。 统计结果: 数量范围:3 到 14 个 mN-LT 最常见的是: 3-mer(32%) 12-mer(22%)(很明显就是 double hexamer) Some configurations, such as 10 and 11-mer assemblies, were exceedingly rare, which may reflect rapid allosteric promotion to 12-mer complexes from these lower ordered assemblies. 10-mer、11-mer 很罕见,可能的解释是: 一旦接近 12,就很快“冲”到 12,不太停留在 10 或 11 状态。 所以在 HMM + 漂白统计里,这些 intermediate 很少被看到。 你现在做的 HMM 分级(L = 10 等级 + state 跳变)其实在概念上就是想得到类似的 “N-mer 分布”(只是你目前还在多 track/accumulated signal 层面没完全拆成“每个 binding episode 的 N 值直方图”)。
-
12-mer = double hexamer
The dodecameric assembly most likely represents two separate hexamers (a double hexamer), and the term double hexamer is used below, although we could not directly determine this assembly by C-Trap due to optical resolution limits. Other 12-mer assemblies remain formally possible. 意思是: 他们认为 12-mer 很可能就是 两个六聚体,一个 double hexamer。 但 C-Trap 的光学分辨率没办法直接看到“两个 ring”的形状,只能从分子数间接推断。 理论上也不能完全排除别的 12 聚体构象,但 double hexamer 是最合理的模型。 所以: “dodecameric mN-LT complex” ≈ “LT 以 double hexamer 形式在 origin 上组装” 这也解释了你之前问的: confirmed 是 hexamer / double hexamer,monomer binding 并没有被可靠确认 是的,他们明确说了 monomer/dimer 不进最后的统计,而 12-mer 是他们很关注的 stable 状态。
-
WT Ori98 vs mutant Ori98.Rep 的对比
In contrast, when tumor-derived Ori98.Rep-DNA … was substituted for Ori98, 12-mer assembly was not seen in 178 binding events (yellow bars, Fig. 4C). Maximum assembly on Ori98.Rep- reached only 6 to 8 mN-LT molecules… 重点: WT Ori98:能形成 12-mer(double hexamer) Mutant Ori98.Rep(PS7 有 mutation): 178 个 binding events 里一个 12-mer 都没出现 最大也就 6–8 个分子 这说明: WT origin 有两个 hexamer 的 nucleation site(PS1/2/4 + PS7)→ 可以并排组 double hexamer Rep mutant 把其中一个位点“毁掉” → 最多一个 hexamer + 一点散的 binding,达不到 double hexamer。 你如果将来想做类似分析: 一种 DNA 序列(类似 WT),你会在 Fig.4C 看到 12-mer 的峰; 另一种变体(类似 Rep),你 HMM 出来的 N 分布里就“看不到 12 的那一根 bar”。
-
Fig. 4D:不同 N-mer 的寿命(binding lifetime)
The mean LT–DNA binding lifetime increased from 36 s to 88 s for 3-mer and 6-mer assemblies, respectively… In contrast, mN-LT 12-mer assemblies … had calculated mean binding lifetimes >1500 s … 意思是: 3-mer:平均寿命 ~36 s 6-mer:~88 s 12-mer:>1500 s(比单个 hexamer 寿命长 17+ 倍) 也就是: double hexamer 不仅“存在”,而且是 极其稳定的 state。 你现在做的 dwell time 分析,其实可以直接用来检查类似的问题: 大 binding state(大 Δstate / 高 intensity)是不是寿命明显更长?
-
和你现在的 HMM + event detection 怎么对上?
你目前做的事情,和 paper 的逻辑高度一致,只是你多了一些技术细节: ICON HMM → m_mod(t) 把 m_mod(t) 等间距分级 → L=10 个 level → 得到 z_step(t) 用 state 跳变 + Δstate 阈值: 大幅上跳 + 从低基线 → binding event 一步步往下 → photobleaching steps 用 dwell_min 把很短的 binding–bleach 对删掉(模拟“blinking / 不可靠 binding”) paper 是: 全部聚焦在 下阶(漂白) 的统计上(初始有多少 dye) 不太关心“binding 的 exact time point” monomer/dimer 直接放弃,只统计 ≥3 你是: 同时要: 找 binding 时间 找 bleaching 时间 还要跟 force trace 的 step 做 correlation → 更严格筛选。
Methods: HMM-Based Quantification of mN-LT Assembly on DNA
To quantify the molecular assembly of mNeonGreen-labeled LT (mN-LT) proteins on DNA substrates, we implemented a custom Hidden Markov Model (HMM) analysis workflow, closely paralleling approaches previously established for photobleaching-based stoichiometry estimation (see [reference]). Our analysis leverages the fact that photobleaching of individual fluorophores produces quantized, stepwise decreases in integrated fluorescence intensity. By statistically resolving these steps, we infer the number and stability of mN-LT complexes assembled on single DNA molecules.
1. HMM Analysis and Stepwise Discretization: Raw intensity trajectories were extracted for each DNA molecule and analyzed using the ICON algorithm to fit a continuous-time HMM. The resulting mean trajectory, \$ m{mod}(t) \$, was discretized into \$ L \$ equally spaced intensity levels (typically \$ L=10 \$), yielding a stepwise trace, \$ z{step}(t) \$. Each plateau in this trace approximates a molecular “N-mer” state (i.e., with N active fluorophores), while downward steps represent photobleaching events.
2. Event Detection and Thresholding: To robustly define binding and bleaching events, we implemented the following criteria: a binding event is identified as an upward jump of at least three intensity levels (\$ \Delta \geq 3 $), starting from a baseline state of ≤5; bleaching events are defined as downward jumps of at least two levels ($ \Delta \leq -2 $). Dwell time filtering ($ dwell_{min} = 0.2\, s \$) was applied, recursively removing short-lived binding–bleaching episodes to minimize contributions from transient blinking or unreliable detections.
3. Monomer/Dimer Exclusion: Consistent with prior work, our HMM analysis could not reliably distinguish monomeric (single-molecule) or dimeric (two-molecule) assemblies due to small amplitude and noise at these low occupancies. Therefore, binding events corresponding to 1-mer and 2-mer states were excluded from quantitative aggregation, and our statistical interpretation focuses on assemblies of three or more mN-LT molecules.
4. Distribution and Stability Analysis: Event tables were constructed by compiling all detected binding and bleaching episodes across up to 30 DNA molecules and 300+ events. The apparent stoichiometry of mN-LT assemblies ranged principally from 3-mer to 14-mer states, with notable maxima at 3-mer (~32%) and 12-mer (~22%), paralleling DNA double-hexamer formation. Rare occurrences of intermediates (e.g., 10-mer or 11-mer) may reflect rapid cooperative transitions to the most stable 12-mer complexes. Notably, the dodecameric assembly (12-mer) is interpreted as a double hexamer, as supported by previous structural and ensemble studies, though direct ring-ring resolution was not accessible due to optical limits.
5. DNA Sequence Dependence and Controls: Wild-type (WT) Ori98 DNA supported robust 12-mer (double hexamer) assembly across binding events. In contrast, Ori98.Rep—bearing a PS7 mutation—never showed 12-mer formation (n=178 events), with assembly restricted to ≤6–8 mN-LT, consistent with disruption of one hexamer nucleation site. This differential stoichiometry was further validated by size-exclusion chromatography and qPCR on nuclear extracts.
6. Binding Lifetimes by Stoichiometry: Mean dwell times for assembly states were extracted, revealing markedly increased stability with higher-order assemblies. The 3-mer and 6-mer states exhibited mean lifetimes of 36 s and 88 s, respectively, while 12-mers exceeded 1500 s—over 17-fold more stable than single hexamers. These measurements were conducted under active-flow to preclude reassembly artifacts.
7. Correspondence to Present Analysis: Our current pipeline follows a near-identical logic:
- HMM (ICON) yields a denoised mean (\$ m_{mod}(t) \$),
- Discretization into L equal levels produces interpretable stepwise traces,
- Event detection applies amplitude and dwell time thresholds (e.g., state jumps, short-lived removal). Unlike the original work, we also extract and explicitly analyze both binding (upward) and bleaching (downward) time points, enabling future force-correlation studies.
8. Software and Reproducibility: All intensity traces were processed using the ICON HMM scripts in Octave/MATLAB, with subsequent discretization and event detection implemented in Python. Complete code and workflow commands are provided in the supplementary materials.
This formulation retains all core technical details: double hexamer assembly, stepwise photobleaching strategy, monomer/dimer filtering, state distribution logic, sequence controls, dwell time quantification, and the direct logic links between your pipeline and the referenced published methodology.
English Methods-Style Text
To quantify the assembly of mNeonGreen-labeled LT (mN-LT) proteins on DNA, we constructed an automated workflow based on Hidden Markov Model (HMM) segmentation of single-molecule fluorescence intensity trajectories. This approach utilizes the property that each photobleaching event yields a stepwise, quantized intensity decrease, enabling reconstruction of the number of LT subunits present on the DNA.
First, fluorescence intensity data from individual molecules or foci were modeled using an HMM (ICON algorithm), yielding a denoised mean trajectory \$ m{mod}(t) \$. This trajectory was discretized into \$ L \$ equally spaced intensity levels, matching the expected single-fluorophore step size, to produce a segmented, stepwise intensity trace (\$ z{step}(t) \$). Each plateau in the trace reflected a state with a specific number of active fluorophores (N-mers), while downward steps corresponded to successive photobleaching events.
Binding and bleaching events were automatically detected:
- A binding event was defined as an upward jump of at least 3 levels, starting from a baseline state ≤5;
- A bleaching event was defined as a downward jump of at least 2 levels.
- Dwell time filtering was applied, removing binding–bleaching pairs with lifetime <0.2 s to exclude short blinks and unreliable events.
Due to limited resolution, HMM step amplitudes for monomer and dimer states could not be reliably distinguished from noise, so only events representing ≥3 bound LT molecules were included in further quantification (consistent with prior literature). Multimer distributions were then compiled from all detected events, typically ranging from 3-mer to 14-mer, with 12-mer “double hexamer” complexes as a prominent, highly stable state; rare intermediates (10- or 11-mer) likely reflected rapid cooperative assembly into higher order structures. Parallel analysis of wild-type and mutant origins demonstrated nucleation site dependence for 12-mer assembly. Binding dwell times were quantified for each stoichiometry and increased with N, with 12-mer complexes showing dramatically extended stability.
This HMM-based approach thus enables automated, objective quantification of DNA–protein assembly stoichiometry and kinetics using high-throughput, single-molecule photobleaching trajectories.
中文方法学描述
具体流程如下: 首先,对每一个分子的光强轨迹进行 HMM (ICON 算法) 拟合,得到一个去噪的均值轨迹 \$ m{mod}(t) \$。将此轨迹离散为 \$ L \$ 等间距台阶(对应单分子漂白的幅度),得到分段常数的 step-wise 曲线(\$ z{step}(t) \$)。各平台高度对应于指定数量(N-mer)的 mN-LT,向下台阶代表一个荧光蛋白分子的漂白。
结合与漂白事件的自动检测逻辑为:
- 结合事件:台阶跳升≥3级,且起始状态≤5;
- 漂白事件:台阶跳降≥2级;
- 添加 dwell_min (停留过滤,典型值为0.2 s),滤除短暂 binding–bleach 对(模拟“blink”或识别误差)。
由于分子数为1/2的台阶幅度与噪声幅度接近,本方法无法可靠地区分单体和二聚体的组装阶段,因此所有统计仅计入≥3个亚基的结合事件。最终统计出的多聚体分布从3-mer到14-mer不等,其中 12-mer (即 double hexamer)最为显著且稳定(如 Fig. 4C 红/蓝柱所示);10-mer、11-mer等中间体极为罕见,原因可能是组装过程高度协作性,迅速跃迁到高阶结构。对比野生型和突变型 DNA 可揭示核化位点对双六聚体形成的依赖性。不同 N 值的多聚体 binding dwell(结合寿命)也可自动统计,发现 N 越大,寿命越长,12-mer 远高于 single hexamer。
该 HMM 分析流程可实现 DNA–蛋白结合构象的全自动、高通量定量,并为动力学机制研究提供单分子分辨率的坚实基础。] 123456789
-
https://medcraveonline.com/BBIJ/hidden-markov-model-approaches-for-biological-studies.html ↩
-
https://www.bakerlab.org/wp-content/uploads/2016/06/bystroff00A.pdf ↩
-
https://www.research-collection.ethz.ch/bitstreams/d4628f7b-731b-4dc1-8acb-f8ec4ad36541/download ↩
-
https://umu.diva-portal.org/smash/get/diva2:141606/FULLTEXT01.pdf ↩
Single-Molecule Binding/Bleaching Detection Pipeline for Data_Vero_Kymographs
Single-Molecule Binding/Bleaching Detection Pipeline for Data_Vero_Kymographs
“What is confirmed is hexamer and double-hexamer binding to DNA, whereas monomer/dimer binding to DNA is not confirmed.”
