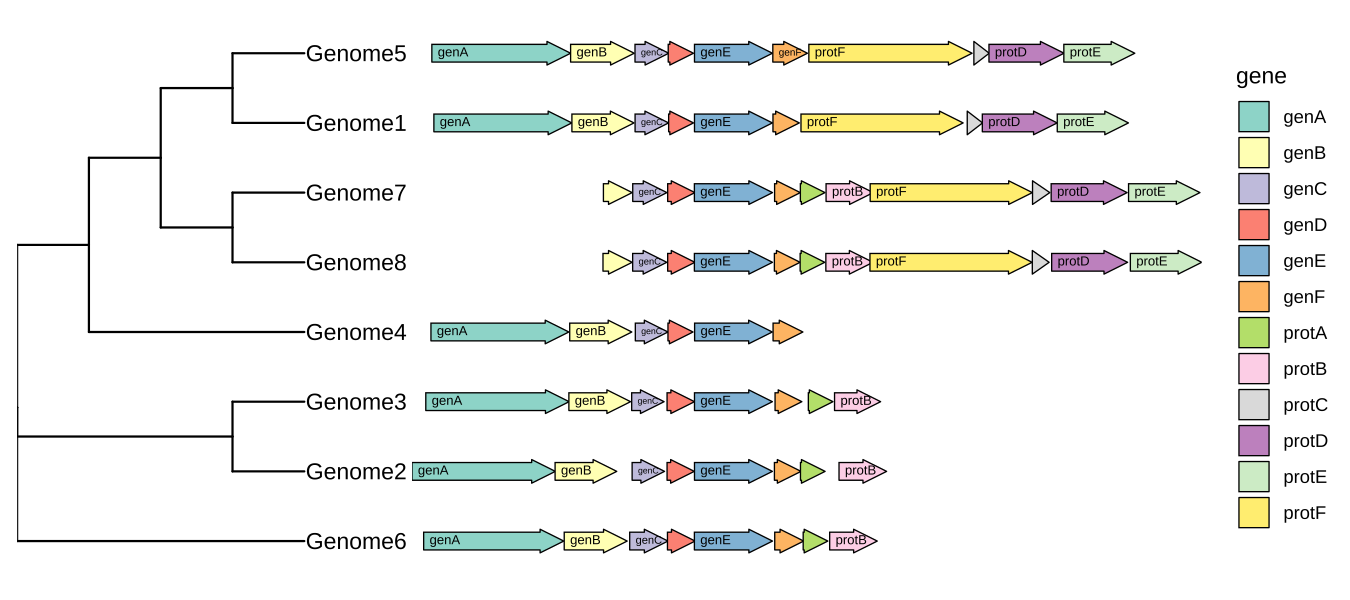
# Load necessary libraries
library(dplyr)
library(ggplot2)
#library(gggenes)
library(ggtree)
library(ape)
# Reconstruct example_genes data
example_genes <- data.frame(
molecule = c(rep("Genome1", 10), rep("Genome2", 8), rep("Genome3", 8), rep("Genome4", 6),
rep("Genome5", 10), rep("Genome6", 8), rep("Genome7", 11), rep("Genome8", 9)),
gene = c("genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF",
"genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD", "protE", "protF",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD"),
start = c(15389, 17301, 18176, 18641, 18999, 20086, 22777, 22986, 24024, 20474,
8345, 10327, 11394, 11878, 12258, 13365, 13726, 14260,
-67849, -65867, -64997, -64507, -64127, -63011, -62550, -62187,
-47353, -45431, -44522, -44070, -43701, -42614,
405113, 407035, 407927, 408387, 408751, 409836, 412621, 412830, 413867, 410335,
65751, 67698, 68605, 69128, 69501, 70614, 71008, 71375,
-9390, -8984, -8500, -8130, -7019, -6662, -6306, -3446, -3188, -2116, -5695,
2, 413, 898, 1268, 2376, 2733, 3089, 5949, 6217),
end = c(17299, 18161, 18640, 18985, 20078, 20451, 22989, 24023, 25010, 22720,
10330, 11181, 11843, 12255, 13337, 13733, 14067, 14919,
-65864, -65013, -64548, -64127, -63048, -62640, -62209, -61549,
-45443, -44571, -44070, -43723, -42625, -42201,
407035, 407916, 408394, 408737, 409830, 410315, 412833, 413870, 414850, 412596,
67691, 68570, 69135, 69511, 70583, 71015, 71349, 72034,
-8992, -8511, -8123, -7048, -6663, -6321, -5653, -3207, -2136, -1127, -3449,
406, 886, 1275, 2350, 2732, 3074, 3742, 6182, 7269),
strand = c("reverse", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse",
"reverse", "reverse", "reverse", "forward", "reverse", "reverse", "reverse", "reverse",
"reverse", "reverse", "forward", "reverse", "forward", "forward",
"forward", "forward", "forward", "reverse", "forward", "forward", "forward", "forward", "forward", "reverse",
"forward", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward",
"reverse", "forward", "reverse", "reverse", "forward", "reverse", "forward", "reverse", "reverse", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse", "reverse"),
orientation = c(1, 0, 1, 0, 1, 1, 1, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1,
0, 1, 0, 0, 1, 1, 1, 0,
1, 1, 1, 0, 0, 0,
0, 0, 0, 0, 1, 0, 1, 0, 0, 1,
0, 1, 0, 0, 1, 1, 0, 0,
1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0,
1, 1, 1, 0, 0, 0, 0, 1, 1)
)
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
#for (i in 1:n) {
# for (j in 1:i) {
# jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
# length(union(genes[[i]], genes[[j]]))
# d[j, i] <- d[i, j] <- 1 - jaccard_sim
# }
#}
#tree <- ape::bionj(d)
# Define the tree structure in Newick format
newick <- "((Genome1:0.1, Genome2:0.2):0.3, (Genome3:0.4, Genome4:0.5):0.6, Genome5:0.7, Genome6:0.8, Genome7:0.9, Genome8:1.0);"
tree <- read.tree(text = newick)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))
#TODO: plot the image combining tree and SNP!!!!
# Load necessary libraries
library(dplyr)
library(ggplot2)
#library(gggenes)
library(ggtree)
library(ape)
# Reconstruct example_genes data
example_genes <- data.frame(
molecule = c(rep("Genome1", 10), rep("Genome2", 8), rep("Genome3", 8), rep("Genome4", 6),
rep("Genome5", 10), rep("Genome6", 8), rep("Genome7", 11), rep("Genome8", 9)),
gene = c("genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF",
"genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD", "protE", "protF",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD"),
start = c(15389, 17301, 18176, 18641, 18999, 20086, 22777, 22986, 24024, 20474,
8345, 10327, 11394, 11878, 12258, 13365, 13726, 14260,
-67849, -65867, -64997, -64507, -64127, -63011, -62550, -62187,
-47353, -45431, -44522, -44070, -43701, -42614,
405113, 407035, 407927, 408387, 408751, 409836, 412621, 412830, 413867, 410335,
65751, 67698, 68605, 69128, 69501, 70614, 71008, 71375,
-9390, -8984, -8500, -8130, -7019, -6662, -6306, -3446, -3188, -2116, -5695,
2, 413, 898, 1268, 2376, 2733, 3089, 5949, 6217),
end = c(17299, 18161, 18640, 18985, 20078, 20451, 22989, 24023, 25010, 22720,
10330, 11181, 11843, 12255, 13337, 13733, 14067, 14919,
-65864, -65013, -64548, -64127, -63048, -62640, -62209, -61549,
-45443, -44571, -44070, -43723, -42625, -42201,
407035, 407916, 408394, 408737, 409830, 410315, 412833, 413870, 414850, 412596,
67691, 68570, 69135, 69511, 70583, 71015, 71349, 72034,
-8992, -8511, -8123, -7048, -6663, -6321, -5653, -3207, -2136, -1127, -3449,
406, 886, 1275, 2350, 2732, 3074, 3742, 6182, 7269),
strand = c("reverse", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse",
"reverse", "reverse", "reverse", "forward", "reverse", "reverse", "reverse", "reverse",
"reverse", "reverse", "forward", "reverse", "forward", "forward",
"forward", "forward", "forward", "reverse", "forward", "forward", "forward", "forward", "forward", "reverse",
"forward", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward",
"reverse", "forward", "reverse", "reverse", "forward", "reverse", "forward", "reverse", "reverse", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse", "reverse"),
orientation = c(1, 0, 1, 0, 1, 1, 1, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1,
0, 1, 0, 0, 1, 1, 1, 0,
1, 1, 1, 0, 0, 0,
0, 0, 0, 0, 1, 0, 1, 0, 0, 1,
0, 1, 0, 0, 1, 1, 0, 0,
1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0,
1, 1, 1, 0, 0, 0, 0, 1, 1)
)
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
#for (i in 1:n) {
# for (j in 1:i) {
# jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
# length(union(genes[[i]], genes[[j]]))
# d[j, i] <- d[i, j] <- 1 - jaccard_sim
# }
#}
#tree <- ape::bionj(d)
# Define the tree structure in Newick format
newick <- "((Genome1:0.1, Genome2:0.2):0.3, (Genome3:0.4, Genome4:0.5):0.6, Genome5:0.7, Genome6:0.8, Genome7:0.9, Genome8:1.0);"
tree <- read.tree(text = newick)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))
#----
# Install and load necessary packages
install.packages(c("Biostrings", "ape", "ggplot2", "gggenes", "ggtree", "dplyr", "patchwork"))
library(Biostrings)
library(ape)
library(ggplot2)
library(gggenes)
library(ggtree)
library(dplyr)
library(patchwork)
# Function to extract SCCmec/ACME regions
extract_regions <- function(genome, regions_of_interest) {
# Example function to simulate extraction of regions based on provided coordinates
extracted_regions <- genome[genome$start %in% regions_of_interest$start & genome$end %in% regions_of_interest$end,]
return(extracted_regions)
}
# Hypothetical function to read genome data
read_genome <- function(filepath) {
# Read the genome data from file
# Here you would implement code to read your genomic data
# Example placeholder for genomic data
genome <- data.frame(
molecule = rep("Genome1", 10),
gene = paste0("gene", 1:10),
start = seq(100, 1000, by = 100),
end = seq(200, 1100, by = 100),
strand = rep(c("forward", "reverse"), 5),
orientation = sample(0:1, 10, replace = TRUE)
)
return(genome)
}
# Example file paths (replace with your actual file paths)
file_paths <- c("path/to/genome1.fasta", "path/to/genome2.fasta", ..., "path/to/genome10.fasta")
# Example regions of interest (replace with actual coordinates)
regions_of_interest <- data.frame(
start = c(100, 300, 500),
end = c(200, 400, 600)
)
# Extract SCCmec/ACME regions for each genome
all_extracted_regions <- lapply(file_paths, function(file) {
genome <- read_genome(file)
extracted_regions <- extract_regions(genome, regions_of_interest)
return(extracted_regions)
})
# Combine all extracted regions into a single data frame
combined_regions <- do.call(rbind, all_extracted_regions)
# Add a 'genome' column to identify the source genome
combined_regions$genome <- rep(paste0("Genome", 1:length(file_paths)), each = nrow(regions_of_interest))
# Display the combined data frame
print(combined_regions)
#install.packages("gggenes")
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
for (i in 1:n) {
for (j in 1:i) {
jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
length(union(genes[[i]], genes[[j]]))
d[j, i] <- d[i, j] <- 1 - jaccard_sim
}
}
tree <- ape::bionj(d)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))
#adapt the following code to use a tree file and a vcf-file to draw the figure.