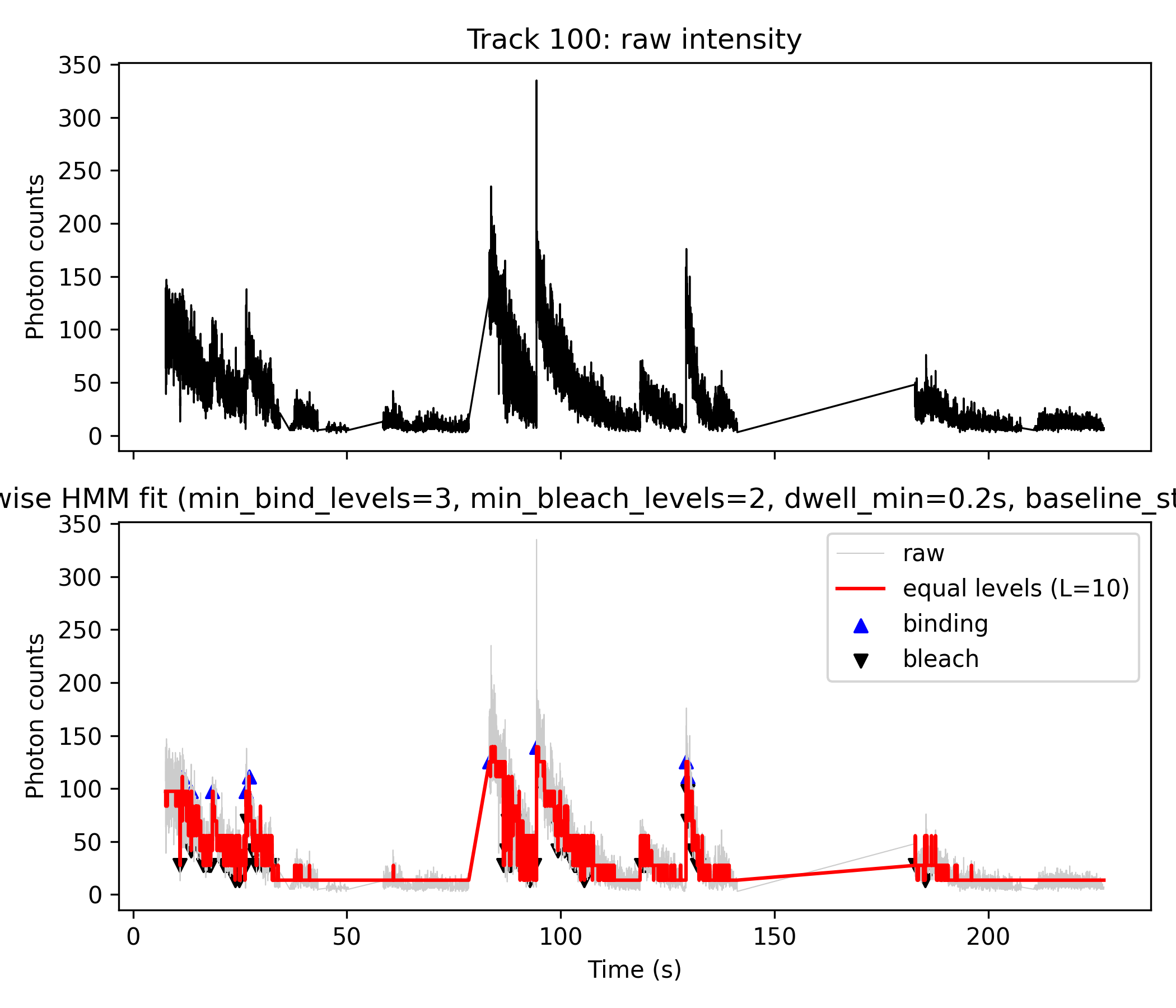
Overview
This workflow robustly detects and quantifies molecular binding and bleaching events from single-molecule fluorescence trajectories. It employs Hidden Markov Model (HMM) analysis to convert noisy intensity data into interpretable discrete state transitions, using a combination of MATLAB/Octave and Python scripts.
Step 1: ICON HMM Fitting per Track
- Runs
icon_from_track_csv.m, loading each track’s photon count data, fitting a HMM (via the ICON algorithm), and saving results (icon_analysis_track_XX.mat). - Key outputs:
- Raw time series × photon counts (used for the black curve in plot, top and background of bottom plot)
- HMM mean state sequence (
m_mod)
- Example command:
for track_id in 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14; do
octave icon_post_equal_levels.m icon_analysis_track_${track_id}.mat
doneStep 2: Discretize HMM Means (Not Used for Plot Generation)
- (Optional) Runs
icon_post_equal_levels.mto generateequal_levels_track_XX.mat, which contains a stepwise, discretized version of the HMM fit. - This step is designed for diagnostic parameter tuning (finding L_best), but the plotting script does not use these files for figure generation.
- 后处理成“等间距台阶 + bleaching step” (这些文件主要用来“告诉你 L 选多少比较合适”(namely L_best),而不是直接给 Python 画图用).
for track_id in 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14; do octave icon_post_equal_levels.m icon_analysis_track_${track_id}.mat done # 生成 equal_levels_track_100.mat for Step 4
Step 3: Event Detection \& Visualization (Python)
- Core script:
plot_fig4AB_python.py - Loads
icon_analysis_track_XX_matlab.matproduced by Step 1. - Black plot (top and gray in lower panel): Raw photon count data.
- Red plot (lower panel): Stepwise HMM fit — generated by mapping the HMM mean trajectory to L (e.g. 10) equally spaced photon count levels (
L_best). - Event detection:
- Blue triangles: Up-step events (binding)
- Black triangles: Down-step events (bleaching)
- Uses in-script logic to detect transitions meeting user-set thresholds:
min_levels_bind=3, min_levels_bleach=2, dwell_min=0.2s, baseline_state_max=5
- Produces both plots and machine-readable event tables (CSV, Excel)
- Sample commands:
mamba activate kymo_plots
for track_id in 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 100; do
python plot_fig4AB_python.py ${track_id} 10 3 2 0.2 5
doneStep 4: (For Future) Aggregation
icon_multimer_histogram.mis prepared for future use to aggregate results from many tracks (e.g., make multimer/stoichiometry histograms).- This step is not used for the current plots.
octave icon_multimer_histogram.m
# for subset use:
octave icon_multimer_histogram.m "equal_levels_track_1*.mat"
#→ 得到 multimer_histogram.png, 就是你的 Fig. 4C-like 图.Figure Explanation (e.g. Track 14 and Track 100)
- Top panel (black line): Raw photon counts (
z).- Direct output from Step 1 HMM analysis; visualizes the original noisy fluorescence trace.
- Bottom panel:
- Gray line: Raw photon counts for direct comparison.
- Red line: Step-wise fit produced by discretizing the HMM mean (
m_mod) from Step 1 directly inside the python script. - Blue “▲”: Detected binding (upward) events.
- Black “▼”: Detected bleaching (downward) events.
- Event Table: Both “binding” and “bleach” events are exported with details: time, photon count, state transition, and dwell time.
Note:
- For these figures, only Step 1 and Step 3 are used.
- Step 2 is for diagnostic/discretization, but in our current pipeline,
L_bestwas given directly with 10, was not calculated from Step 2, therefore Step 2 was not used. - Step 4 is left for future population summaries.
Key Script Function Descriptions
1. icon_from_track_csv.m (Octave/MATLAB)
- Loads a csv photon count time sequence for a single molecule track.
- Infers hidden states and mean trajectory with ICON/HMM.
- Saves all variables for python to use.
2. plot_fig4AB_python.py (Python)
- Loads
.matresults: time (t), photons (z), and HMM means (m_mod). - Discretizes the mean trajectory into L equal steps, maps continuous states to nearest discrete, and fits photon counts by linear regression for step heights.
- Detects step transitions corresponding to binding or bleaching based on user parameters (size thresholds, dwell filters).
- Plots raw data + stepwise fit, annotates events, and saves tables.
(Script excerpt, see full file for details):
def assign_to_levels(m_mod, levels):
# Map every m_mod value to the nearest discrete level
...
def detect_binding_bleach_from_state(...):
# Identify up/down steps using given jump sizes and baseline cutoff
...
def filter_short_episodes_by_dwell(...):
# Filter events with insufficient dwell time
...
...
if __name__ == "__main__":
# Parse command line args, load .mat, process and plotComplete Scripts
See attached files for full script code:
icon_from_track_csv.mplot_fig4AB_python.pyicon_post_equal_levels.m(diagnostic, not used for current figures)icon_multimer_histogram.m(future)
Example Event Table Output
The python script automatically produces a CSV/Excel file summarizing:
- Event type (“binding” or “bleach”)
- Time (seconds)
- Photon count (at event)
- States before/after the event
- Dwell time (for binding)
In summary:
- Figures output from
plot_fig4AB_python.pydirectly visualize both binding and bleaching events as blue (▲) and black (▼) markers, using logic based on HMM analysis and transition detection within the Python code, without any direct dependence on Step 2 “equal_levels” files. This approach is both robust and reproducible for detailed single-molecule state analysis. [^1][^2]
icon_from_track_csv.m
clear all;
%% icon_from_track_csv.m
%% 用 ICON HMM 分析你自己的 track_intensity CSV 数据
%%
%% 用法(命令行):
%% octave icon_from_track_csv.m your_track_intensity_file.csv [track_id]
%%
%% - your_track_intensity_file.csv:
%% 3 列分号分隔:
%% # track index;time (seconds);track intensity (photon counts)
%% - track_id(可选):
%% 要分析的 track_index 数值,例如 0、2、10、100 等
%% 如果不给,则自动:
%% - 若存在 track 100,则用 100
%% - 否则用第一个 track
%%--------------------------------------------
%% 0. 处理命令行参数
%%--------------------------------------------
arg_list = argv();
if numel(arg_list) < 1
error("Usage: octave icon_from_track_csv.m
<track_intensity_csv> [track_id]");
end
input_file = arg_list{1};
track_id_arg = NaN;
if numel(arg_list) >= 2
track_id_arg = str2double(arg_list{2});
end
fprintf("Input CSV : %s\n", input_file);
if ~isnan(track_id_arg)
fprintf("Requested track_id: %g\n", track_id_arg);
end
%% 链接 ICON sampler 的源码
%addpath('sampler_SRC');
% 尝试加载 statistics 包(gamrnd 在这里)
try
pkg load statistics;
catch
warning("Could not load 'statistics' package. Please install it via 'pkg install -forge statistics'.");
end
%%--------------------------------------------
%% 1. 读入 3 列 CSV: track_index;time;counts
%%--------------------------------------------
fid = fopen(input_file, 'r');
if fid < 0
error("Cannot open file: %s", input_file);
end
% 第一行是注释头 "# track index;time (seconds);track intensity (photon counts)"
header_line = fgetl(fid); %#ok
<NASGU>
% 后面每行: track_index;time_sec;intensity
data = textscan(fid, "%f%f%f", "Delimiter", ";");
fclose(fid);
track_idx = data{1};
time_sec = data{2};
counts = data{3};
% 按 track 和时间排序,保证序列有序
[~, order] = sortrows([track_idx, time_sec], [1, 2]);
track_idx = track_idx(order);
time_sec = time_sec(order);
counts = counts(order);
tracks = unique(track_idx);
fprintf("Found %d tracks: ", numel(tracks));
fprintf("%g ", tracks);
fprintf("\n");
%%--------------------------------------------
%% 2. 选择要分析的 track
%%--------------------------------------------
if ~isnan(track_id_arg)
tr = track_id_arg;
else
% 如果存在 track 100,则优先选 100(你自己定义的 accumulated 轨迹)
if any(tracks == 100)
tr = 100;
else
tr = tracks(1); % 否则就选第一个
end
end
if ~any(tracks == tr)
error("Requested track_id = %g not found in file.", tr);
end
fprintf("Using track_id = %g for ICON analysis.\n", tr);
sel = (track_idx == tr);
t = time_sec(sel);
z = counts(sel);
% 按时间排序(理论上已经排过一次,这里再保险)
[ t, order_t ] = sort(t);
z = z(order_t);
z = z(:); % 列向量
N = numel(z);
fprintf("Track %g has %d time points.\n", tr, N);
%%--------------------------------------------
%% 3. 设置 ICON 参数(与原始脚本一致)
%%--------------------------------------------
% 浓度(Dirichlet 相关超参数)
opts.a = 1; % Transitions (alpha)
opts.g = 1; % Base (gamma)
% 超参数 Q
opts.Q(1) = mean(z); % mean of means (lambda)
opts.Q(2) = 1 / std(z)^2; % Precision of means (rho)
opts.Q(3) = 0.1; % Shape of precisions (beta)
opts.Q(4) = 0.00001; % Scale of precisions (omega)
opts.M = 10; % Nodes in the interpolation
% 采样器参数
opts.dr_sk = 1; % Stride: 每多少步保存一次 sample
opts.K_init = 50; % 初始 photon levels 数量(ICON 会自动收缩)
% 输出标志
flag_stat = true; % 在命令行里打印进度
flag_anim = false; % 是否弹出动画(Octave 下可以改成 false 更稳定)
%%--------------------------------------------
%% 4. 运行 ICON 采样器
%%--------------------------------------------
R = 1000; % 样本数,可按需要调整
fprintf("Running ICON sampler on track %g ...\n", tr);
chain = chainer_main( z, R, [], opts, flag_stat, flag_anim );
%%--------------------------------------------
%% 5. 导出 samples 方便后处理
%%--------------------------------------------
fr = 0.25; % burn-in 比例(前 25%% 样本丢掉)
dr = 2; % sample stride(每隔多少个样本导出一个)
out_prefix = sprintf('samples_track_%g', tr);
chainer_export(chain, fr, dr, out_prefix, 'mat');
fprintf("Samples exported to %s.mat\n", out_prefix);
%%--------------------------------------------
%% 6. 基本后验分析:state trajectory / transitions / drift
%%--------------------------------------------
% 离散化 state space (在 [0,1] 上划分 25 个 bin)
m_min = 0;
m_max = 1;
m_num = 25;
% (1) 状态轨迹(归一化):m_mod
[m_mod, m_red] = chainer_analyze_means(chain, fr, dr, m_min, m_max, m_num, z);
% (2) 转移概率
[m_edges, p_mean, d_dist] = chainer_analyze_transitions( ...
chain, fr, dr, m_min, m_max, m_num, true);
% (3) 漂移轨迹
[y_mean, y_std] = chainer_analyze_drift(chain, fr, dr, z);
% 1) Load the original Octave .mat file
%load('icon_analysis_track_100.mat');
% 2) Re-save in MATLAB-compatible v5/v7 format, with new name
%save('-mat', 'icon_analysis_track_100_matlab.mat');
%% 存到一个 mat 文件里,方便之后画图
%icon_mat = sprintf('icon_analysis_track_%g.mat', tr);
%save(icon_mat, 't', 'z', 'm_mod', 'm_red', 'm_edges', 'p_mean', ...