# Install required packages if they are not already installed
if (!requireNamespace("ggtree", quietly = TRUE)) {
BiocManager::install("ggtree")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("vcfR", quietly = TRUE)) {
install.packages("vcfR")
}
# Load libraries
library(tidyverse)
library(vcfR)
library(ggtree)
#library(TDbook)
library(cowplot)
# Define input and output file paths CP133676
#cp raxml-ng/snippy.core.aln.raxml.bestTree raxml-ng/snippy.core.aln.raxml.bestTree_
tree_file <- "raxml-ng/snippy.core.aln.raxml.bestTree_"
vcf_file <- "variants/snippy.core.vcf"
output_image <- "output_image.png"
# Function to load VCF file and extract SNP data
load_vcf <- function(vcf_file) {
vcf <- read.vcfR(vcf_file, verbose = FALSE)
gt <- extract.gt(vcf)
snp_data <- as.data.frame(t(gt))
colnames(snp_data) <- sub("X", "", colnames(snp_data))
rownames(snp_data) <- NULL
snp_data <- snp_data %>%
rownames_to_column("pos") %>%
gather(key = "name", value = "allele", -pos) %>%
mutate(pos = as.numeric(pos), allele = as.character(allele))
return(snp_data)
}
# Function to plot the tree and SNP data
plot_variants <- function(tree_file, vcf_file, output_image) {
# Load the tree from a Newick file
tree <- read.tree(tree_file)
# Load the VCF file and extract SNP data
snp_data <- load_vcf(vcf_file)
# Debug: Print the first few rows of SNP data
print("SNP data:")
print(head(snp_data))
# Visualize the tree
tree_plot <- ggtree(tree)
# Debug: Print the tree plot object
print(tree_plot)
# Filter SNP data to include only variable positions
gapChar <- "?"
snp_filtered <- snp_data %>%
group_by(pos) %>%
filter(any(allele != first(allele) & allele != gapChar & first(allele) != gapChar))
# Debug: Print the first few rows of filtered SNP data
print("Filtered SNP data:")
print(head(snp_filtered))
if (nrow(snp_filtered) == 0) {
warning("No variable SNP positions found.")
} else {
# Plot SNPs
snp_plot <- ggplot(snp_filtered, aes(x = pos, y = name, color = allele)) +
geom_point(shape = '|', size = 5) +
scale_color_manual(values = c("0" = "blue", "1" = "red")) +
theme_minimal() +
theme(axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank())
# Combine the plots
combined_plot <- plot_grid(tree_plot, snp_plot, ncol = 1, align = "v", rel_heights = c(2, 1))
# Save the plot
ggsave(output_image, plot = combined_plot, height = 10, width = 8)
}
}
# Plot the tree and variants
plot_variants(tree_file, vcf_file, output_image)
#------
## load `tree_nwk`, `df_info`, `df_alleles`, and `df_bar_data` from 'TDbook'
tree <- tree_nwk
snps <- df_alleles
snps_strainCols <- snps[1,]
snps<-snps[-1,] # drop strain names
colnames(snps) <- snps_strainCols
gapChar <- "?"
snp <- t(snps)
lsnp <- apply(snp, 1, function(x) {
x != snp[1,] & x != gapChar & snp[1,] != gapChar
})
lsnp <- as.data.frame(lsnp)
lsnp$pos <- as.numeric(rownames(lsnp))
lsnp <- tidyr::gather(lsnp, name, value, -pos)
snp_data <- lsnp[lsnp$value, c("name", "pos")]
## visualize the tree
p <- ggtree(tree)
## attach the sampling information data set
## and add symbols colored by location
p <- p %<+% df_info + geom_tippoint(aes(color=location))
## visualize SNP and Trait data using dot and bar charts,
## and align them based on tree structure
p + geom_facet(panel = "SNP", data = snp_data, geom = geom_point,
mapping=aes(x = pos, color = location), shape = '|') +
geom_facet(panel = "Trait", data = df_bar_data, geom = geom_col,
aes(x = dummy_bar_value, color = location,
fill = location), orientation = 'y', width = .6) +
theme_tree2(legend.position=c(.05, .85))
#------
# Install required packages if they are not already installed
if (!requireNamespace("ggtree", quietly = TRUE)) {
BiocManager::install("ggtree")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("vcfR", quietly = TRUE)) {
install.packages("vcfR")
}
if (!requireNamespace("cowplot", quietly = TRUE)) {
install.packages("cowplot")
}
# Load libraries
library(ggtree)
library(tidyverse)
library(vcfR)
library(cowplot)
# Define input and output file paths
#cp raxml-ng/snippy.core.aln.raxml.bestTree raxml-ng/snippy.core.aln.raxml.bestTree_
tree_file <- "raxml-ng/snippy.core.aln.raxml.bestTree_"
vcf_file <- "variants/snippy.core.vcf"
output_image <- "output_image.png"
# Function to load VCF file and extract SNP data
load_vcf <- function(vcf_file) {
vcf <- read.vcfR(vcf_file, verbose = FALSE)
gt <- extract.gt(vcf)
snp_data <- as.data.frame(t(gt))
colnames(snp_data) <- sub("X", "", colnames(snp_data))
rownames(snp_data) <- NULL
snp_data <- snp_data %>%
rownames_to_column("pos") %>%
gather(key = "name", value = "allele", -pos) %>%
mutate(pos = as.numeric(pos), allele = as.character(allele))
return(snp_data)
}
# Function to plot the tree and SNP data
plot_variants <- function(tree_file, vcf_file, output_image) {
# Load the tree from a Newick file
tree <- read.tree(tree_file)
# Load the VCF file and extract SNP data
snp_data <- load_vcf(vcf_file)
# Debug: Print the first few rows of SNP data
print("SNP data:")
print(head(snp_data))
# Sampling information, ensuring the length of locations matches the number of tree tips
df_info <- data.frame(
name = c("HDRNA_01_K01", "HDRNA_01_K02", "HDRNA_01_K03", "HDRNA_01_K04", "HDRNA_01_K05", "HDRNA_01_K06", "HDRNA_01_K07", "HDRNA_01_K08", "HDRNA_01_K09", "HDRNA_01_K10"),
location = c("Location1", "Location2", "Location3", "Location4", "Location5", "Location6", "Location7", "Location8", "Location9", "Location10")
)
# Debug: Print the df_info data frame
print("Sampling information:")
print(df_info)
# Visualize the tree
p <- ggtree(tree)
# Attach the sampling information data set and add symbols colored by location
p <- p %<+% df_info + geom_tippoint(aes(color = location))
# Debug: Print the tree plot object
print(p)
# Filter SNP data to include only variable positions
gapChar <- "?"
snp_filtered <- snp_data %>%
group_by(pos) %>%
filter(any(allele != first(allele) & allele != gapChar & first(allele) != gapChar))
# Debug: Print the first few rows of filtered SNP data
print("Filtered SNP data:")
print(head(snp_filtered))
if (nrow(snp_filtered) == 0) {
warning("No variable SNP positions found.")