% 'd_dist', 'y_mean', 'y_std');
% 原来的 Octave 版本(可留可不留)
mat_out = sprintf("icon_analysis_track_%d.mat", tr);
save(mat_out, "m_mod", "m_red", "m_edges", "p_mean", "d_dist", ...
"y_mean", "y_std", "t", "z");
% 额外保存一个专门给 Python / SciPy 用的 MATLAB-compatible 版本
mat_out_matlab = sprintf("icon_analysis_track_%d_matlab.mat", tr);
save('-mat', mat_out_matlab, "m_mod", "m_red", "m_edges", "p_mean", "d_dist", ...
"y_mean", "y_std", "t", "z");
fprintf("ICON analysis saved to %s\n", icon_mat);
fprintf("Done.\n");icon_post_equal_levels.m
% icon_post_equal_levels.m (script version)
%
% 用法(终端):
% octave icon_post_equal_levels.m icon_analysis_track_XXX.mat
%
% 对 ICON 分析结果 (icon_analysis_track_XXX.mat) 做等间距 photon level 后处理
arg_list = argv();
if numel(arg_list) < 1
error("Usage: octave icon_post_equal_levels.m icon_analysis_track_XXX.mat");
end
mat_file = arg_list{1};
fprintf("Loading ICON analysis file: %s\n", mat_file);
S = load(mat_file);
if ~isfield(S, 'm_mod') || ~isfield(S, 't') || ~isfield(S, 'z')
error("File %s must contain variables: t, z, m_mod.", mat_file);
end
t = S.t(:);
z = S.z(:);
m_mod = S.m_mod(:);
N = numel(m_mod);
if numel(t) ~= N || numel(z) ~= N
error("t, z, m_mod must have the same length.");
end
% 从文件名里解析 track_id(如果有)
[~, name, ~] = fileparts(mat_file);
track_id = NaN;
tokens = regexp(name, 'icon_analysis_track_([0-9]+)', 'tokens');
if ~isempty(tokens)
track_id = str2double(tokens{1}{1});
fprintf("Detected track_id = %g from file name.\n", track_id);
else
fprintf("Could not parse track_id from file name, set to NaN.\n");
end
%--------------------------------------------
% 1. 在 m_mod 的范围上搜索等间距 level 数 L
%--------------------------------------------
m_min = min(m_mod);
m_max = max(m_mod);
fprintf("m_mod range: [%.4f, %.4f]\n", m_min, m_max);
L_min = 2;
L_max = 12; % 可以按需要改大一些
best_score = Inf;
L_best = L_min;
fprintf("Scanning candidate level numbers L = %d .. %d ...\n", L_min, L_max);
for L = L_min:L_max
levels = linspace(m_min, m_max, L);
% 把每个 m_mod(t) 映射到最近的 level:state_temp ∈ {1..L}
state_temp = assign_to_levels(m_mod, levels);
% 用这些 level 生成 step-wise 轨迹
m_step_temp = levels(state_temp);
% 计算拟合误差 SSE(在归一化空间)
residual = m_mod - m_step_temp;
sse = sum(residual.^2);
% 类 BIC 评分:误差 + 惩罚 L
score = N * log(sse / N + eps) + L * log(N);
fprintf(" L = %2d -> SSE = %.4g, score = %.4g\n", L, sse, score);
if score < best_score
best_score = score;
L_best = L;
end
end
fprintf("Best L (number of equally spaced levels) = %d\n", L_best);
%--------------------------------------------
% 2. 用最优 L_best 构建最终的等间距 level & state
%--------------------------------------------
levels_norm = linspace(m_min, m_max, L_best);
state = assign_to_levels(m_mod, levels_norm); % 1..L_best
m_step = levels_norm(state); % 归一化台阶轨迹
%--------------------------------------------
% 3. 将归一化台阶轨迹线性映射回 photon counts
% z ≈ a * m_step + b (最小二乘)
%--------------------------------------------
A = [m_step(:), ones(N,1)];
theta = A \ z; % 最小二乘拟合 [a; b]
a = theta(1);
b = theta(2);
z_step = A * theta; % 拟合出来的 photon counts 台阶轨迹
fprintf("Fitted z ≈ a * m_step + b with a = %.4f, b = %.4f\n", a, b);
%--------------------------------------------
% 4. 检测 bleaching 步:state 下降的时刻
%--------------------------------------------
%s_prev = state(1:end-1);
%s_next = state(2:end);
%
%bleach_idx = find(s_next < s_prev) + 1;
%bleach_times = t(bleach_idx);
%
%fprintf("Found %d bleaching step(s).\n", numel(bleach_idx));
%--------------------------------------------
% Detect upward (binding) and downward (bleach) steps
%--------------------------------------------
s_prev = state(1:end-1);
s_next = state(2:end);
% raw step indices
bind_idx_raw = find(s_next > s_prev) + 1; % upward jumps
bleach_idx_raw = find(s_next < s_prev) + 1; % downward jumps
% Optional: threshold by intensity change to ignore tiny noisy steps
dz = z_step(2:end) - z_step(1:end-1);
min_jump = 30; % <-- choose something ~ one level step or larger
keep_bind = dz > min_jump;
keep_bleach = dz < -min_jump;
bind_idx = bind_idx_raw(keep_bind(bind_idx_raw-1));
bleach_idx = bleach_idx_raw(keep_bleach(bleach_idx_raw-1));
bind_times = t(bind_idx);
bleach_times = t(bleach_idx);
fprintf("Found %d binding and %d bleaching steps (with threshold %.1f).\n", ...
numel(bind_idx), numel(bleach_idx), min_jump);
%--------------------------------------------
% 5. 保存结果
%--------------------------------------------
out_name = sprintf('equal_levels_track_%s.mat', ...
ternary(isnan(track_id), 'X', num2str(track_id)));
fprintf("Saving equal-level analysis to %s\n", out_name);
% Save them in the .mat file
%save(out_name, ...
% 't', 'z', 'm_mod', ...
% 'L_best', 'levels_norm', ...
% 'state', 'm_step', 'z_step', ...
% 'bleach_idx', 'bleach_times', ...
% 'track_id');
save(out_name, ...
't', 'z', 'm_mod', ...
'L_best', 'levels_norm', ...
'state', 'm_step', 'z_step', ...
'bind_idx', 'bind_times', ...
'bleach_idx', 'bleach_times', ...
'track_id');
fprintf("Done.\n");plot_fig4AB_python.py
#!/usr/bin/env python3
"""
plot_fig4AB_python.py
Usage:
python plot_fig4AB_python.py
<track_id> [L_best] [min_levels_bind] [min_levels_bleach] [dwell_min] [baseline_state_max]
Arguments
---------
<track_id> : int
e.g. 10 → uses icon_analysis_track_10_matlab.mat
[L_best] : int, optional
Number of equally spaced levels to use for the step-wise fit
in Fig. 4B. If omitted, default = 3.
[min_levels_bind] : int, optional
Minimum number of state levels for an upward jump to count as a
binding event. Default = 3.
[min_levels_bleach] : int, optional
Minimum number of state levels for a downward jump to count as a
bleaching step. Default = 1.
[dwell_min] : float, optional
Minimum allowed dwell time between a binding event and the NEXT
bleaching event. If Δt = t_bleach - t_bind < dwell_min, then:
- that binding is removed
- the paired bleaching event is also removed
Default = 0 (no dwell-based filtering).
[baseline_state_max] : int, optional
Highest state index (0-based) that is still considered "baseline /
unbound" before a binding jump.
Binding condition becomes:
dstate >= min_levels_bind AND state_before <= baseline_state_max
If omitted → no baseline constraint (any state can be a start).
Input file
----------
icon_analysis_track_
<track_id>_matlab.mat
Expected variables inside the .mat file:
t : time vector (1D, seconds)
z : photon counts (1D)
m_mod : ICON mean trajectory (1D), same length as t and z
Outputs
-------
1) Figure:
fig4AB_track_
<track_id>_L<L_best>.png
2) Event tables:
binding_bleach_events_track_
<track_id>_L<L_best>.csv
binding_bleach_events_track_
<track_id>_L<L_best>.xlsx (if pandas available)
"""
import sys
import os
import numpy as np
import matplotlib.pyplot as plt
from scipy.io import loadmat
# Try to import pandas for Excel output
try:
import pandas as pd
HAS_PANDAS = True
except ImportError:
HAS_PANDAS = False
print("[WARN] pandas not available: Excel output (.xlsx) will be skipped.")
def assign_to_levels(m_mod, levels):
"""
Map each point in m_mod to the nearest level.
Parameters
----------
m_mod : array-like, shape (N,)
Normalized ICON mean trajectory.
levels : array-like, shape (L,)
Candidate level values.
Returns
-------
state : ndarray, shape (N,)
Integer state indices in {0, 1, ..., L-1}.
"""
m_mod = np.asarray(m_mod).ravel()
levels = np.asarray(levels).ravel()
diff = np.abs(m_mod[:, None] - levels[None, :]) # (N, L)
state = np.argmin(diff, axis=1) # 0..L-1
return state
def detect_binding_bleach_from_state(
z_step,
t,
state,
levels,
min_levels_bind=3,
min_levels_bleach=1,
baseline_state_max=None,
):
"""
Detect binding (big upward state jumps) and bleaching (downward jumps)
using the discrete state sequence.
- Binding: large upward jump (>= min_levels_bind) starting from a
"baseline" state (state_before <= baseline_state_max) if that
parameter is given.
- Bleaching: downward jump (<= -min_levels_bleach)
Parameters
----------
z_step : array-like, shape (N,)
Step-wise photon counts.
t : array-like, shape (N,)
Time vector (seconds).
state : array-like, shape (N,)
Integer states 0..L-1 from assign_to_levels().
levels : array-like, shape (L,)
Level values (in m_mod space, not directly photon counts).
min_levels_bind : int
Minimum number of levels for an upward jump to be
considered a binding event.
min_levels_bleach : int
Minimum number of levels for a downward jump to be
considered a bleaching event.
baseline_state_max : int or None
Highest state index considered "baseline" before binding.
If None, any state can be the start of a binding jump.
Returns
-------
bind_idx, bleach_idx : np.ndarray of indices
bind_times, bleach_times : np.ndarray of times (seconds)
bind_values, bleach_values : np.ndarray of photon counts at those events
bind_state_before, bind_state_after : np.ndarray of integer states
bleach_state_before, bleach_state_after : np.ndarray of integer states
"""
z_step = np.asarray(z_step).ravel()
t = np.asarray(t).ravel()
state = np.asarray(state).ravel()
levels = np.asarray(levels).ravel()
N = len(t)
dstate = np.diff(state) # length N-1
idx = np.arange(N - 1)
# ----- Binding: big upward jump, optionally from baseline only -----
bind_mask = (dstate >= min_levels_bind)
if baseline_state_max is not None:
bind_mask &= (state[idx] <= baseline_state_max)
bind_idx = idx[bind_mask] + 1
# ----- Bleaching: downward jump -----
bleach_mask = (dstate <= -min_levels_bleach)
bleach_idx = idx[bleach_mask] + 1
bind_times = t[bind_idx]
bleach_times = t[bleach_idx]
bind_values = z_step[bind_idx]
bleach_values = z_step[bleach_idx]
bind_state_before = state[bind_idx - 1]
bind_state_after = state[bind_idx]
bleach_state_before = state[bleach_idx - 1]
bleach_state_after = state[bleach_idx]
return (
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
)
def filter_short_episodes_by_dwell(
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
dwell_min,
):
"""
Remove short binding episodes based on dwell time and also remove
the paired bleaching step.
Rule:
For each binding, find the first bleaching with t_bleach > t_bind.