} else {
# Plot tree
tree_plot <- ggtree(tree) + geom_tippoint(aes(color = location))
# Plot SNPs
snp_plot <- ggplot(snp_filtered, aes(x = pos, y = name, color = allele)) +
geom_point(shape = '|', size = 5) +
scale_color_manual(values = c("0" = "blue", "1" = "red")) +
theme_minimal() +
theme(axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank())
# Combine the plots
combined_plot <- plot_grid(tree_plot, snp_plot, ncol = 1, align = "v", rel_heights = c(2, 1))
# Save the plot
ggsave(output_image, plot = combined_plot, height = 10, width = 8)
}
}
# Plot the tree and variants
plot_variants(tree_file, vcf_file, output_image)Author Archives: gene_x
Bacterial Genome Assembly – Analysis Method
- Raw nanopore sequencing reads are assessed for quality and filtering.
- Filtlong v0.2.1 [1] is used to remove short and low-quality reads from the raw nanopore sequencing data.
- The high-quality nanopore sequencing reads are then used for de novo assembly of the bacterial genome.
- The assembly is performed using Flye v2.9.3 [2] with parameters optimized for bacterial genomes.
- The resulting contigs are further polished using Medaka v1.8 [3] to improve base accuracy.
- The assembled genomes are annotated using Bakta v.1.8.2 [4].
- The annotation includes the prediction of coding sequences, tRNAs, rRNAs, and other genomic features based on published databases such as RefSeq and UniProt.
- The quality of the assembled genomes is assessed using various quality assessment tools (QUAST v5.2 [5], CheckM2 v1.0.1 [6], Mash v2.3 [7]).
- Genome completeness, contiguity, and accuracy are evaluated to ensure the reliability of the assemblies.
- A final QC step is included to ensure the purity of samples, which uses minimap2 v2.24 [8] to map the sequence-cleaned reads onto the assembly and finally employs Clair3 v1.0.4 [9] to call variations (SNPs and INDELS) within the assembled genome.
- Any variants detected along with the location in the bacterial assembly are reported.
References
- Filtlong: Ryan R Wick, https://github.com/rrwick/Filtlong
- Flye: Kolmogorov, Mikhail, Jeffrey Yuan, Yu Lin, und Pavel A. Pevzner. “Assembly of Long, Error-Prone Reads Using Repeat Graphs”. Nature Biotechnology 37, Nr. 5 (Mai 2019): 540–46. https://doi.org/10.1038/s41587-019-0072-8.
- Medaka: ONT Research, https://github.com/nanoporetech/medaka
- Bakta: Schwengers, Oliver, Lukas Jelonek, Marius Alfred Dieckmann, Sebastian Beyvers, Jochen Blom, und Alexander Goesmann. “Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification”. Microbial Genomics 7, Nr. 11 (5. November 2021): 000685. https://doi.org/10.1099/mgen.0.000685.
- Quast: Gurevich, Alexey, Vladislav Saveliev, Nikolay Vyahhi, und Glenn Tesler. “QUAST: Quality Assessment Tool for Genome Assemblies”. Bioinformatics (Oxford, England) 29, Nr. 8 (15. April 2013): 1072–75. https://doi.org/10.1093/bioinformatics/btt086.
- CheckM2: Chklovski, Alex, Donovan H. Parks, Ben J. Woodcroft, und Gene W. Tyson. “CheckM2: A Rapid, Scalable and Accurate Tool for Assessing Microbial Genome Quality Using Machine Learning”. Nature Methods 20, Nr. 8 (August 2023): 1203–12. https://doi.org/10.1038/s41592-023-01940-w.
- Mash: Ondov, Brian D., Todd J. Treangen, Páll Melsted, Adam B. Mallonee, Nicholas H. Bergman, Sergey Koren, und Adam M. Phillippy. “Mash: Fast Genome and Metagenome Distance Estimation Using MinHash”. Genome Biology 17, Nr. 1 (20. Juni 2016): 132. https://doi.org/10.1186/s13059-016-0997-x.
- Minimap2: Li, Heng. “Minimap2: Pairwise Alignment for Nucleotide Sequences”. Bioinformatics (Oxford, England) 34, Nr. 18 (15. September 2018): 3094–3100. https://doi.org/10.1093/bioinformatics/bty191.
- Clair3: Zheng, Zhenxian, Shumin Li, Junhao Su, Amy Wing-Sze Leung, Tak-Wah Lam, und Ruibang Luo. “Symphonizing Pileup and Full-Alignment for Deep Learning-Based Long-Read Variant Calling”. Nature Computational Science 2, Nr. 12 (Dezember 2022): 797–803. https://doi.org/10.1038/s43588-022-00387-x.
To compare long Nanopore sequencing data with a reference genome, you can follow these steps:
* Align the Nanopore reads to the reference genome.
* Convert and sort the alignment file.
* Call variants.
* Analyze the variants to understand the differences.Here’s a detailed step-by-step guide:
Step 1: Align Nanopore Reads to the Reference Genome
Use a long-read aligner such as minimap2 to align your Nanopore FASTQ reads to the reference genome.
Install minimap2 (if not already installed):
sudo apt-get install minimap2Align the reads:
minimap2 -ax map-ont reference.fasta nanopore_reads.fastq > aligned.samStep 2: Convert SAM to BAM and Sort
Convert the SAM file to BAM format and sort it using samtools.
Install samtools (if not already installed):
sudo apt-get install samtoolsConvert SAM to BAM:
samtools view -S -b aligned.sam > aligned.bamSort the BAM file:
samtools sort aligned.bam -o aligned_sorted.bamIndex the sorted BAM file:
samtools index aligned_sorted.bamStep 3: Call Variants
Call variants using bcftools.
Install bcftools (if not already installed):
sudo apt-get install bcftoolsGenerate a VCF file:
bcftools mpileup -f reference.fasta aligned_sorted.bam | bcftools call -mv -Oz -o variants.vcf.gzIndex the VCF file:
tabix -p vcf variants.vcf.gzStep 4: Analyze Variants
Analyze the variants to understand the differences between your sequencing data and the reference genome.
View the VCF file:
bcftools view variants.vcf.gzFilter the variants (if needed):
bcftools filter -i 'QUAL>20' variants.vcf.gz -Oz -o filtered_variants.vcf.gztabix -p vcf filtered_variants.vcf.gz
Compare with another VCF file (if you have another VCF file for comparison):
bcftools isec -p output_prefix -n=2 -c all variants.vcf.gz another_variants.vcf.gzStep 5: Visualize and Interpret
Use visualization tools like IGV (Integrative Genomics Viewer) to visualize the alignments and variants.
Download and install IGV from IGV's official website.
Load your reference genome in IGV.
Load your sorted BAM file (aligned_sorted.bam) and the VCF file (variants.vcf.gz) in IGV.By following these steps, you can align your long Nanopore reads to the reference genome, call and analyze variants, and visualize the results to identify the differences between your sequencing data and the reference. This process will help you determine if the sequencing data matches the reference genome and identify any variations.
To compare your reassembled contigs with a reference genome to determine if they are from the same sample, you can use various bioinformatics tools and approaches. Here’s a step-by-step guide:
Step 1: Align Contigs to the Reference Genome
First, align your reassembled contigs to the reference genome. Tools like BWA, Bowtie2, or minimap2 are commonly used for this purpose. Using BWA:
Index the reference genome:
bwa index CP052959-CP052961.fasta
Align the contigs to the reference genome:
bwa mem CP052959-CP052961.fasta 11108975687_HD46_1_Wt.assembly.fasta > aligned.samStep 2: Convert SAM to BAM and Sort
Convert the resulting SAM file to BAM format and sort it using samtools.
Convert SAM to BAM:
samtools view -S -b aligned.sam > aligned.bam
Sort the BAM file:
samtools sort aligned.bam -o aligned_sorted.bam
Index the sorted BAM file:
samtools index aligned_sorted.bamStep 3: Variant Calling
Call variants using tools like bcftools or GATK.
Using bcftools:
Generate a VCF file:
bcftools mpileup -f CP052959-CP052961.fasta aligned_sorted.bam | bcftools call -mv -Oz -o variants.vcf.gz
Index the VCF file:
tabix -p vcf variants.vcf.gzStep 4: Analyze Variants
Compare the variants in your reassembled contigs with the reference genome. You can use tools like bcftools to filter and compare these variants.
View and filter the VCF file:
bcftools view variants.vcf.gz
Compare VCF files (if you have another VCF file for a different sample for comparison):
bcftools isec -p output_prefix -n=2 -c all variants1.vcf.gz variants2.vcf.gzStep 5: Visualize and Interpret
Use visualization tools like IGV (Integrative Genomics Viewer) to visualize the alignments and variants. This can help you manually inspect regions of interest and ensure that your contigs align well with the reference genome.
Load BAM and VCF files in IGV:
Open IGV and load your reference genome.
Load the sorted BAM file (aligned_sorted.bam).
Load the VCF file (variants.vcf.gz).By following these steps, you can align your reassembled contigs to the reference genome, call and analyze variants, and visualize the results to determine if they are from the same sample. If your contigs align well and have similar variants as the reference genome, it is likely they are from the same sample.