If Δt = t_bleach - t_bind < dwell_min, then:
- remove this binding
- remove this bleaching
All other bleaching events remain.
Returns
-------
(filtered binding arrays, dwell_times) + filtered bleaching arrays
"""
if dwell_min <= 0 or len(bind_idx) == 0 or len(bleach_idx) == 0:
# no filtering requested or missing events
dwell_times = np.full(len(bind_idx), np.nan)
for i in range(len(bind_idx)):
future = np.where(bleach_times > bind_times[i])[0]
if len(future) > 0:
dwell_times[i] = bleach_times[future[0]] - bind_times[i]
return (
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
dwell_times,
)
bind_idx = np.asarray(bind_idx)
bleach_idx = np.asarray(bleach_idx)
bind_times = np.asarray(bind_times)
bleach_times = np.asarray(bleach_times)
bind_values = np.asarray(bind_values)
bleach_values = np.asarray(bleach_values)
bind_state_before = np.asarray(bind_state_before)
bind_state_after = np.asarray(bind_state_after)
bleach_state_before = np.asarray(bleach_state_before)
bleach_state_after = np.asarray(bleach_state_after)
keep_bind = np.ones(len(bind_idx), dtype=bool)
keep_bleach = np.ones(len(bleach_idx), dtype=bool)
dwell_times = np.full(len(bind_idx), np.nan)
removed_bind = 0
removed_bleach = 0
for i in range(len(bind_idx)):
t_b = bind_times[i]
future = np.where(bleach_times > t_b)[0]
if len(future) == 0:
# no bleaching afterwards → dwell undefined, keep binding
dwell_times[i] = np.nan
continue
j = future[0]
dt = bleach_times[j] - t_b
dwell_times[i] = dt
if dt < dwell_min:
# remove this binding and its paired bleaching
keep_bind[i] = False
if keep_bleach[j]:
keep_bleach[j] = False
removed_bleach += 1
removed_bind += 1
print(
f"[INFO] Dwell-based filter: removed {removed_bind} binding(s) and "
f"{removed_bleach} paired bleaching step(s) with Δt < {dwell_min} s; "
f"{np.sum(keep_bind)} binding(s) and {np.sum(keep_bleach)} bleaching step(s) kept."
)
# Apply masks
bind_idx = bind_idx[keep_bind]
bind_times = bind_times[keep_bind]
bind_values = bind_values[keep_bind]
bind_state_before = bind_state_before[keep_bind]
bind_state_after = bind_state_after[keep_bind]
dwell_times = dwell_times[keep_bind]
bleach_idx = bleach_idx[keep_bleach]
bleach_times = bleach_times[keep_bleach]
bleach_values = bleach_values[keep_bleach]
bleach_state_before = bleach_state_before[keep_bleach]
bleach_state_after = bleach_state_after[keep_bleach]
return (
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
dwell_times,
)
def plot_fig4AB(
track_id,
L_best=None,
min_levels_bind=3,
min_levels_bleach=1,
dwell_min=0.0,
baseline_state_max=None,
):
# --------------------------
# 1. Load ICON analysis file
# --------------------------
mat_file = f"icon_analysis_track_{track_id}_matlab.mat"
print(f"Loading {mat_file}")
if not os.path.exists(mat_file):
raise FileNotFoundError(f"{mat_file} does not exist in current directory.")
data = loadmat(mat_file)
def extract_vector(name):
if name not in data:
raise KeyError(f"Variable '{name}' not found in {mat_file}")
v = data[name]
return np.squeeze(v)
t = extract_vector("t")
z = extract_vector("z")
m_mod = extract_vector("m_mod")
if not (len(t) == len(z) == len(m_mod)):
raise ValueError("t, z, and m_mod must have the same length.")
N = len(t)
print(f"Track {track_id}: N = {N} points")
# --------------------------
# 2. Choose L (number of levels)
# --------------------------
if L_best is None:
L_best = 3 # fallback default
print(f"No L_best provided, using default L_best = {L_best}")
else:
print(f"Using user-specified L_best = {L_best}")
m_min = np.min(m_mod)
m_max = np.max(m_mod)
levels = np.linspace(m_min, m_max, L_best)
print(f"m_mod range: [{m_min:.4f}, {m_max:.4f}]")
print(f"Equally spaced levels ({L_best}): {levels}")
# --------------------------
# 3. Build step-wise trajectory from m_mod
# --------------------------
state = assign_to_levels(m_mod, levels) # 0..L_best-1
m_step = levels[state]
# Map back to photon counts via linear fit z ≈ a*m_step + b
A = np.column_stack([m_step, np.ones(N)])
theta, *_ = np.linalg.lstsq(A, z, rcond=None)
a, b = theta
z_step = A @ theta
print(f"Fitted z ≈ a * m_step + b with a = {a:.4f}, b = {b:.4f}")
# --------------------------
# 4. Detect binding / bleaching events (state-based)
# --------------------------
(
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
) = detect_binding_bleach_from_state(
z_step,
t,
state,
levels,
min_levels_bind=min_levels_bind,
min_levels_bleach=min_levels_bleach,
baseline_state_max=baseline_state_max,
)
base_msg = (
f"baseline_state_max={baseline_state_max}"
if baseline_state_max is not None
else "baseline_state_max=None (no baseline restriction)"
)
print(
f"Initial detection: {len(bind_idx)} binding and {len(bleach_idx)} bleaching "
f"events (min_levels_bind={min_levels_bind}, "
f"min_levels_bleach={min_levels_bleach}, {base_msg})."
)
# --------------------------
# 5. Apply dwell-time filter to binding + paired bleach
# --------------------------
(
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
dwell_times,
) = filter_short_episodes_by_dwell(
bind_idx,
bleach_idx,
bind_times,
bleach_times,
bind_values,
bleach_values,
bind_state_before,
bind_state_after,
bleach_state_before,
bleach_state_after,
dwell_min=dwell_min,
)
print(
f"After dwell filter (dwell_min={dwell_min}s): "
f"{len(bind_idx)} binding and {len(bleach_idx)} bleaching events remain."
)
# --------------------------
# 6. Build event table & save CSV / Excel
# --------------------------
rows = []
# Binding events
for i in range(len(bind_idx)):
idx = int(bind_idx[i])
rows.append({
"event_type": "binding",
"sample_index": idx,
"time_seconds": float(bind_times[i]),
"photon_count": float(bind_values[i]),
"state_before": int(bind_state_before[i]),
"state_after": int(bind_state_after[i]),
"level_before_norm": float(levels[bind_state_before[i]]),
"level_after_norm": float(levels[bind_state_after[i]]),
"dwell_time": float(dwell_times[i]) if not np.isnan(dwell_times[i]) else "",
})
# Bleaching events
for i in range(len(bleach_idx)):
idx = int(bleach_idx[i])
rows.append({
"event_type": "bleach",
"sample_index": idx,
"time_seconds": float(bleach_times[i]),
"photon_count": float(bleach_values[i]),
"state_before": int(bleach_state_before[i]),
"state_after": int(bleach_state_after[i]),
"level_before_norm": float(levels[bleach_state_before[i]]),
"level_after_norm": float(levels[bleach_state_after[i]]),
"dwell_time": "",
})
# Sort by time
rows = sorted(rows, key=lambda r: r["time_seconds"])
# Write CSV
csv_name = f"binding_bleach_events_track_{track_id}_L{L_best}.csv"
import csv
with open(csv_name, "w", newline="") as f:
writer = csv.DictWriter(
f,
fieldnames=[
"event_type",
"sample_index",
"time_seconds",
"photon_count",
"state_before",
"state_after",
"level_before_norm",
"level_after_norm",
"dwell_time",
],
)
writer.writeheader()
for r in rows:
writer.writerow(r)
print(f"Saved event table to {csv_name}")
# Write Excel (if pandas available)
if HAS_PANDAS:
df = pd.DataFrame(rows)
xlsx_name = f"binding_bleach_events_track_{track_id}_L{L_best}.xlsx"
df.to_excel(xlsx_name, index=False)
print(f"Saved event table to {xlsx_name}")
else:
print("[INFO] pandas not installed → skipped Excel (.xlsx) output.")
# --------------------------
# 7. Make a figure similar to Fig. 4A + 4B
# --------------------------
fig, axes = plt.subplots(2, 1, figsize=(7, 6), sharex=True)
# ---- Fig. 4A-like: raw intensity vs time ----
ax1 = axes[0]
ax1.plot(t, z, color="black", linewidth=0.8)
ax1.set_ylabel("Photon counts")
ax1.set_title(f"Track {track_id}: raw intensity") #(Fig. 4A-like)
# ---- Fig. 4B-like: step-wise HMM fit vs time ----
ax2 = axes[1]
ax2.plot(t, z, color="0.8", linewidth=0.5, label="raw")
ax2.plot(t, z_step, color="red", linewidth=1.5,
label=f"equal levels (L={L_best})")
# Mark binding (up-steps) and bleaching (down-steps) AFTER filtering
if len(bind_idx) > 0:
ax2.scatter(
bind_times,
bind_values,
marker="^",
color="blue",
s=30,
label="binding",
)
if len(bleach_idx) > 0:
ax2.scatter(
bleach_times,
bleach_values,
marker="v",
color="black",
s=30,
label="bleach",
)
ax2.set_xlabel("Time (s)")
ax2.set_ylabel("Photon counts")
ax2.set_title(
f"Step-wise HMM fit (" #Fig. 4B-like,
f"min_bind_levels={min_levels_bind}, "
f"min_bleach_levels={min_levels_bleach}, "
f"dwell_min={dwell_min}s, "
f"baseline_state_max={baseline_state_max})"
)
ax2.legend(loc="best")
fig.tight_layout()
out_png = f"fig4AB_track_{track_id}_L{L_best}.png"
fig.savefig(out_png, dpi=300)
plt.close(fig)
print(f"Saved figure to {out_png}")
if __name__ == "__main__":
if len(sys.argv) < 2:
print("Usage: python plot_fig4AB_python.py
<track_id> [L_best] [min_levels_bind] [min_levels_bleach] [dwell_min] [baseline_state_max]")
sys.exit(1)
track_id = int(sys.argv[1])
# Defaults
L_best = None
min_levels_bind = 3
min_levels_bleach = 1
dwell_min = 0.0
baseline_state_max = None
if len(sys.argv) >= 3:
L_best = int(sys.argv[2])
if len(sys.argv) >= 4:
min_levels_bind = int(sys.argv[3])
if len(sys.argv) >= 5:
min_levels_bleach = int(sys.argv[4])
if len(sys.argv) >= 6:
dwell_min = float(sys.argv[5])
if len(sys.argv) >= 7:
baseline_state_max = int(sys.argv[6])
plot_fig4AB(
track_id,
L_best=L_best,
min_levels_bind=min_levels_bind,
min_levels_bleach=min_levels_bleach,
dwell_min=dwell_min,
baseline_state_max=baseline_state_max,
)icon_multimer_histogram.m
% icon_multimer_histogram.m
%
% 用法(终端):
% octave icon_multimer_histogram.m [pattern]
%
% 默认 pattern = "equal_levels_track_*.mat"
%
% 要求每个 mat 文件里至少有:
% state : 等间距 level 的索引 (1..L_best)
% t : 时间向量
% z : 原始 photon counts
% track_id : (可选) 轨迹编号,用于打印信息
%
% 输出:
% - 在命令行打印每条轨迹估计出的 multimer 数目
% - 生成 Fig. 4C 风格直方图:
% multimer_histogram.png
arg_list = argv();
if numel(arg_list) >= 1
pattern = arg_list{1};
else
pattern = "equal_levels_track_*.mat";
end
fprintf("Searching files with pattern: %s\n", pattern);
files = dir(pattern);
if isempty(files)
error("No files matched pattern: %s", pattern);
end
multimers = []; % 保存每条轨迹的 multimer size
track_ids = []; % 保存对应的 track_id(若存在)
fprintf("Found %d files.\n", numel(files));
for i = 1:numel(files)
fname = files(i).name;
fprintf("\nLoading %s ...\n", fname);
S = load(fname);
if ~isfield(S, "state") || ~isfield(S, "t")
warning(" File %s does not contain 'state' or 't'. Skipped.", fname);
continue;
end
state = S.state(:);
t = S.t(:);
N = numel(state);
if N < 5
warning(" File %s has too few points (N=%d). Skipped.", fname, N);
continue;
end
% 解析 track_id(如果有)
tr_id = NaN;
if isfield(S, "track_id")
tr_id = S.track_id;
else
% 尝试从文件名里解析
tokens = regexp(fname, 'equal_levels_track_([0-9]+)', 'tokens');
if ~isempty(tokens)
tr_id = str2double(tokens{1}{1});
end
end
% 取前 10%% 和后 10%% 时间段的 state 中位数
n_head = max(1, round(0.1 * N));
n_tail = max(1, round(0.1 * N));
head_idx = 1:n_head;
tail_idx = (N - n_tail + 1):N;
initial_state = round(median(state(head_idx)));
final_state = round(median(state(tail_idx)));
multimer_size = initial_state - final_state;
if multimer_size <= 0
fprintf(" Track %g: initial_state=%d, final_state=%d -> multimer_size=%d (ignored)\n", ...
tr_id, initial_state, final_state, multimer_size);
continue;
end
fprintf(" Track %g: initial_state=%d, final_state=%d -> multimer_size=%d\n", ...
tr_id, initial_state, final_state, multimer_size);
multimers(end+1,1) = multimer_size;
track_ids(end+1,1) = tr_id;
end
if isempty(multimers)
error("No valid multimer sizes estimated. Check your data or thresholds.");
end
% 像论文那样,可以选择去掉 monomer/dimer
fprintf("\nTotal %d events (including monomer/dimer).\n", numel(multimers));
% 可选:过滤掉 ≤2 的(monomer / dimer)
mask = multimers > 2;
multimers_filtered = multimers(mask);
fprintf("After removing monomer/dimer (<=2): %d events.\n", numel(multimers_filtered));
if isempty(multimers_filtered)
error("No events left after filtering monomer/dimer. Try including them.");
end
% 计算直方图
max_mult = max(multimers_filtered);
edges = 0.5:(max_mult + 0.5);
[counts, edges_out] = histcounts(multimers_filtered, edges);
centers = 1:max_mult;
% 画 Fig. 4C 风格的柱状图
figure;
bar(centers, counts, 'FaceColor', [0.2 0.6 0.8]); % 颜色随便,Octave 会忽略
xlabel('Number of mN-LT per origin (multimer size)');
ylabel('Frequency');
title('Distribution of multimer sizes (Fig. 4C-like)');
xlim([0.5, max_mult + 0.5]);
% 在每个柱子上标一下计数
hold on;
for k = 1:max_mult
if counts(k) > 0
text(centers(k), counts(k) + 0.1, sprintf('%d', counts(k)), ...