Short-Read Sequencing vs Long-Read Sequencing
When dealing with sequencing libraries, particularly when working with short-read (e.g., Illumina) and long-read (e.g., Nanopore or PacBio) technologies, understanding their error profiles, and how to process and analyze the data is crucial. Below is an explanation of these concepts and some practical steps for managing and analyzing the data. Error Rates in Sequencing Technologies
* Short-Read Sequencing (e.g., Illumina):
- Error Rates: Generally low, around 0.1% to 1%.
- Advantages: High accuracy, high throughput, and good for variant detection.
- Disadvantages: Short read lengths, which can make it challenging to resolve repetitive regions and complex structural variations.
* Long-Read Sequencing (e.g., Nanopore, PacBio):
- Error Rates: Higher, ranging from 5% to 20% for individual reads.
- Advantages: Long reads, which can span entire genes or large structural variations, making assembly and complex variant detection easier.
- Disadvantages: Higher error rates and lower throughput compared to short-read technologies.Practical Steps for Data Processing
* Data Preprocessing:
- Quality Control: Use tools like FastQC to assess the quality of sequencing data.
- Trimming: Remove low-quality bases and adapters using tools like Trimmomatic (short-read) or Porechop (long-read).
* Assembly and Alignment:
- Short-Read Assembly: Use assemblers like SPAdes or Velvet.
- Long-Read Assembly: Use assemblers like Canu, Flye, or Shasta.
- Hybrid Assembly: Combine both short and long reads using tools like Unicycler or MaSuRCA.
* Error Correction:
- Short-Read Correction: Generally not needed due to low error rates.
- Long-Read Correction: Use tools like Nanocorrect or FMLRC to correct long-read data using short reads.
* Variant Calling:
- Short-Read Variant Calling: Use tools like GATK or FreeBayes.
- Long-Read Variant Calling: Use tools like Medaka (Nanopore) or Longshot (PacBio).
- Integrative Analysis: Combine data using WhatsHap for phasing and DeepVariant for accurate variant calling.Pacbio Sequel 20Kb (Microorganism)
Pacbio Sequel 10Kb (Microorganism)
<=800bp
Nanopore (Microorganism)
PacBio barcode library (Microorganism)
PacBio Revio library
Cyclone normal long library
Sequencing services for microorganisms:
-
PacBio Sequel 20Kb (Microorganism)
PacBio Sequel: This is a sequencing platform developed by Pacific Biosciences, known for generating long reads. 20Kb: Refers to the average length of the DNA fragments (20,000 base pairs) that are sequenced. Longer reads are particularly useful for de novo assembly, resolving complex regions, and identifying structural variations. Microorganism: Indicates that this service is optimized for sequencing microbial genomes, which can be challenging due to their diverse and complex genetic content.
-
PacBio Sequel 10Kb (Microorganism)
PacBio Sequel: Same platform as above. 10Kb: Refers to a shorter average read length of 10,000 base pairs. These reads are still long compared to other technologies and useful for similar applications, but might be chosen for different balance of throughput and read length depending on the project needs. Microorganism: Again, optimized for microbial genomes.
-
<=800bp
<=800bp: This likely refers to a sequencing service that generates reads of up to 800 base pairs in length. This could be indicative of Sanger sequencing or certain targeted sequencing applications where short reads are sufficient and high accuracy is required.
-
Nanopore (Microorganism)
Nanopore: Refers to Oxford Nanopore Technologies (ONT) sequencing, which can produce very long reads (up to several megabases) but with higher error rates compared to short-read technologies. Microorganism: Tailored for microbial genome sequencing. ONT is useful for its ability to sequence long stretches of DNA, providing comprehensive insights into genome structure and function.
-
PacBio barcode library (Microorganism)
PacBio barcode library: A library preparation method that includes barcoding (adding unique sequences to DNA fragments). This allows multiplexing of multiple samples in a single sequencing run, distinguishing them bioinformatically afterward. Microorganism: Optimized for microbial samples. Barcoding is particularly useful in high-throughput studies where multiple microbial genomes are sequenced simultaneously.
-
PacBio Revio library
PacBio Revio: Refers to a newer or advanced library preparation method from PacBio, possibly associated with the Revio system (or similar advanced sequencers). The details might be specific to the latest improvements in sequencing chemistry and protocols that enhance read length, accuracy, or throughput. Library: Refers to the prepared DNA ready for sequencing on the PacBio platform.
-
Cyclone normal long library
Cyclone: This term is not widely recognized in the current sequencing technologies or literature, which suggests it might be a proprietary or specific method/service offered by BGI. It could be a specialized library preparation method that BGI has developed, focusing on certain aspects of long-read sequencing. Normal long library: Likely indicates that this service involves preparing long-read sequencing libraries (similar to those used in PacBio or Nanopore sequencing) but with a “normal” protocol that might be standard or default for general long-read sequencing projects.
Summary
PacBio Sequel 20Kb and 10Kb: Long-read sequencing options for microbial genomes, with average read lengths of 20Kb and 10Kb, respectively.
<=800bp: Short-read or targeted sequencing, possibly high accuracy for specific applications.
Nanopore (Microorganism): Long-read sequencing from Oxford Nanopore, tailored for microbial genomes.
PacBio barcode library (Microorganism): Barcoded sequencing library preparation for multiplexing microbial samples.
PacBio Revio library: Likely refers to advanced or newer library preparation methods for PacBio sequencing.
Cyclone normal long library: Likely a BGI-specific or proprietary long-read sequencing library preparation method.Comparison of the precision of three popular sequencing technologies: PacBio, Nanopore, and Illumina.
-
PacBio (Pacific Biosciences)
- Technology: Single Molecule, Real-Time (SMRT) Sequencing
- Read Length: Long reads, often exceeding 10,000 base pairs, with some reads over 100,000 base pairs.
- Accuracy:
- Raw Read Accuracy: Approximately 85-90%
- Consensus Accuracy: Greater than 99.9% after error correction through multiple reads
- Strengths:
- Excellent for detecting structural variants and large insertions/deletions. High-quality assembly of genomes with complex regions. Limitations: Higher error rates in raw reads compared to Illumina. More expensive per base compared to other technologies.
-
Nanopore (Oxford Nanopore Technologies)
- Technology: Nanopore Sequencing
- Read Length: Ultra-long reads, theoretically limited only by the length of the DNA molecule, with some reads over 2 million base pairs.
- Accuracy:
- Raw Read Accuracy: Approximately 90-95%
- Consensus Accuracy: Greater than 99% with sufficient coverage and error correction
- Strengths:
- Ability to produce very long reads.
- Portable and scalable devices (e.g., MinION, PromethION).
- Limitations:
- Higher raw read error rate compared to Illumina.
- Requires high coverage for accurate consensus sequences.
-
Illumina
- Technology: Sequencing by Synthesis (SBS)
- Read Length: Short reads, typically 150-300 base pairs.
- Accuracy:
- Raw Read Accuracy: Greater than 99.9%
- Consensus Accuracy: Very high due to low error rates in raw reads
- Strengths:
- Extremely high accuracy and throughput.
- Cost-effective for large-scale projects.
- Limitations:
- Short read length can make it challenging to resolve complex regions of the genome.
- Limited ability to detect large structural variants.
-
Summary
- PacBio: Best for long-read sequencing with high consensus accuracy after error correction, ideal for complex genomes and structural variant analysis.
- Nanopore: Offers ultra-long reads and portable sequencing options, with improving accuracy, making it versatile for various applications.
- Illumina: Provides the highest raw read accuracy and throughput, perfect for applications requiring short reads and high precision.
Each technology has its unique strengths and is chosen based on the specific requirements of the sequencing project.
Standard Bioinformatics Service and Custom Bioinformatics Solutions
https://www.genewiz.com/en-GB/Public/Services/Next-Generation-Sequencing https://www.lexogen.com/services/bioinformatics/
-
Gene Expression Profiling
-
Whole Transcriptome Sequencing
-
FFPE RNA Sequencing
-
Small RNA Sequencing
-
Single-cell RNA Sequencing
-
Ultra-low Input RNA Sequencing
-
DNA Sequencing
-
Standard Bioinformatics Service: Next-generation sequencing (NGS) technologies are invaluable in academic research, biotechnology, biomedical and clinical research, and the pharmaceutical industry. As NGS technologies rapidly expand and develop, it is imperative to correctly interpret increasingly complex data sets and relate them to biological functions. More than ever, it is necessary to approach NGS data analysis with tailored and creative data analysis workflows to extract the most from the datasets obtained. Our team consists of genomic data analysis experts with experience in various NGS data analysis pipelines who are passionate about developing novel, customized workflows and solutions while keeping biology at the forefront. We have extensive experience in handling diverse genomic data analysis pipelines, including various types of RNA-Seq data analysis, DNA-Seq data analysis or epigenetics. Indeed, we can adapt and customize pipelines specifically to your project needs or even develop a new data analysis pipeline for you.
-
Standard pipelines:
- Differential Gene Expression analysis: Differential Gene Expression analysis, also known as DGE analysis is used to identify changes in gene expression levels between different biological conditions, such as different genotypes, treated and untreated samples, or in disease-related research. Differentially expressed genes provide valuable insights into the underlying drivers and affected processes of studied phenotypes.
- Functional enrichment analysis: Functional enrichment analysis transforms a list of differentially expressed genes into meaningful biological insights. It helps to understand pathways and processes underlying studied phenotypes.