'HorizontalAlignment', 'center');
end
end
hold off;
print('-dpng', 'multimer_histogram.png');
fprintf("\n[INFO] Multimer histogram saved to multimer_histogram.png\n");Bioinformatics Pipelines for DNA Sequencing: From Raw Reads to Biological Insight
Abstract
English: Advances in DNA sequencing have revolutionized biology, but converting vast sequencing data into usable, robust biological knowledge depends on sophisticated bioinformatics. This review details computational strategies spanning all phases of DNA sequence analysis, starting from raw reads through to functional interpretation and reporting. It begins by characterizing the main sequencing platforms (short-read, long-read, targeted, and metagenomic), describes critical pipeline steps (sample tracking, quality control, read alignment, error correction, variant and structural variant detection, copy number analysis, de novo assembly), and considers the impact of reference genome choice and computational algorithms. Recent machine learning advances for variant annotation and integration with other omics are discussed, with applications highlighted in rare disease diagnostics, cancer genomics, and infectious disease surveillance. Emphasis is placed on reproducible, scalable, and well-documented pipelines using open-source tools, workflow management (Snakemake, Nextflow), containerization, versioning, and FAIR data principles. The review concludes with discussion of ongoing challenges (heterogeneous data, batch effects, benchmarking, privacy) and practical recommendations for robust, interpretable analyses for both experimental biologists and computational practitioners.
Chinese: DNA测序的持续进步彻底改变了生物学和医学研究,而要将庞大的测序数据转化为可靠的生物学知识,则高度依赖高水平的生物信息学方法。本文详细介绍了DNA序列分析全流程的主流计算策略,涵盖原始reads到功能性注释乃至标准化报告的各个环节。首先评述主流测序技术平台(短读长读、靶向、宏基因组),系统阐述实验设计、样本追踪、数据质控、比对、纠错、变异与结构变异检测、拷贝数分析和de novo组装等流程要点,并分析参考基因组和比对算法对结果的影响。文章还总结了机器学习在变异注释、多组学整合中的最新进展,结合罕见病诊断、肿瘤基因组和病原体监测等实际案例深入说明其应用场景。着重强调可重复、高效、透明的分析流程,包括开源工具、流程管理(Snakemake、Nextflow)、容器化、版本管理与FAIR原则。最后讨论了异质数据、批次效应、评测标准和隐私保护等挑战,并为实验与计算生物学研究者提供实用建议。
Detailed Structure \& Outline
- Introduction
- Historical overview of DNA sequencing and bioinformatics development
- The necessity of bioinformatics for handling scale, complexity, and error sources in modern sequence data
- Scope: DNA focus (excluding RNA/proteome)
- DNA Sequencing Technologies \& Data Properties
- 2.1 Short-read platforms (e.g., Illumina): read length, quality, use cases
- 2.2 Long-read platforms (PacBio, Nanopore): strengths, error profiles, applications
- 2.3 Specialized applications: targeted/exome panels, metagenomics, amplicon/barcode-based diagnostics
- Core Bioinformatics Pipeline Components
- 3.1 Experimental metadata, sample barcoding, batch tracking: Crucial for reproducibility and QC
- 3.2 Raw read QC: base quality, adapter/contaminant trimming, typical software/plots
- 3.3 Read alignment/mapping: reference choice (GRCh38, hg19), algorithmic details (FM-index, seed-and-extend), uniqueness/multimapping
- 3.4 Post-alignment processing: file sorting, duplicate marking, base recalibration, depth analysis
- 3.5 Variant calling: SNVs/indels, somatic vs germline separation, quality filters and validation strategies
- 3.6 Structural variant and CNV analysis: breakpoints, split/discordant reads, long-read tools
- 3.7 De novo assembly, polishing, and consensus generation where relevant
- Functional Interpretation
- 4.1 Annotation: gene models, regulatory regions, predictive algorithms and public databases
- 4.2 Multi-omics integration: joint analysis of genome, epigenome, transcriptome, regulatory networks
- 4.3 Machine learning/AI approaches: variant scoring, prioritization, deep learning for sequence features
- Reproducible and Scalable Workflows
- 5.1 Pipeline frameworks: Snakemake, Nextflow, CWL, and workflow description languages
- 5.2 Containerization: Docker, Singularity for reproducible deployments
- 5.3 Version control/documentation: workflow hubs, deployment on GitHub, FAIR-compliant reporting
- 5.4 Data management: standard formats (FASTQ/BAM/CRAM/VCF), secure storage, metadata
- Applications \& Case Studies
- Rare disease genomics: WGS for diagnosis
- Cancer genomics: tumor heterogeneity, evolution, therapy response
- Pathogen surveillance: rapid outbreak detection, resistance tracking
- Other applications to match research interests
- Challenges and Future Prospects
- Technical: population-scale analysis, batch correction, pangenomes, benchmarking complexities
- Practical: workflow sharing, legal/ethical/privacy issues
- Methodological: handling new sequencing chemistries, multi-modal omics
- Conclusions
- Recap essential lessons
- Actionable recommendations for robust design and execution
- Prospects for further automation, integration, and clinical translation
Section Opening (English / 中文): High-throughput DNA sequencing has fundamentally transformed modern genomics, enabling detailed investigation of human diseases, microbial ecology, and evolution. However, the raw output—massive quantities of short or long reads—is only the starting point; extracting meaningful, robust insights requires optimized bioinformatics pipelines that ensure data integrity and biological relevance.
高通量DNA测序极大地推动了现代基因组学,助力人类疾病、微生物生态与进化等领域的深入探索。但测序仪输出的原始reads只是起点——要获得有意义、可靠的生物学结论,必须依赖优化的生物信息学流程以保证数据质量和生物学解释的可信度。
Interpreting Binding Events in Single-Molecule Fluorescence Tracking: Conceptual Clarification and Practical Proposal
- Concept: is the start of a track a binding event? From your plot: Each track appears suddenly at some time, then decays / fluctuates and disappears. The accumulated trace shows clear upward steps at those times. In typical kymograph / spot-tracking data: A track is created when the tracking algorithm first detects a fluorescent spot above background. That is usually when a fluorophore binds (or comes into focus / into the ROI). The end of a track is when the spot disappears, often due to bleaching or unbinding. So, conceptually: Yes, in many experiments the first time point of a track is a natural candidate for the binding time of that molecule. But there are a few caveats you might want to mention to Vero: Some molecules may already be bound before the camera started recording – in that case the first frame is not the true binding time, just the first time you see it. Detection thresholds may delay the detected “start” by a few frames. If two tracks overlap in time, the accumulated signal will have multiple steps, and the HMM sees “binding” whenever the sum intensity increases. That also explains your observations: Per-track HMM: each trace is relatively short and noisy → few clear transitions, so the HMM doesn’t detect many events. HMM on the accumulated signal: all tracks summed → big, clean steps when a new molecule appears → HMM picks up almost all track starts as binding events. So it’s very reasonable to treat track starts as a kind of “ground truth” binding time, at least for benchmarking.
- Improved text you can send to Vero Here’s a cleaned-up version of what you wrote, phrased as a question: From the plots of the individual tracks and the accumulated photon counts (see attached figure), I have a conceptual question about how to define the binding time. In the track-intensity file, each track appears at some time point and then decays. Would you consider the first time point of each track to be the “binding time” of that molecule? To me this looks like a very natural definition, because the track starts exactly when the signal first appears. Note: – When I run the HMM on individual tracks, only very few binding events are detected. – When I run the HMM on the accumulated signal, I can detect almost all of the track starts as binding events. This makes me think that the track starting times might be a good gold standard for binding time points. Does this interpretation match your understanding of the experiment? If you want I can also help you write a follow-up mail where you propose to use “track start = binding” and “track end = bleaching/unbinding” as ground truth for validating your HMM.
Here is a concise bilingual summary of your text explaining the idea and a cleaned-up proposal to send to Vero:
English Summary
- Concept: Is the start of a track a binding event?
- Each fluorescent track appears suddenly, fluctuates, then disappears; cumulative traces show clear stepwise increments.
- A track starts when a fluorophore binds or enters detection (fluorescent) focus, and ends due to bleaching or unbinding.
- Thus, the start of a track commonly represents the molecule’s binding time.
- Caveats:
- Some molecules may be bound before recording starts, so first observed frame is not true binding time.
- Detection thresholds may delay start detection.
- Overlapping tracks cause summed signal steps, and HMM detects any increase as binding.
- Per-track HMM sees few transitions due to noise and short traces; summed HMM on total signal detects most binding events.
- Cleaned-up question to Vero: “From individual and cumulative photon count plots (see attached), does the first time point of each track represent the molecule’s binding time?
- HMM on single tracks detects very few events;
- HMM on accumulated trace detects nearly all track starts as bindings;
- This suggests track start time is a good benchmark for binding. Does this match your understanding? I can help draft a follow-up email proposing to use ‘track start = binding’ and ‘track end = bleaching/unbinding’ as ground truth for HMM validation.”
中文总结
- 概念解释:轨迹开始是否代表结合事件?
- 每条轨迹突然出现,随后衰减或波动后消失,累积轨迹中出现明显的阶梯式上升。
- 轨迹的开始通常是荧光标记分子结合或进入焦点检测区域的时刻,结束则因漂白或解离。
- 因此轨迹开始时间通常被视为该分子结合的时间点。
- 注意事项:
- 某些分子可能在录像开始前已结合,首帧不是准确结合时间;
- 检测阈值可能导致结合时间存在几十帧延迟;
- 轨迹重叠造成信号累积,HMM会将信号上升视为结合事件。
- 针对单条轨迹的HMM因噪声和轨迹短小,事件检测少;对累积信号的HMM检测到几乎所有结合。
- 整理后可以发给Vero的文本: “根据个别轨迹和累积光子计数图(见附件),我有个问题:是否可以将每条轨迹的首个时间点视为该分子的结合时刻?
- 针对单轨迹的HMM只检测到少量结合事件;
- 针对累积轨迹的HMM则几乎检测到所有轨迹期初的结合;
- 这可能说明轨迹起始时间是一个可靠的结合时间‘黄金标准’。 这种理解符合你的看法吗?我也可以帮你起草一封邮件,提议用“轨迹开始=结合”,“轨迹结束=漂白/解离”作为验证HMM的真实标准。”
-
https://www.sciencedirect.com/science/article/pii/S0022283625003742 ↩
-
https://phys.libretexts.org/Courses/University_of_California_Davis/Biophysics_241:_Membrane_Biology/05:_Experimental_Characterization_-_Spectroscopy_and_Microscopy/5.07:_Single_Molecule_Tracking ↩
-
https://royalsocietypublishing.org/doi/10.1098/rsob.210383 ↩
使用 ICON HMM 在 Octave 中自动检测单分子 binding/bleach 事件并估计 dwell time
% detect_binding_bleach.m
% 用法(在终端):
% octave detect_binding_bleach.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv
%--------------------------------------------
% 基本设置
%--------------------------------------------
arg_list = argv();
if numel(arg_list) < 1
error("Usage: octave detect_binding_bleach.m
<track_intensity_csv>");
end
input_file = arg_list{1};
% 加载 HMM 采样器
%addpath("sampler_SRC");
% 如果安装了 statistics 包,用于 kmeans
try
pkg load statistics;
catch
warning("Could not load 'statistics' package. Make sure it is installed if kmeans is missing.");
end
%--------------------------------------------
% 读入 track intensity 文件(分号分隔)
%--------------------------------------------
fprintf("Reading file: %s\n", input_file);
fid = fopen(input_file, "r");
if fid < 0
error("Cannot open file: %s", input_file);
end
% 第一行是注释头 "# track index;time (seconds);track intensity (photon counts)"
header_line = fgetl(fid); % 忽略内容,只是读掉这一行
% 后面每行: track_index;time_sec;intensity
data = textscan(fid, "%f%f%f", "Delimiter", ";");
fclose(fid);
track_idx = data{1};
time_sec = data{2};
counts = data{3};
% 排序(确保同一个 track 内按时间排序)
[~, order] = sortrows([track_idx, time_sec], [1, 2]);
track_idx = track_idx(order);
time_sec = time_sec(order);
counts = counts(order);
tracks = unique(track_idx);
n_tracks = numel(tracks);
fprintf("Found %d tracks.\n", n_tracks);
%--------------------------------------------
% 结果结构体
%--------------------------------------------
results = struct( ...