- Transcriptome assembly: Transcriptome assembly is a process of de novo transcriptome reconstruction directly from RNA-Seq reads. This method is often employed to study non-model organisms for which no reliable genome or transcriptome assembly exists.
- Small RNA data analysis: Small RNAs (sRNAs) are short non-coding RNA molecules, typically ranging in size from 20 to 200 nucleotides. sRNAs play a crucial role in regulating gene expression and are involved in essential cellular processes such as mRNA turnover, translational regulation, and chromatin compaction. microRNAs (miRNAs) are the most extensively studied, and their aberrant expressions have been reported in various diseases, particularly cancer. miRNA-Seq analysis includes differential gene expression analysis of mature and premature miRNAs, discovery of new miRNAs and analysis of verified and predicted miRNA targets.
- Alternative splicing analysis: Alternative splicing is a process that gives rise to several different transcripts from a single gene, thus enhancing transcriptome and, consequently, proteome diversity. Alternative splicing is cell-type and developmental stage-specific. Abnormal splicing variants or altered isoform levels have been connected to various diseases and cancers. Alternative splicing analysis involves the detection and quantification of new and known isoforms and the evaluation of new splicing events.
- Circular RNA analysis: Circular RNAs (circRNAs) are single-stranded RNAs that form covalently closed loops. While the biological function of most circRNAs remains unclear, they have been found to act as transcriptional regulators and microRNA sponges and even have the ability to code proteins. Moreover, circRNAs exhibit unique expression signatures and have been linked to various diseases, suggesting their potential as diagnostic biomarkers and therapeutic targets.
- Alternative polyA site analysis: Alternative polyadenylation (APA) leads to the generation of multiple matured mRNA molecules with variable 3′ ends originating from a single gene. As a key posttranscriptional regulation, APA widely affects RNA metabolism including mRNA maturation, RNA stability, cellular RNA decay and protein diversification. Consequently, APA plays a significant role in various cellular processes and its dysregulation has been described in cancer.
- Internal priming filtering: 3′ end targeting libraries predominantly rely on oligo dT primers, which can mis-prime in A-rich regions located across the genome. Internal priming filter helps to identify and retain reads that genuinely originate at the 3′ end polyA sites.
- Shape-Seq: Shape-Seq (selective 2′-hydroxyl acylation analyzed by primer extension sequencing) is a technique that allows researchers to study RNA structure. In Shape-Seq, RNA molecules are selectively modified in a structure-dependent manner and these modifications induce mutations in the cDNA during the reverse transcription. Consequently, analysis of the mutation profile in Shape-Seq data provides insights into the structural features of RNA.
- ChIP-Seq: ChIP-seq (Chromatin Immunoprecipitation followed by sequencing) is a technique used to investigate protein-DNA interactions on a genome-wide scale. In ChIP-seq protocol, DNA is co-immunoprecipitated with the protein of interest using an antibody specific to that protein. The isolated DNA is then fragmented and subjected to Next-Generation Sequencing (NGS), which allows for the global detection of the protein-DNA binding sites. Thus ChIP-Seq provides valuable insights into gene regulation, biological pathways and their deregulation, which often leads to diseases.
- ATAC-Seq: ATAC-Seq (Assay for Transposase-Accessible Chromatin with sequencing) is a technique that is used to detect accessible chromatin landscapes associated with certain cell types. Hyperactive Tn5 transposase enzyme fragments DNA and simultaneously adds sequencing adapter preferably in open chromatin regions, that are more accessible for Tn5 binding. These regions typically correspond to regulatory elements such as promoters, enhancers, and transcription factor binding sites. Thus, ATAC-Seq data provides valuable insights into gene regulation in complex biological processes and diseases.
- Germline and somatic variant calling: Variant calling is a method for the detection of changes in DNA, ranging from single nucleotide polymorphism (SNPs) to larger rearrangements like insertions and deletions. Germline variants impact all cells in the body, including germ cells, and can be passed on to the next generation. In contrast, somatic mutations occur in somatic cells during an individual’s lifetime and are not passed on to offspring. Accurate identification of these variants is essential for understanding the genetic basis of diseases, enabling the development of targeted therapies, and advancing personalized medicine.
- Single-cell RNA-Seq Analysis (10x and proprietary Luthor technology): Single‐cell RNA sequencing (scRNA‐seq) is the leading technique for studying transcriptome heterogeneity within individual cells. It enables cell characterization at the transcriptome level, identification of rare but functionally significant cell populations, and addresses complex experimental questions that bulk analysis cannot answer. We offer analysis of several scRNA-Seq data types, including 10x and our proprietary Luthor technology. scRNA-Seq data analysis can be used for cell-type clustering, marker identification, trajectory analysis, and many other types of analysis.
- SLAMseq data analysis: SLAMseq (thiol (SH)Linked Alkylation for the Metabolic Sequencing of RNA) enables transcriptome-wide analysis of RNA synthesis and turnover by measuring nascent RNA expression and transcript stability. SLAMseq combines the labeling of newly synthesized RNA transcripts with RNA-Seq readout. SLAMseq technique can be coupled with whole transcriptome sequencing or 3’ mRNA-Seq for cost-efficient, high-throughput screening setups. SLAMseq data allows transcript half-live estimation and analysis of differential RNA production or decay. Thus SLAMseq is a great tool for studying the effects of fast-acting drugs or drug candidates on RNA kinetics.
- Whole-exome and whole-genome sequencing: Whole-exome sequencing (WES) is a targeted sequencing approach that focuses on analyzing protein-coding regions of the genome, known as the exome. This method provides valuable insights into the genetic basis of diseases, facilitates the identification of pathogenic mutations, and supports personalized medicine approaches. Since the exome represents only about 1-2% of the entire genome, WES is more cost-effective compared to whole-genome sequencing (WGS). On the other hand, WGS is the method of choice when a comprehensive analysis of the entire genome is required, including coding and non-coding regions, as well as mitochondrial and chloroplast DNA, or for identification of novel genomic variants.
- De-novo genome assembly: De-novo genome assembly is used for organisms without known genome references or for organisms with highly dynamic genomes. The process usually begins with assembling short reads into longer sequences – contigs that are subsequently organized into scaffolds and, in the end, into chromosomes.
- Primer design for rRNA depletion: Total RNA is comprised of large amounts of ribosomal RNA (rRNA) which can make up between ~80-98 % of all RNA molecules in a sample. rRNA depletion removes these undesired transcripts to access transcripts of interest. If you work with less common species and struggle with rRNA depletion for your RNA-Seq experiment, we offer the primer design using our proprietary advanced algorithm.
-
Working process:
- Introductory Consultations & Project Planning: We start with an introductory consultation. This is an opportunity to get to know each other, and that we understand your research questions and get to the heart of your project. Only then can we select and design data analysis workflow and deliverables that best meet your needs. Based on this initial consultation we will plan the project, and as a result, you will receive a list of deliverables, along with cost and timeline information for project completion.
- Project Initiation & Data Transfer: After you send us the purchase order, we start the project and wait for your data! You can choose from several options for data transfer, including FTP server transfer, cloud bucket sharing or a hard drive shipment.
- Data Analysis, Delivery of Results & Report: Upon completion of the data analysis, we will generate a report based on all deliverables, and send you results data and the report, including detailed methodology, using your preferred data transfer method.
- Discussion & Conclusion Meeting: We want to make sure that you have a clear understanding of the provided results and that they meet your expectations. Therefore, we will review the results and the report together and answer any questions or concerns you may have.
- Follow-up Support: We provide follow-up support for your current or future projects, even after our collaboration is completed. We are here for you if you have any questions or need additional assistance!
-
-
Custom Bioinformatics Solutions: With decades of experience in transcriptomics, genomics, and related fields of biology, our bioinformatics team is empowered to tailor new bioinformatics developments to your unique project. We offer various Custom Bioinformatics Solutions, including new tool and pipeline developments, by using state-of-the-art algorithms and computational methods. With our personal and customized approach, we deliver most reliable and accurate outcomes enabling you to differentiate and drive innovation. Post-project, we provide detailed documentation and reporting, as well as secure and full transfer of custom pipeline.
- new algorithm development,
- pipeline development from scratch,
- tailored tool development & optimization, or
- flexible realization using state-of-the art open-source, ad-hoc, and proprietary solutions.
博氏疏螺旋体在宿主细胞内的摄取与处理机制
https://grk2771.de/phd-students-2/
Project 2 Summary (Interplay between Yersinia enterocolitica and the autophagosomal/lysosomal system in epithelial host cells and organoids, 耶尔森菌与上皮宿主细胞和类器官中的自噬体/溶酶体系统的相互作用)
-
Yersinia enterocolitica engages a cell-invasive strategy to colonize the host intestinal tissue.
-
The intracellular, invasive phenotype depends on the Yersinia outer membrane protein invasin which binds and activates eukaryotic beta-1-integrin receptors to promote uptake of the bacteria into infected host cells.
-
Our previous studies have shown that internalization of Yersinia by epithelial cells results in the formation of two distinct populations of intravacuolar bacteria.
-
One part of the ingested bacteria is subjected to the endosomal / phagolysosomal pathway in which the Yersinia-containing vacuoles (YCVs) fuse with lysosomes and the enclosed bacteria are eliminated.
-
The second half of the bacteria ends up in vacuoles with autophagy-related characteristics, displaying recruitment of autophagosomes, phagophore formation, and xenophagy.