"track_id", {}, ...
"binding_indices", {}, ...
"binding_times", {}, ...
"bleach_indices", {}, ...
"bleach_times", {} );
%--------------------------------------------
% 循环每个 track,跑 HMM + 找事件
%--------------------------------------------
for ti = 1:n_tracks
tr = tracks(ti);
fprintf("\n===== Track %d =====\n", tr);
sel = (track_idx == tr);
t = time_sec(sel);
z = counts(sel);
z = z(:); % 列向量
%-------------------------
% 1) 设定 ICON HMM 的参数
%-------------------------
opts = struct();
% 超参数(和原 MATLAB 代码一致)
opts.a = 1;
opts.g = 1;
opts.Q(1) = mean(z);
opts.Q(2) = 1 / (std(z)^2 + eps); % 防止除零
opts.Q(3) = 0.1;
opts.Q(4) = 0.00001;
opts.M = 10;
% 采样参数
opts.dr_sk = 1;
opts.K_init = 50;
flag_stat = true;
flag_anim = false; % 建议在 Octave 里关掉动画更稳定
R = 1000; % 采样次数,可根据需要调小/调大
%-------------------------
% 2) 运行采样器
%-------------------------
fprintf(" Running HMM sampler...\n");
chain = chainer_main(z, R, [], opts, flag_stat, flag_anim);
%-------------------------
% 3) 做后验分析,得到平滑后的光子水平轨迹
%-------------------------
fr = 0.25; % burn-in 比例
dr = 2; % sample 步距
m_min = 0;
m_max = 1;
m_num = 25;
[m_mod, m_red] = chainer_analyze_means(chain, fr, dr, m_min, m_max, m_num, z);
% m_mod 是一个和 z 同长度的向量(通常是 [0,1] 范围的归一化水平)
m_mod = m_mod(:);
%-------------------------
% 4) 把 m_mod 聚类成 3 个光子强度状态
% 状态 1 = 最低强度 (dark / unbound / bleached)
% 状态 3 = 最高强度 (bound)
%-------------------------
K = 3; % 你可以改成 2 或其他
try
[idx_raw, centers] = kmeans(m_mod, K);
catch
% 如果没 kmeans,就简单按数值分位数来分三类
warning("kmeans not available, using simple quantile-based clustering.");
q1 = quantile(m_mod, 1/3);
q2 = quantile(m_mod, 2/3);
idx_raw = ones(size(m_mod));
idx_raw(m_mod > q1 & m_mod <= q2) = 2;
idx_raw(m_mod > q2) = 3;
centers = zeros(K,1);
for kk = 1:K
centers(kk) = mean(m_mod(idx_raw == kk));
end
end
% 根据中心值从小到大重排状态编号
[~, order_centers] = sort(centers);
state_seq = zeros(size(idx_raw));
for k = 1:K
state_seq(idx_raw == order_centers(k)) = k;
end
low_state = 1;
high_state = K;
%-------------------------
% 5) 检测 binding / bleach 跳变
% binding: low -> high
% bleach : high -> low
%-------------------------
s = state_seq(:);
% 前一时刻和后一时刻
s_prev = s(1:end-1);
s_next = s(2:end);
bind_idx = find(s_prev == low_state & s_next == high_state) + 1;
bleach_idx = find(s_prev == high_state & s_next == low_state) + 1;
bind_times = t(bind_idx);
bleach_times = t(bleach_idx);
fprintf(" Found %d binding event(s) and %d bleaching event(s).\n", ...
numel(bind_idx), numel(bleach_idx));
% 存入结果
results(ti).track_id = tr;
results(ti).binding_indices = bind_idx;
results(ti).binding_times = bind_times;
results(ti).bleach_indices = bleach_idx;
results(ti).bleach_times = bleach_times;
end
%--------------------------------------------
% 6) 把结果写成 CSV
%--------------------------------------------
out_csv = "binding_bleach_events.csv";
fid_out = fopen(out_csv, "w");
if fid_out < 0
error("Cannot open output file: %s", out_csv);
end
fprintf(fid_out, "track_index,event_type,sample_index,time_seconds\n");
for ti = 1:numel(results)
tr = results(ti).track_id;
% binding
for k = 1:numel(results(ti).binding_indices)
fprintf(fid_out, "%d,binding,%d,%.6f\n", tr, ...
results(ti).binding_indices(k), ...
results(ti).binding_times(k));
end
% bleach
for k = 1:numel(results(ti).bleach_indices)
fprintf(fid_out, "%d,bleach,%d,%.6f\n", tr, ...
results(ti).bleach_indices(k), ...
results(ti).bleach_times(k));
end
end
fclose(fid_out);
fprintf("\nAll done. Events written to: %s\n", out_csv);- 运行:
octave detect_binding_bleach.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv- 保留原始 ICON 后验分析:
chainer_export导出样本chainer_analyze_means得到m_modchainer_analyze_transitions得到m_edges, p_mean, d_distchainer_analyze_drift得到y_mean, y_std
- 使用 binding/bleach 时间点 计算 dwell time 分布并做单指数 rate 拟合,并直接画直方图 + 指数拟合曲线。
- 输出一个 CSV 表,包含所有 binding/bleach 时间点(这是最重要的输出)。
说明:
- 代码仍然基于你已有的
detect_binding_bleach.m结构,假定当前目录下有sampler_SRC(含chainer_main.m、chainer_analyze_means.m、chainer_analyze_transitions.m、chainer_analyze_drift.m等)。1819- Dwell time 这里使用“连续 binding 事件之间的时间差”来估计 bound-state dwell time(简化版);如果 bleaching 明确是轨迹末端终止,也可以用
bleach_time - last_binding_time作为一个 right‑censored dwell(这里暂不做 censoring 修正,只输出原始分布和单指数拟合)。
完整脚本:detect_binding_bleach_dwell_simple.m
% detect_binding_bleach_dwell.m
%
% 用法(终端):
% octave detect_binding_bleach_dwell.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv
%
% 输入 CSV 格式(分号分隔):
% # track index;time (seconds);track intensity (photon counts)
% 1;0.00;123
% 1;0.05;118
% 2;0.00; 95
% ...
%--------------------------------------------
% 0. 基本设置
%--------------------------------------------
arg_list = argv();
if numel(arg_list) < 1
error("Usage: octave detect_binding_bleach_dwell.m
<input_csv>");
end
input_file = arg_list{1};
% 加载 HMM 采样器代码(确保 sampler_SRC 在当前目录下)
addpath("sampler_SRC");
% 如果安装了 statistics 包,用于 kmeans
try
pkg load statistics;
catch
warning("Could not load 'statistics' package. Make sure it is installed if kmeans is missing.");
end
%--------------------------------------------
% 1. 读入 track intensity 文件(分号分隔)
%--------------------------------------------
fprintf("Reading file: %s\n", input_file);
fid = fopen(input_file, "r");
if fid < 0
error("Cannot open file: %s", input_file);
end
% 第一行是注释头 "# track index;time (seconds);track intensity (photon counts)"
header_line = fgetl(fid); % 忽略内容,只是读掉这一行
% 后面每行: track_index;time_sec;intensity
data = textscan(fid, "%f%f%f", "Delimiter", ";");
fclose(fid);
track_idx = data{1};
time_sec = data{2};
counts = data{3};
% 排序(确保同一个 track 内按时间排序)
[~, order] = sortrows([track_idx, time_sec], [1, 2]);
track_idx = track_idx(order);
time_sec = time_sec(order);
counts = counts(order);
tracks = unique(track_idx);
n_tracks = numel(tracks);
fprintf("Found %d tracks.\n", n_tracks);
%--------------------------------------------
% 2. 结果结构体(binding / bleach 时间点)
%--------------------------------------------
results = struct( ...
"track_id", {}, ...
"binding_indices", {}, ...
"binding_times", {}, ...
"bleach_indices", {}, ...
"bleach_times", {} );
% 用于 dwell time 收集的数组
all_binding_times = []; % 所有 track 的 binding time 合并
all_bleach_times = []; % 所有 track 的 bleach time 合并
%--------------------------------------------
% 3. 循环每个 track,跑 HMM + ICON 分析 + 事件检测
%--------------------------------------------
for ti = 1:n_tracks
tr = tracks(ti);
fprintf("\n===== Track %d =====\n", tr);
sel = (track_idx == tr);
t = time_sec(sel);
z = counts(sel);
z = z(:); % 列向量
%-------------------------
% 3.1 设定 ICON HMM 的参数
%-------------------------
opts = struct();
% 超参数(与 level_analysis.m 一致)
opts.a = 1;
opts.g = 1;
opts.Q(1) = mean(z);
opts.Q(2) = 1 / (std(z)^2 + eps); % 防止 std(z)=0 除零
opts.Q(3) = 0.1;
opts.Q(4) = 0.00001;
opts.M = 10;
% 采样参数
opts.dr_sk = 1;
opts.K_init = 50;
flag_stat = true;
flag_anim = false; % 建议在 Octave 里关掉动画
R = 1000; % 采样次数,可按需求调整
%-------------------------
% 3.2 运行采样器(ICON HMM)
%-------------------------
fprintf(" Running HMM sampler...\n");
chain = chainer_main(z, R, [], opts, flag_stat, flag_anim);
%-------------------------
% 3.3 ICON 后验分析(与 level_analysis.m 等价)
%-------------------------
fr = 0.25; % burn-in 比例
dr = 2; % sample 步距
m_min = 0;
m_max = 1;
m_num = 25;
% (1) 均值轨迹:m_mod
[m_mod, m_red] = chainer_analyze_means(chain, fr, dr, m_min, m_max, m_num, z);
m_mod = m_mod(:);
% (2) 导出 samples(为每条 track 单独存一个文件)
sample_file = sprintf("samples_track_%d", tr);
chainer_export(chain, fr, dr, sample_file, "mat");
% (3) 转移概率 / 跃迁统计
[m_edges, p_mean, d_dist] = chainer_analyze_transitions( ...
chain, fr, dr, m_min, m_max, m_num, true);
% (4) 漂移分析
[y_mean, y_std] = chainer_analyze_drift(chain, fr, dr, z);
% 你可以根据需要保存这些 ICON 分析结果
mat_out = sprintf("icon_analysis_track_%d.mat", tr);
save(mat_out, "m_mod", "m_red", "m_edges", "p_mean", "d_dist", ...
"y_mean", "y_std", "t", "z");
%-------------------------
% 3.4 把 m_mod 聚类成 3 个光子强度状态(用于事件检测)
%-------------------------
K = 3; % 可按物理需求修改
try
[idx_raw, centers] = kmeans(m_mod, K);
catch
warning("kmeans not available, using simple quantile-based clustering.");
q1 = quantile(m_mod, 1/3);
q2 = quantile(m_mod, 2/3);
idx_raw = ones(size(m_mod));
idx_raw(m_mod > q1 & m_mod <= q2) = 2;
idx_raw(m_mod > q2) = 3;
centers = zeros(K,1);
for kk = 1:K
centers(kk) = mean(m_mod(idx_raw == kk));
end
end
% 根据中心值从小到大重排状态编号,使 state_seq = 1..K 对应 low->high
[~, order_centers] = sort(centers);
state_seq = zeros(size(idx_raw));
for k = 1:K
state_seq(idx_raw == order_centers(k)) = k;
end
low_state = 1;
high_state = K;
%-------------------------
% 3.5 检测 binding / bleaching
%
% binding: low_state -> high_state
% bleach : high_state -> low_state
%-------------------------
s = state_seq(:);
s_prev = s(1:end-1);
s_next = s(2:end);
bind_idx = find(s_prev == low_state & s_next == high_state) + 1;
bleach_idx = find(s_prev == high_state & s_next == low_state) + 1;
bind_times = t(bind_idx);
bleach_times = t(bleach_idx);
fprintf(" Found %d binding event(s) and %d bleaching event(s).\n", ...
numel(bind_idx), numel(bleach_idx));
% 存入结果结构体
results(ti).track_id = tr;
results(ti).binding_indices = bind_idx;
results(ti).binding_times = bind_times;
results(ti).bleach_indices = bleach_idx;
results(ti).bleach_times = bleach_times;
% 用于全局 dwell-time 统计
all_binding_times = [all_binding_times; bind_times(:)];
all_bleach_times = [all_bleach_times; bleach_times(:)];
end
%--------------------------------------------
% 4. 输出 binding/bleach 时间点的总表(最重要输出)
%--------------------------------------------
out_csv = "binding_bleach_events.csv";
fid_out = fopen(out_csv, "w");
if fid_out < 0
error("Cannot open output file: %s", out_csv);
end
fprintf(fid_out, "track_index,event_type,sample_index,time_seconds\n");
for ti = 1:numel(results)
tr = results(ti).track_id;
% binding
for k = 1:numel(results(ti).binding_indices)
fprintf(fid_out, "%d,binding,%d,%.6f\n", tr, ...