-
Importantly, fusion of these autophagic vacuoles with lysosomes is actively prevented by Yersinia which results in intracellular survival and proliferation of Yersinia in the non-acidified, autophagic compartments.
-
This indicates that Y. enterocolitica takes advantage of the macroautophagy pathway to gain access to an intracellular niche that enables bacterial replication in a protected compartment.
-
Eventually, this leads to bacterial egress from the infected cells.
-
The molecular mechanisms that direct Yersinia to autophagosomes and prevent fusion with lysosomes are, however, presently unclear.
-
Our project consequently aims to explore the intracellular lifestyle of Yersinia in order to uncover novel principles in bacterial pathogenesis and in the regulation of vesicle function and trafficking pathways in infected epithelial host cells.
-
The autophagosomal [‘fægәsәum] and lysosomal [‘laisəsəum] pathways to which Yersinia is sorted are differentially characterized by proteomic and co-localization studies.
-
Furthermore, the Yersinia factors that manipulate the physiological endocytic-lysosomal pathway are investigated by evaluating a transposon-based Yersinia mutation library.
-
Functional approaches in molecularly modified host cells will then specify the roles of the identified pathways for Yersinia survival, replication and release from infected cells.
-
We expect the results of this work to be of general importance for cell and human infection biology.
-
Yersinia enterocolitica 通过细胞侵入策略定殖宿主肠组织。
-
这种细胞内侵入表型依赖于耶尔森菌外膜蛋白入侵素(invasin),该蛋白与真核β-1整合素(beta-1-integrin)受体结合并激活,从而促进细菌进入受感染的宿主细胞。
-
我们之前的研究表明,耶尔森菌被上皮细胞内化后,会形成两个不同的细胞内空泡细菌群体。
-
部分吞噬的细菌会被引导至内体/吞噬溶酶体途径(endosomal / phagolysosomal pathway),其中含有耶尔森菌的空泡(YCVs)与溶酶体(lysosomes)融合,包裹的细菌被消灭。
-
另一部分细菌最终进入具有自噬相关特征的空泡,表现出自噬体招募、吞噬泡形成和异噬现象(autophagy-related characteristics, displaying recruitment of autophagosomes, phagophore formation, and xenophagy)。
-
重要的是,耶尔森菌主动阻止这些自噬空泡与溶酶体的融合,从而在未酸化的自噬隔室中存活并增殖。
-
这表明耶尔森菌利用巨自噬途径获得一个使细菌在受保护隔室内复制的细胞内生态位。
-
最终,这导致细菌从受感染的细胞中释放出来。
-
然而,指导耶尔森菌进入自噬体并防止其与溶酶体融合的分子机制目前尚不清楚。
-
因此,我们的项目旨在探索耶尔森菌的细胞内生活方式,以揭示细菌致病机制和受感染上皮宿主细胞中囊泡功能及运输途径调控的新原理。
-
通过蛋白质组学和共定位研究,分别表征耶尔森菌被引导至的自噬体和溶酶体途径(autophagosomal [‘fægәsәum] and lysosomal [‘laisəsəum] pathways)。
-
此外,通过评估一个基于转座子的耶尔森菌突变文库 (transposon-based Yersinia mutation library),研究操纵生理性内吞-溶酶体途径的耶尔森菌因子。
-
然后,通过在分子上修改的宿主细胞中进行功能性方法研究,明确这些途径在耶尔森菌存活、复制和从受感染细胞中释放的角色。
-
我们预计该研究的结果将对细胞和人类感染生物学具有普遍的重要性。
-
endocytic[ˏendәu’sitik]生内吞作用的
Project 1 Summary
-
The T3SS/injectisome of Yersinia enterocolitica is a molecular machine that injects effector proteins into host cells to support the bacterial infection strategy.
-
The injectisome directly connects to host cells via a translocon/pore complex, that serves as an entry gate for the effectors.
-
Simultaneously attached to the T3SS needle tip and integrated into the host cell membrane, the translocon is subject to multi-layered regulation by bacterial and host cell factors. YopQ/YopK is a Yersinia effector that controls both, activity and immune recognition of the translocon from within the host cells.
-
Despite the critical role of YopQ/YopK in function and immune recognition of the Yersinia translocon, its direct interaction partners and molecular mode of action are unknown.
-
The overall goal of P1 is to determine the molecular mode by which Y. enterocolitica YopQ controls activity and immune recognition of the T3SS translocon. To this end, i) the dynamics of bacterially translocated YopQ in host cells and its correlation with function, formation, degradation and/or immune sensing of the translocon will be visualized at the highest spatial and temporal resolution; ii) the macrophage interaction partners of YopQ/YopK will be identified by affinity purification and verified in functional assays, and iii) the structure-function relationship of YopQ in terms of its intracellular spatiotemporal dynamics, interaction with binding partners, and effects on translocon activity and immune recognition will be investigated.
-
Methods to be applied include: sCRISPR/Cas mediated mutagenesis and gene editing of Yersinia; affinity purification and mass spectrometric analysis of host-pathogen protein complexes; in vitro protein-protein interaction; super-resolution fluorescence microscopy of bacterial and host proteins in fixed and living macrophages.
-
耶尔森菌的T3SS/注射体是一种分子机器,可以将效应蛋白注入宿主细胞,以支持细菌的感染策略。
-
注射体通过转位子/孔复合物直接与宿主细胞连接,作为效应物进入的入口。
-
同时附着在T3SS针尖并整合到宿主细胞膜中的转位子受到细菌和宿主细胞因素的多层调控。YopQ/YopK是控制转位子在宿主细胞内活动和免疫识别的耶尔森菌效应物。
-
尽管YopQ/YopK在耶尔森菌转位子功能和免疫识别中起着关键作用,其直接的相互作用伙伴和分子作用机制尚不清楚。 P1的总体目标是确定耶尔森菌YopQ控制T3SS转位子活动和免疫识别的分子机制。为此,i) 将以最高的空间和时间分辨率可视化细菌转移到宿主细胞内的YopQ的动态及其与转位子的功能、形成、降解和/或免疫感知的相关性;ii) 通过亲和纯化识别YopQ/YopK的巨噬细胞相互作用伙伴,并在功能性测定中验证;iii) 调查YopQ在其细胞内时空动态、与结合伙伴的相互作用以及对转位子活动和免疫识别的影响方面的结构-功能关系。
-
应用的方法包括:sCRISPR/Cas介导的耶尔森菌突变和基因编辑;宿主-病原体蛋白复合物的亲和纯化和质谱分析;体外蛋白-蛋白相互作用;固定和活巨噬细胞中细菌和宿主蛋白的超分辨率荧光显微镜。
Project 7 Summary
-
Human Polyomaviruses (PyV) are highly prevalent and establish a lifelong asymptomatic persistence in the healthy immunocompetent host1,2.
-
However, under immunosuppression, these viruses can reactivate, causing life-threatening infections (e.g. BKV caused PyV associated nephropathy, PVAN) due to uncontrolled viral replication1.
-
Currently, no specific antiviral treatment is available. This lack of effective therapeutics is partly due to the lack of small animal models and the availability of only poor surrogate in vitro/in vivo systems.
-
We have recently identified 16 small molecule inhibitors, C1-16, against BKV using a phenotypic high throughput screen (Kraus et al., unpublished).
-
For the further development of these inhibitors, it is essential to have an understanding of their cellular target structure and/or which part of the viral life cycle they inhibit.
-
Within this project we will gain a better understanding of the BKV the life cycle in relevant infection systems (e.g. primary cells and organoids).
-
We will use these previously identified antiviral compounds in terms of their interference with essential host structures for viral reproduction (transport vesicles, nuclear uptake, replication compartments or vesicle dependent egress).
-
Furthermore, we will characterize specific viral inhibitors at the molecular and structural level.
-
The project uses organoids and primary cells as infection models and BKV inhibitor characterization.
-
It applies confocal live cell microscopy to follow BKV entry/spread.
-
Furthermore, the project takes advantage of X-ray crystallography to characterize inhibitor/target interaction.
-
人类多瘤病毒(Polyomaviruses,简称PyV)具有高度流行性,并在健康的免疫健全宿主中建立终生的无症状持续感染。
-
然而,在免疫抑制的情况下,这些病毒可能会重新激活,导致由于病毒失控复制而引起的致命感染(例如,由BKV引起的与多瘤病毒相关的肾病,PVAN)。
-
目前,没有特定的抗病毒治疗方法。这种缺乏有效治疗手段的部分原因是缺乏小型动物模型以及仅有不良替代的体外/体内系统。
-
我们最近通过表型高通量筛选(Kraus等,未发表)鉴定出16种针对BKV的小分子抑制剂,C1-16。
-
为了进一步开发这些抑制剂,了解其细胞靶结构和/或其抑制病毒生命周期的哪一部分是至关重要的。
-
在这个项目中,我们将更好地理解BKV在相关感染系统中的生命周期(例如,原代细胞和类器官)。
-
我们将使用这些先前鉴定的抗病毒化合物,研究它们对病毒复制所需的关键宿主结构(如运输小泡、核摄取、复制区或依赖小泡的出芽)的干扰。
-
此外,我们将在分子和结构水平上表征特定的病毒抑制剂。
-
该项目使用类器官和原代细胞作为感染模型,并对BKV抑制剂进行表征。
-
它应用共聚焦活细胞显微镜来追踪BKV的进入/传播。
-
此外,该项目利用X射线晶体学来表征抑制剂/靶标的相互作用。
Project 4 Summary
The Role of CEP55 in exosomal section by ovarian cancer cells
-
Borrelia burgdorferi is the causative agent of Lyme disease, a multisystemic disorder affecting skin, nervous system and joints.