results(ti).binding_indices(k), ...
results(ti).binding_times(k));
end
% bleach
for k = 1:numel(results(ti).bleach_indices)
fprintf(fid_out, "%d,bleach,%d,%.6f\n", tr, ...
results(ti).bleach_indices(k), ...
results(ti).bleach_times(k));
end
end
fclose(fid_out);
fprintf("\n[INFO] Binding/bleach events written to: %s\n", out_csv);
%--------------------------------------------
% 5. Dwell-time 直方图 + rate 拟合
%
% 简化假设:
% - 把每个 track 中连续 binding 事件之间的时间差视为
% bound state dwell time(中间没有 bleaching,就重新 binding)。
% - 如果有明确的最后一个 binding 到 bleaching 的时间,
% 你也可以扩展这里把它们当作 dwell time 加进去。
%--------------------------------------------
% 5.1 收集 bound-state dwell times(简单用 binding 时间差)
dwell_times = [];
for ti = 1:numel(results)
bt = sort(results(ti).binding_times(:)); % 单个 track 的 binding times
if numel(bt) >= 2
dt = diff(bt); % 相邻 binding 之间的时间差
dwell_times = [dwell_times; dt];
end
% 可选:如果你想把最后一个 binding -> 第一个 bleach 也算入 dwell:
% if ~isempty(results(ti).binding_times) && ~isempty(results(ti).bleach_times)
% last_binding = max(results(ti).binding_times);
% first_bleach = min(results(ti).bleach_times);
% if first_bleach > last_binding
% dwell_times = [dwell_times; first_bleach - last_binding];
% end
% end
end
if isempty(dwell_times)
fprintf("[WARN] No sufficient binding events to compute dwell-time distribution.\n");
else
% 5.2 绘制 dwell-time 直方图
figure;
hold on;
nbins = max(10, round(sqrt(numel(dwell_times)))); % 简单经验 bin 数
[counts_hist, edges] = histcounts(dwell_times, nbins);
centers = (edges(1:end-1) + edges(2:end)) / 2;
bar(centers, counts_hist, "hist");
xlabel("Dwell time (s)");
ylabel("Counts");
title("Bound-state dwell-time histogram");
% 5.3 拟合单指数: p(t) ~ (1/tau) * exp(-t/tau)
% 使用简单的最小二乘拟合 log(count) 对 t
valid = counts_hist > 0;
t_fit = centers(valid);
y_fit = log(counts_hist(valid));
% 线性拟合: log(N) = a + b * t, 其中 b ~= -1/tau
p = polyfit(t_fit, y_fit, 1);
a = p(1);
b = p(2);
tau_est = -1 / b;
fprintf("[INFO] Fitted single-exponential dwell time tau = %.4f s\n", tau_est);
% 画出拟合曲线(归一到 histogram 的总 counts 约束)
t_plot = linspace(min(dwell_times), max(dwell_times), 200);
% 预测密度 ~ exp(a + b t),但我们要匹配 histogram 的离散计数
y_model = exp(a + b * t_plot);
plot(t_plot, y_model, "r-", "LineWidth", 2);
legend("Histogram", "Single-exp fit");
hold off;
% 可选:保存图像
print("-dpng", "dwell_time_histogram_fit.png");
fprintf("[INFO] Dwell-time histogram and fit saved to dwell_time_histogram_fit.png\n");
end
fprintf("\nAll done.\n");使用与扩展建议
- 运行:
octave detect_binding_bleach_dwell_simple.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv会生成:
binding_bleach_events.csv:所有 track 的 binding/bleach 时间点(你最关心的表格)。icon_analysis_track_X.mat:每个 track 的 ICON 分析结果(m_mod, m_edges, p_mean, d_dist, y_mean, y_std等)。- 如果有足够事件,生成
dwell_time_histogram_fit.png,包含 dwell-time 直方图和单指数拟合曲线。 - dwell time 定义:
当前用的是“相邻 binding 时间点之差”作为 dwell;你如果有更明确的物理模型(例如每条轨迹只有 1 次 binding + 1 次 bleach),可以把注释里那段
last_binding -> first_bleach的逻辑打开或进一步改写成更严格的两态 dwell 定义。 - rate constant:
对于单指数 $p(t) \propto \exp(-t/\tau)$,
tau_est是平均 dwell time,解离 rate 大致是 $k{\text{off}} = 1/\tau{\text{est}}$。如果你需要可以在脚本里再打印一条k_off_est。
如果你希望 dwell-time 统计严格区分 “bound state dwell vs unbound state dwell”,或者想直接从 p_mean / d_dist 中做 rate 矩阵估计,可以在这份脚本基础上再往前一步;可以一起设计一个更“教科书版”的 HMM‑rate 推断流程。
特性概览:
- 使用 ICON HMM(
chainer_main)分析每个 track。18 - 做原始
level_analysis.m的四类后验分析:chainer_export、chainer_analyze_means、chainer_analyze_transitions、chainer_analyze_drift。18 - 基于
m_mod聚类成 3 个强度状态,检测 binding / bleaching 事件。19 - 输出
binding_bleach_events.csv,包含所有 binding/bleach 时间点。19 - 用 binding 时间点计算 dwell-time 分布,做单指数拟合;绘图部分只在内存中生成,用
print + close保存 PNG(类似 R 的dev.off()),并使用hist而不是histcounts。19
将以下内容保存为 detect_binding_bleach_dwell.m 即可直接运行。
detect_binding_bleach_dwell.m 完整代码
% detect_binding_bleach_dwell.m
%
% 用法(终端):
% octave detect_binding_bleach_dwell.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv
%
% 输入 CSV 格式(分号分隔):
% # track index;time (seconds);track intensity (photon counts)
% 1;0.00;123
% 1;0.05;118
% 2;0.00; 95
% ...
%--------------------------------------------
% 0. 基本设置
%--------------------------------------------
arg_list = argv();
if numel(arg_list) < 1
error("Usage: octave detect_binding_bleach_dwell.m
<input_csv>");
end
input_file = arg_list{1};
% 加载 HMM 采样器代码(确保 sampler_SRC 在当前目录下)
addpath("sampler_SRC");
% 如果安装了 statistics 包,用于 kmeans
try
pkg load statistics;
catch
warning("Could not load 'statistics' package. Make sure it is installed if kmeans is missing.");
end
%--------------------------------------------
% 1. 读入 track intensity 文件(分号分隔)
%--------------------------------------------
fprintf("Reading file: %s\n", input_file);
fid = fopen(input_file, "r");
if fid < 0
error("Cannot open file: %s", input_file);
end
% 第一行是注释头 "# track index;time (seconds);track intensity (photon counts)"
header_line = fgetl(fid); % 忽略内容,只是读掉这一行
% 后面每行: track_index;time_sec;intensity
data = textscan(fid, "%f%f%f", "Delimiter", ";");
fclose(fid);
track_idx = data{1};
time_sec = data{2};
counts = data{3};
% 排序(确保同一个 track 内按时间排序)
[~, order] = sortrows([track_idx, time_sec], [1, 2]);
track_idx = track_idx(order);
time_sec = time_sec(order);
counts = counts(order);
tracks = unique(track_idx);
n_tracks = numel(tracks);
fprintf("Found %d tracks.\n", n_tracks);
%--------------------------------------------
% 2. 结果结构体(binding / bleach 时间点)
%--------------------------------------------
results = struct( ...
"track_id", {}, ...
"binding_indices", {}, ...
"binding_times", {}, ...
"bleach_indices", {}, ...
"bleach_times", {} );
% 用于 dwell time 收集的数组(跨所有 track)
all_binding_times = []; % 所有 track 的 binding time 合并
all_bleach_times = []; % 所有 track 的 bleach time 合并
%--------------------------------------------
% 3. 循环每个 track,跑 HMM + ICON 分析 + 事件检测
%--------------------------------------------
for ti = 1:n_tracks
tr = tracks(ti);
fprintf("\n===== Track %d =====\n", tr);
sel = (track_idx == tr);
t = time_sec(sel);
z = counts(sel);
z = z(:); % 列向量
%-------------------------
% 3.1 设定 ICON HMM 的参数
%-------------------------
opts = struct();
% 超参数(与 level_analysis.m 一致)
opts.a = 1;
opts.g = 1;
opts.Q(1) = mean(z);
opts.Q(2) = 1 / (std(z)^2 + eps); % 防止 std(z)=0 除零
opts.Q(3) = 0.1;
opts.Q(4) = 0.00001;
opts.M = 10;
% 采样参数
opts.dr_sk = 1;
opts.K_init = 50;
flag_stat = true;
flag_anim = false; % 建议在 Octave 里关掉动画
R = 1000; % 采样次数,可按需求调整
%-------------------------
% 3.2 运行采样器(ICON HMM)
%-------------------------
fprintf(" Running HMM sampler...\n");
chain = chainer_main(z, R, [], opts, flag_stat, flag_anim);
%-------------------------
% 3.3 ICON 后验分析(与 level_analysis.m 等价)
%-------------------------
fr = 0.25; % burn-in 比例
dr = 2; % sample 步距
m_min = 0;
m_max = 1;
m_num = 25;
% (1) 均值轨迹:m_mod
[m_mod, m_red] = chainer_analyze_means(chain, fr, dr, m_min, m_max, m_num, z);
m_mod = m_mod(:);
% (2) 导出 samples(为每条 track 单独存一个文件)
sample_file = sprintf("samples_track_%d", tr);
chainer_export(chain, fr, dr, sample_file, "mat");
fprintf("%s --- Exported\n", [sample_file ".mat"]);
% (3) 转移概率 / 跃迁统计
[m_edges, p_mean, d_dist] = chainer_analyze_transitions( ...
chain, fr, dr, m_min, m_max, m_num, true);
% (4) 漂移分析
[y_mean, y_std] = chainer_analyze_drift(chain, fr, dr, z);
% 保存这些 ICON 分析结果(可选)
mat_out = sprintf("icon_analysis_track_%d.mat", tr);
save(mat_out, "m_mod", "m_red", "m_edges", "p_mean", "d_dist", ...
"y_mean", "y_std", "t", "z");
%-------------------------
% 3.4 把 m_mod 聚类成 3 个光子强度状态(用于事件检测)
%-------------------------
K = 3; % 可按物理需求修改
try
[idx_raw, centers] = kmeans(m_mod, K);
catch
warning("kmeans not available, using simple quantile-based clustering.");
q1 = quantile(m_mod, 1/3);
q2 = quantile(m_mod, 2/3);
idx_raw = ones(size(m_mod));
idx_raw(m_mod > q1 & m_mod <= q2) = 2;
idx_raw(m_mod > q2) = 3;
centers = zeros(K,1);
for kk = 1:K
centers(kk) = mean(m_mod(idx_raw == kk));
end
end
% 根据中心值从小到大重排状态编号,使 state_seq = 1..K 对应 low->high
[~, order_centers] = sort(centers);
state_seq = zeros(size(idx_raw));
for k = 1:K
state_seq(idx_raw == order_centers(k)) = k;
end
low_state = 1;
high_state = K;
%-------------------------
% 3.5 检测 binding / bleaching
%
% binding: low_state -> high_state
% bleach : high_state -> low_state
%-------------------------
s = state_seq(:);
s_prev = s(1:end-1);
s_next = s(2:end);
bind_idx = find(s_prev == low_state & s_next == high_state) + 1;
bleach_idx = find(s_prev == high_state & s_next == low_state) + 1;
bind_times = t(bind_idx);
bleach_times = t(bleach_idx);
fprintf("Found %d binding event(s) and %d bleaching event(s).\n", ...
numel(bind_idx), numel(bleach_idx));
% 存入结果结构体
results(ti).track_id = tr;
results(ti).binding_indices = bind_idx;
results(ti).binding_times = bind_times;
results(ti).bleach_indices = bleach_idx;
results(ti).bleach_times = bleach_times;
% 用于全局 dwell-time 统计
all_binding_times = [all_binding_times; bind_times(:)];
all_bleach_times = [all_bleach_times; bleach_times(:)];
end
%--------------------------------------------
% 4. 输出 binding/bleach 时间点的总表(最重要输出)
%--------------------------------------------
out_csv = "binding_bleach_events.csv";
fid_out = fopen(out_csv, "w");
if fid_out < 0
error("Cannot open output file: %s", out_csv);
end
fprintf(fid_out, "track_index,event_type,sample_index,time_seconds\n");
for ti = 1:numel(results)
tr = results(ti).track_id;
% binding
for k = 1:numel(results(ti).binding_indices)
fprintf(fid_out, "%d,binding,%d,%.6f\n", tr, ...
results(ti).binding_indices(k), ...
results(ti).binding_times(k));
end
% bleach
for k = 1:numel(results(ti).bleach_indices)
fprintf(fid_out, "%d,bleach,%d,%.6f\n", tr, ...
results(ti).bleach_indices(k), ...