-
Uptake and intracellular processing of borreliae by host cells proceed in several steps:
- i) immobilisation of highly motile borreliae by formin-regulated filopodia1,3,
- ii) enwrapment by a coiling pseudopod, regulated by the formin Daam1 and Arp2/3 complex1,2,
- iii) uptake into a Rab22a-positive phagosome, which is contacted by Rab5a-positive vesicles in an ER-dependent manner2,
- iv) compaction of borreliae by reduction of the phagosome surface through membrane tubulation2,4,
- v) degradation in mature phago-lysosomes2,4 (see also Fig.).
-
We also identified lipids such as PI(3)P and phosphatidylserine that play specific roles during uptake and processing of borreliae4,5.
-
Moreover, we recently discovered that membrane tunnels, a novel phagocytic structure, extend deeper into the host cell cytoplasm than the bacteria-containing part of phagosomes, probably due to partial extrication of highly motile borreliae (see Fig.).
-
This shows that internalisation of borreliae by macrophages is a unique and highly dynamic process, whose outcome depends on a tug-of-war between spirochetes and host cells5.
-
Notably, both phagosomes and tunnels form multiple STIM1-positive ER contact sites, pointing to a role of ER-connected processes, including regulation of Ca2+ influx, in the maturation of borreliae containing phagosomes5.
-
However, the roles of ER contact sites and specifically phagosomal Ca2+ influx in borreliae phagosome maturation are currently unclear.
-
To address these questions, and to identify Borrelia-specific regulators of phagosome compaction and maturation, we have established magnetic labelling of borreliae, permitting subsequent purification of phagosomes. Results from these experiments will identify novel targets for modulation of intracellular processing of spirochetes in human immune cells.
-
This project will
- a) analyse the proteome and lipidome of Borrelia-containing phagosomes and tunnels,
- b) characterise ER contact sites at phagosomes and tunnels on a molecular and functional level, and
- c) identify regulators that are specific for compaction of spirochetes, as opposed to bacteria without pronounced phagosomal compaction such as staphylococci and streptococci, or which follow different strategies for intracellular survival or persistence such as Yersinia, Legionella or Salmonella.
-
SILAC labelling of macrophages will be combined with chemical crosslinking, (co-) immunoprecipitation and proximity labelling assays to identify ligands on the Borrelia surface involved in recruiting the molecular machinery driving phagosome/tunnel closure and phagosome compaction. Lattice light sheet microscopy and focused ion beam microscopy will be used to analyse membrane flow at phagosomes and tunnels.
LAMP1(溶酶体相关膜蛋白1)是一个在溶酶体功能和维护中起关键作用的蛋白质。以下是关于LAMP1的详细介绍: 功能: 溶酶体膜蛋白: LAMP1主要位于溶酶体的膜上,溶酶体是细胞内负责降解和回收各种生物分子的细胞器。 细胞内运输: LAMP1帮助溶酶体与其他细胞室的融合和运输,确保大分子的正确处理。 内吞作用和自噬: LAMP1参与内吞作用(细胞摄取外部物质)和自噬(细胞内部成分的降解)。 细胞表面标记物: LAMP1也可以作为细胞研究中的溶酶体标记物,用于追踪溶酶体的活动。 结构: 糖蛋白: LAMP1是糖蛋白,即它具有附加的糖分子,这些糖链对其功能和稳定性至关重要。 跨膜结构: 它通过跨膜域穿越溶酶体膜,细胞质部分朝向溶酶体内部。 临床相关性: 溶酶体储存病: LAMP1的突变或功能失调可能与溶酶体储存病有关,这种病症中溶酶体不能正常降解物质。 癌症研究: LAMP1的表达水平有时可以作为癌症研究中的标记,因为其水平在不同类型的癌症中可能有所不同。 总的来说,LAMP1对溶酶体功能和细胞稳态至关重要。 -
博氏疏螺旋体是引起莱姆病的病原体,这是一种影响皮肤、神经系统和关节的多系统疾病。
-
宿主细胞对疏螺旋体的摄取和细胞内处理过程包括几个步骤:
- i) 通过formin调控的丝状伪足将高度运动的疏螺旋体固定,
- ii) 由formin Daam1和Arp2/3复合物调节的螺旋伪足包裹,
- iii) 被Rab22a阳性吞噬体摄取,后者通过依赖ER的方式与Rab5a阳性小泡接触,
- iv) 通过膜管化减少吞噬体表面使疏螺旋体压缩,
- v) 在成熟的吞噬溶酶体中降解(参见图示)。
-
我们还发现了在疏螺旋体摄取和处理过程中发挥特定作用的脂类,如PI(3)P和磷脂酰丝氨酸。
-
此外,我们最近发现膜隧道(一种新型吞噬结构)深入宿主细胞胞质,比含细菌的吞噬体部分延伸得更深,可能是由于高度运动的疏螺旋体的部分抽离(参见图示)。
-
这表明巨噬细胞对疏螺旋体的内化是一个独特且高度动态的过程,其结果取决于螺旋体和宿主细胞之间的拉锯战。
-
值得注意的是,吞噬体和隧道都形成多个STIM1阳性的ER接触位点,表明ER连接过程(包括Ca2+内流调节)在含疏螺旋体的吞噬体成熟中的作用。
-
然而,ER接触位点的作用及吞噬体Ca2+内流在疏螺旋体吞噬体成熟中的具体作用尚不清楚。
-
为了解决这些问题,并识别疏螺旋体特异性调节吞噬体压缩和成熟的因素,我们已经建立了疏螺旋体的磁标记,从而允许随后纯化吞噬体。这些实验的结果将识别出在人体免疫细胞中调节螺旋体细胞内处理的新靶点。
-
该项目将:
- a) 分析含疏螺旋体的吞噬体和隧道的蛋白质组和脂质组,
- b) 在分子和功能水平上表征吞噬体和隧道的ER接触位点,
- c) 识别特异性调节螺旋体压缩的因子,相对于没有显著吞噬体压缩的细菌(如葡萄球菌和链球菌),或者采取不同细胞内生存或持久性策略的细菌(如耶尔森菌、军团菌或沙门氏菌)。
-
将结合巨噬细胞的SILAC标记与化学交联、(共同)免疫沉淀和邻近标记实验,以识别参与招募驱动吞噬体/隧道关闭和吞噬体压缩的分子机器的疏螺旋体表面配体。晶格光片显微镜和聚焦离子束显微镜将用于分析吞噬体和隧道的膜流动。
疏螺旋体摄取和细胞内处理的分子机制: 疏螺旋体在宿主细胞内的摄取和处理涉及哪些具体的分子通路?特别是formin、Daam1和Arp2/3复合物在这一过程中具体如何调控? 在疏螺旋体摄取和处理过程中,PI(3)P和磷脂酰丝氨酸分别发挥了什么具体作用? 膜隧道结构的功能和形成机制: 新发现的膜隧道结构是如何形成的?其形成机制与疏螺旋体的高运动性之间有什么关系? 这些膜隧道在疏螺旋体与宿主细胞的相互作用中起到了什么具体作用? ER接触位点在吞噬体成熟中的作用: ER接触位点(STIM1阳性)的形成机制是什么?它们在含疏螺旋体吞噬体的成熟过程中起到了什么具体作用? 吞噬体Ca2+内流在疏螺旋体吞噬体成熟中的具体作用是什么?它如何调节吞噬体的功能和疏螺旋体的降解过程? 疏螺旋体特异性吞噬体调节因子: 哪些特异性调节因子参与了疏螺旋体吞噬体的压缩和成熟?这些因子与其他细菌(如葡萄球菌、链球菌、耶尔森菌、军团菌和沙门氏菌)所需的因子有何不同? 在Borrelia感染过程中,哪些特异性调节因子和机制能够调节吞噬体的压缩和成熟,从而影响螺旋体的细胞内存活? 宿主细胞对疏螺旋体的免疫应答机制: 巨噬细胞对疏螺旋体的内化和处理过程中,宿主细胞的免疫应答是如何被调控的? 针对疏螺旋体的高运动性,宿主细胞有哪些特异性免疫防御机制? 药物靶点及治疗策略: 通过对疏螺旋体吞噬体及隧道结构的深入研究,是否可以发现新的药物靶点? 能否基于疏螺旋体特异性调节因子的研究,开发出针对莱姆病的新的治疗策略? Molecular Mechanisms of Borrelia burgdorferi Uptake and Intracellular Processing: What are the specific molecular pathways involved in the uptake and intracellular processing of Borrelia burgdorferi within host cells? Specifically, how do formin, Daam1, and the Arp2/3 complex regulate this process? What specific roles do PI(3)P and phosphatidylserine play during the uptake and processing of Borrelia burgdorferi? Function and Formation Mechanisms of Membrane Tunnels: How are the newly discovered membrane tunnels formed, and what is the relationship between their formation and the high motility of Borrelia burgdorferi? What specific roles do these membrane tunnels play in the interaction between Borrelia burgdorferi and host cells? Roles of ER Contact Sites in Phagosome Maturation: What are the mechanisms of formation for ER contact sites (STIM1-positive) and their specific roles in the maturation of Borrelia-containing phagosomes? What is the specific role of phagosomal Ca2+ influx in the maturation of Borrelia phagosomes, and how does it regulate the function of phagosomes and the degradation process of Borrelia burgdorferi? Borrelia-Specific Regulators of Phagosome Compaction and Maturation: What specific regulators are involved in the compaction and maturation of Borrelia-containing phagosomes? How do these regulators differ from those involved in the processing of other bacteria such as staphylococci, streptococci, Yersinia, Legionella, and Salmonella? During Borrelia infection, which specific regulators and mechanisms control phagosome compaction and maturation, thereby affecting the intracellular survival of Borrelia burgdorferi? Immune Response Mechanisms of Host Cells to Borrelia burgdorferi: How is the immune response of host cells regulated during the internalization and processing of Borrelia burgdorferi by macrophages? What specific immune defense