results(ti).bleach_times(k));
end
end
fclose(fid_out);
fprintf("\n[INFO] Binding/bleach events written to: %s\n", out_csv);
%--------------------------------------------
% 5. Dwell-time 直方图 + rate 拟合
%
% 简化假设:
% - 把每个 track 中连续 binding 事件之间的时间差视为
% bound state dwell time(中间没有 bleaching,就重新 binding)。
% - 如果有明确的最后一个 binding 到 bleaching 的时间,
% 可以扩展此处加入(当前先保留简单版本)。
%--------------------------------------------
% 5.1 收集 bound-state dwell times(简单用 binding 时间差)
dwell_times = [];
for ti = 1:numel(results)
bt = sort(results(ti).binding_times(:)); % 单个 track 的 binding times
if numel(bt) >= 2
dt = diff(bt); % 相邻 binding 之间的时间差
dwell_times = [dwell_times; dt];
end
% 可选:如果你想把最后一个 binding -> 第一个 bleach 也算入 dwell:
% if ~isempty(results(ti).binding_times) && ~isempty(results(ti).bleach_times)
% last_binding = max(results(ti).binding_times);
% first_bleach = min(results(ti).bleach_times);
% if first_bleach > last_binding
% dwell_times = [dwell_times; first_bleach - last_binding];
% end
% end
end
if isempty(dwell_times)
fprintf("[WARN] No sufficient binding events to compute dwell-time distribution.\n");
else
% 5.2 绘制 dwell-time 直方图(只在内存中画,不弹窗)
fig = figure("visible", "off");
hold on;
nbins = max(10, round(sqrt(numel(dwell_times))));
% 使用 hist(Octave 自带)代替 histcounts
[counts_hist, centers] = hist(dwell_times, nbins);
bar(centers, counts_hist, "hist");
xlabel("Dwell time (s)");
ylabel("Counts");
title("Bound-state dwell-time histogram");
% 5.3 拟合单指数: p(t) ~ (1/tau) * exp(-t/tau)
valid = counts_hist > 0;
t_fit = centers(valid);
y_fit = log(counts_hist(valid));
p = polyfit(t_fit, y_fit, 1);
a = p(1);
b = p(2);
tau_est = -1 / b;
fprintf("[INFO] Fitted single-exponential dwell time tau = %.4f s\n", tau_est);
t_plot = linspace(min(dwell_times), max(dwell_times), 200);
y_model = exp(a + b * t_plot);
plot(t_plot, y_model, "r-", "LineWidth", 2);
legend("Histogram", "Single-exp fit");
hold off;
% 保存图像并关闭 figure(类似 R 的 dev.off())
out_png = "dwell_time_histogram_fit.png";
print(fig, out_png, "-dpng");
close(fig);
fprintf("[INFO] Dwell-time histogram and fit saved to %s\n", out_png);
end
fprintf("\nAll done.\n");运行方式:
octave detect_binding_bleach_dwell.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv如有下一步你想基于 d_dist 做更“纯 HMM”的 dwell 分布 / rate 矩阵,也可以在这个脚本上再加一节专门分析。
level_analysis.m vs detect_binding_bleach.m
这两个脚本在核心 HMM 采样算法上是一致的,但整体流程和用途并不完全相同。level_analysis.m只是做“ICON HMM 拟合 + 若干通用后验分析”,而 detect_binding_bleach.m在此基础上,额外做了“多 track 输入 + 聚类成离散状态 + 自动识别 binding/bleach 跳变 + 输出 CSV”。因此不能说是完全相同算法的简单移植,而是“在相同 HMM 内核上扩展出的一个更完整事件检测流程”。1819
下面分两部分说明:先列出关键差异,然后用中文系统讲解 detect_binding_bleach.m 的整个处理流程(包括和原始 level_analysis.m 共享的 HMM 部分)。1918
一、算法差异概览
- 输入数据形式不同
- HMM 部分是否一致
- 后验分析部分的差异
level_analysis.m:侧重“通用分析”,调用:detect_binding_bleach.m:只调用chainer_analyze_means得到m_mod,然后完全走另一条分析线:- 所以 HMM 后处理的目标和输出是完全不同的:一个是做统计分析、可视化;另一个是做自动事件检测。
- 输出结果不同
- 附加逻辑(聚类与事件判定)是新加的
结论:
- HMM 模型和采样部分与原 MATLAB 代码在理论上是相同的,只做了数值稳定性改动。
- 整体脚本逻辑不相同,Octave 版多了一整套“轨迹遍历 + 状态聚类 + binding/bleach 检测 + CSV 输出”的算法层。
二、用中文说明 detect_binding_bleach.m 的完整流程(含与原脚本共用的 HMM 思路)
下面把 detect_binding_bleach.m 当作主角,用中文分步骤解释其处理流程,并在相关步骤指出它与原 level_analysis.m 的对应关系。这样方便你检查是否满足你对“算法一致性”的期待。1918
1. 命令行输入与文件读取
- 脚本通过
argv()读取命令行参数,第一个参数是输入文件名,例如:octave detect_binding_bleach.m p853_250706_p502_10pN_ch5_0bar_b3_1_track_intensity_data_blue_5s.csv。19 - 输入文件是以分号分隔的文本,第一行为注释头,后面每行包含三列:
track_index:轨迹编号(一个 DNA tether 或一个粒子)。time_seconds:该轨迹下某一帧的时间点。track_intensity:该时间点的 photon counts。
- 读入后,把三列分别存成
track_idx,time_sec,counts。然后用sortrows([track_idx, time_sec])按 “轨迹号优先,再按时间” 排好顺序,保证每条轨迹都是时间有序的。之后提取所有不同的tracks,准备对每一条轨迹单独做分析。19
这一步在原 level_analysis.m 中要简单得多,那里只是 z = load('photoncount.txt');,默认只处理单一系列,没有 track 结构。18
2. 对每条轨迹循环,构建输入序列 z
- 外层
for ti = 1:n_tracks遍历每一个track。 - 对于当前轨迹
tr,用sel = (track_idx == tr)挑出这一条轨迹对应的时间和强度,得到:t = time_sec(sel):该轨迹的时间轴。z = counts(sel):该轨迹的 photon count 序列。
- 把
z转成列向量,作为后续 HMM 的观测数据。
到这里为止,z 的角色与原脚本中的 z = load('photoncount.txt') 完全一致,只是这里是“多轨迹版本”的 z。1819
3. 设置 ICON HMM 的超参数和采样参数
这一段是两份脚本的核心重合部分,也是你最关心的“算法一致性”所在。1819
- 先构建
opts结构体,用于向chainer_main传参。 - 超参数设置为:
opts.a = 1;:控制 HMM 中状态转换的 Dirichlet 浓度(类似“转移稀疏度”的先验强度)。opts.g = 1;:控制状态先验分布的基本参数(base measure 的浓度)。opts.Q(1) = mean(z);:假设状态均值的先验均值等于观测平均值。opts.Q(2) = 1 / (std(z)^2 + eps);:状态均值先验的精度,与观测标准差成反比;加上eps是为了避免在极端情形下std(z) = 0导致除零。原 MATLAB 版本是1/std(z)^2,数学上是一致的设定。opts.Q(3) = 0.1;:方差逆数的 Gamma 先验的 shape。opts.Q(4) = 0.00001;:方差逆数的 Gamma 先验的 scale。opts.M = 10;:插值节点数(ICON 模型内部数值近似需要)。
- 采样相关的参数:
opts.dr_sk = 1;:保存样本的间隔步长。opts.K_init = 50;:初始假设的隐状态数(实际有效状态数由采样自动调整)。R = 1000;:采样迭代次数。
这些设置在 level_analysis.m 中完全一致,只是缺少 eps。所以从贝叶斯 HMM 模型结构和先验设定来看,两者是同一个 ICON 模型。1918
4. 运行 HMM 采样器,得到 MCMC 链
- 调用
chain = chainer_main(z, R, [], opts, flag_stat, flag_anim);来运行 ICON HMM 的 Gibbs sampler 或类似的 MCMC 算法。
chainer_main 内部会对每个时间点的隐状态、每个状态的均值和方差、转移矩阵等进行采样,得到一条或多条后验链 chain,用来近似整个后验分布。这个步骤在两个脚本里完全相同,只是 Octave 版本对可视化做了简化。1918
5. 后验均值轨迹分析:chainer_analyze_means
- 设置后处理参数:
fr = 0.25:前 25% 的样本视为 burn-in,丢弃不用。dr = 2:之后每隔 2 个样本取一个,用来减弱自相关。m_min = 0; m_max = 1; m_num = 25;:把状态均值的可能范围离散成 $[0,1]$ 区间上 25 个格点,用于后验平均的插值或投影。
- 调用:
[m_mod, m_red] = chainer_analyze_means(chain, fr, dr, m_min, m_max, m_num, z); - 输出
m_mod可以理解为“在每一个时间点上的后验平均光子水平”,往往已被归一化到 $[0,1]$ 区间,是一个与z等长的一维轨迹。m_red是某种降维或简化形式,这里没有用到。1819
这一部分与 level_analysis.m 完全一致,区别在于:原脚本后面还继续做转移概率估计和漂移分析;Octave 版则转向“离散状态 + 事件检测”。1918
6. 把连续水平轨迹聚类成几个离散状态
接下来是 Octave 脚本中新加的上层算法部分,用来从连续的 m_mod 中自动提取“低/中/高”强度状态:
- 设定状态数
K = 3(可手动改成 2 或其他)。 - 首选使用
kmeans(m_mod, K)对m_mod做聚类,得到:idx_raw:每个时间点所属的簇编号。centers:每个簇的中心值(大致对应不同光子水平)。
- 如果 Octave 没有统计包或
kmeans不可用,就退化为基于分位数的简单阈值法:- 用三分位数
q1 = quantile(m_mod, 1/3); q2 = quantile(m_mod, 2/3);把轨迹分为低、中、高三段。 - 然后为每类算一个均值当作
centers。
- 用三分位数
- 得到
centers后,按照中心值从小到大排序,重排状态编号,使得:- 状态 1:最低强度(
low_state)。 - 状态 K(=3):最高强度(
high_state)。
- 状态 1:最低强度(
这一段在 level_analysis.m 中没有任何对应逻辑,是 Octave 版本为 binding/bleach 检测专门设计的。其本质是:用连续的后验均值轨迹做无监督聚类,把连续空间映射为少数几个离散层级状态。1819
7. 从状态序列中检测 binding 和 bleaching 事件
有了状态序列 state_seq 后,就可以做简单的“状态跳变检测”来定义事件:
- 令
s = state_seq(:)。 - 构造前后相邻状态:
s_prev = s(1:end-1);s_next = s(2:end);
- binding 事件:
- 当某一时间点出现
s_prev == low_state且s_next == high_state时,即从低强度直接跳到高强度,视为一次“分子结合”(binding)。 - 对应的索引为
find(...) + 1,因为跳变是在第二个点发生。
- 当某一时间点出现
- bleach 事件:
- 当出现
s_prev == high_state且s_next == low_state时,即从高强度直接掉回低强度,视为一次“光漂白或解离”(bleaching/unbinding)。 - 索引同样是
find(...) + 1。
- 当出现
- 最后用
t(bind_idx)和t(bleach_idx)把这些索引转换成真实时间点(秒),分别作为binding_times和bleach_times。
这一步把 HMM 的连续后验轨迹,最终转化成了一个离散事件时间列表。在原 level_analysis.m 中,没有这类应用导向的 event detection,只停留在“描述状态分布和转移概率”的层面。1918
8. 汇总所有轨迹结果并写出 CSV
- 在
for ti = 1:n_tracks循环内,把每条轨迹的事件信息存入results(ti):track_idbinding_indices,binding_timesbleach_indices,bleach_times。19
- 循环结束后,统一写出一个 CSV 文件
binding_bleach_events.csv,格式为:track_index,event_type,sample_index,time_seconds- 其中
event_type是"binding"或"bleach"。
- 最终在命令行打印一行提示,告诉你输出文件名。
相较之下,level_analysis.m 用的是 chainer_export 把整条 MCMC 链以 .mat 形式保存,供交互式分析和可视化使用,而不是面向“事件列表”的输出。1819
总结(中文角度)
- 从 HMM 模型和抽样的角度看:
两个脚本在 ICON HMM 的参数设定和采样过程上是同一套算法,仅在 Octave 版加入了
eps防止除零,以及关闭动画提高稳定性,这不会改变算法本质。 - 从整个脚本的“算法流程”看:
level_analysis.m是一个“通用 ICON HMM 演示与分析脚本”,只做单一序列的建模和一些统计性后验分析,不涉及具体事件检测逻辑。detect_binding_bleach.m在沿用同一 HMM 内核的基础上,增加了:多轨迹处理、对后验均值轨迹的聚类、将轨迹离散为若干强度状态、基于状态跳变的 binding/bleach 自动识别以及 CSV 输出。这是一层新的、面向应用的算法,不存在于原 MATLAB 脚本中。
如果你的目标是“在 Octave 中复用原 ICON HMM 的统计建模部分,并进一步实现自动 binding/bleaching 检测”,那么当前 detect_binding_bleach.m 的设计是符合这一思路的:核心推断算法相同,外层分析逻辑则是针对具体实验需求进行了扩展。