mechanisms do host cells employ in response to the high motility of Borrelia burgdorferi? Drug Targets and Therapeutic Strategies: Can in-depth studies of Borrelia-containing phagosomes and membrane tunnels lead to the discovery of new drug targets? Based on research into Borrelia-specific regulators, is it possible to develop new therapeutic strategies for Lyme disease? These questions aim to delve deeply into the interaction mechanisms between Borrelia burgdorferi and host cells, potentially leading to the development of new treatment methods and improving the prevention and control of Lyme disease and related conditions. Mechanisms of Host-Pathogen Interaction: What are the key steps and molecular mechanisms involved in the uptake and intracellular processing of Borrelia burgdorferi by host cells? How do specific host cell structures and proteins, such as formins, phospholipids, and ER contact sites, contribute to the handling and degradation of Borrelia burgdorferi? Dynamics of Intracellular Pathogen Processing: What roles do newly discovered structures, like membrane tunnels, play in the intracellular journey of Borrelia burgdorferi? How does the motility of Borrelia burgdorferi influence its interaction with host cell phagosomes and subsequent intracellular processing? Cellular Defense Mechanisms: How do host immune cells, particularly macrophages, recognize, internalize, and destroy Borrelia burgdorferi? What are the cellular responses and defense mechanisms triggered by Borrelia burgdorferi infection? Phagosome Maturation and Pathogen Survival: What factors regulate the maturation of Borrelia-containing phagosomes, and how do these processes differ from those involving other pathogens? How do intracellular pathogens like Borrelia burgdorferi evade or manipulate host cell processes to ensure their survival and replication? Novel Therapeutic Targets: What potential drug targets can be identified from the molecular mechanisms involved in the uptake and degradation of Borrelia burgdorferi? How can understanding the specific interactions between Borrelia burgdorferi and host cells lead to new therapeutic strategies for Lyme disease? Role of Host Cell Structures in Infection: What is the significance of endoplasmic reticulum (ER) contact sites in the context of Borrelia burgdorferi infection, and how do they affect phagosome function and maturation? How do specific lipids and proteins on the host cell surface facilitate or hinder the intracellular processing of Borrelia burgdorferi? These questions aim to explore the broader aspects of host-pathogen interactions, intracellular processing of pathogens, and potential avenues for therapeutic intervention.
DNA virus: Papalloni-virus, Polyomavirus, Herpes-virus; RNA virus: HIV
男性性别决定了实验性中风后IL-17抗体治疗的长期神经保护效果
男性性别决定了实验性中风后IL-17抗体治疗的长期神经保护效果 (Male sex determines long-term neuroprotection of an Interleukin 17 antibody treatment after experimental stroke in aged mice)
引言
免疫系统的激活显著影响实验性中风模型中的缺血性组织损伤。激活的炎症级联反应中,γδ T细胞在缺血后脑中快速释放IL-17A,对于年轻雄性小鼠早期有害中性粒细胞浸润的放大至关重要。然而,其对长期中风结果的影响以及年龄和性别的效应仍然未知。因此,我们研究的目的是分析性别和年龄是否影响IL-17抗体(Ab)治疗在实验性中风中的效果。
方法
为了研究早期IL-17中和对长期结果的影响,我们在短暂性大脑中动脉阻塞(tMCAO)后1小时给12-14个月大的雄性或雌性小鼠施用IL-17Ab或IgG对照(20µg/kg体重),并在中风后的第3天评估病灶体积以及行为结果,跟踪至90天。我们通过流式细胞术和免疫组织化学分析了缺血后的炎症反应。为了揭示肠道微生物群对IL-17A+ γδ T细胞的影响,我们进行了微生物16S rRNA测序和粪便微生物移植(FMT)。
结果
IL-17中和在tMCAO后1小时显著减少了中风后的第3天的梗死体积,这在年长雄性小鼠中伴随着死亡率降低和90天后的神经功能改善,但在年长雌性小鼠中没有观察到类似效果。因此,年长雄性小鼠的大脑浸润的γδ T细胞在中风后第3天的IL-17A水平较年长雌性小鼠更高,并且在IL-17Ab治疗后,中性粒细胞浸润仅在年长雄性小鼠中减少。此外,16S rRNA测序揭示了一个不同的微生物谱,包括年长雄性小鼠的短链脂肪酸生产相关途径的减少。最后,年长雄性小鼠的粪便FMT在年轻雄性受体小鼠中诱导了更高的IL-17A水平,而年长雌性小鼠的粪便移植则没有这种效果,突显了微生物群的调节作用。
结论
IL-17中和改善了年长雄性小鼠的90天长期结果,但未对年长雌性小鼠产生相同效果,这与年长雄性小鼠的微生物谱强化了IL-17A轴的情况相关。这些发现突显了以IL-17A为中心的中风治疗的潜力,并强调了未来中风研究中考虑性别差异的重要性。
参考文献
- [1] Endres M, Moro MA, Nolte CH, Dames C, Buckwalter MS, Meisel A. Immune Pathways in Etiology, Acute Phase, and Chronic Sequelae of Ischemic Stroke. Circ Res. 2022;130(8):1167-86.
- [2] Shichita T, Sugiyama Y, Ooboshi H, Sugimori H, Nakagawa R, Takada I, Iwaki T, Okada Y, Iida M, Cua DJ, Iwakura Y, Yoshimura A. Pivotal role of cerebral interleukin-17-producing gammadeltaT cells in the delayed phase of ischemic brain injury. Nat Med. 2009;15(8):946-50.
- [3] Gelderblom M, Koch S, Strecker J-K, Jørgensen C, Garcia-Bonilla L, Ludewig P, Schädlich IS, Piepke M, Degenhardt K, Bernreuther C, Pinnschmidt H, Arumugam TV, Thomalla G, Faber C, Sedlacik J, Gerloff C, Minnerup J, Clausen BH, Anrather J, Magnus T. A preclinical randomized controlled multicenter trial of anti-interleukin-17A treatment for acute ischemic stroke. Brain Communications. 2023.
图表说明: 性别差异在IL-17A轴中决定了年长小鼠的中风后结果
- A:梗死体积(3天),B:生存率,C:神经评分(90天)
- D:IL-17A+ CNS γδ T细胞百分比(3天)
- E:IL-17Ab或IgG处理的年长雄性或雌性小鼠的大脑中性粒细胞计数/mm²(1天)
- F:年长雄性或雌性小鼠粪便内容物的16S rRNA测序主成分分析(PCA)
- G:16S rRNA测序途径分析
- H:年轻雄性小鼠在接受年长雄性、雌性或年轻雄性粪便FMT后的IL-17A+ CNS γδ T细胞百分比(3天). // P<0.05/0.01/0.001,
- A, D, E 采用未配对的Student’s t检验,B 采用χ²检验,C 采用Mann-Whitney U检验,H 采用单因素方差分析(Sidak多重比较)。
Introduction
The activation of the immune system significantly impacts ischemic tissue damage in experimental stroke models1. Among the activated inflammatory cascades, the rapid release of IL-17A by γδ T cells in the post-ischemic brain is pivotal for the amplification of the early detrimental neutrophil infiltration in young male mice2,3. However, its influence on long-term stroke outcomes and the effects of age and sex remain unknown. Thus, the objectives of our study were to analyze whether sex and age influence the effect of an IL-17-Antibody (Ab) treatment in experimental stroke.
Methods
To investigate the effect of early IL-17 neutralization on long-term outcomes, we administered IL-17Ab or IgG control (20µg/kg body weight) to 12-14 month-old male or female mice 1h following transient middle cerebral artery occlusion (tMCAO) and assessed lesion volume at 3 days (d) and behavioral outcomes up to 90d post-stroke. We analyzed the post-ischemic inflammatory response using flow cytometry and immunohistochemistry. To decipher the influence of the gut microbiota on IL-17A+ γδ T cells, we performed microbial 16S rRNA sequencing and Fecal Microbial Transplantations (FMTs).
Results
IL-17-neutralization 1h post-tMCAO significantly reduces infarct volume 3d after stroke, which is accompanied by reduced mortality and improved neurological outcomes after 90d in aged male, but not in aged female mice (A-C). Accordingly, brain infiltrating γδ T cells from aged male mice show increased IL-17A levels 3d post-stroke compared to aged female mice, correlating with decreased neutrophil infiltration seen only in aged male mice after IL-17Ab treatment (D, E). Furthermore, 16S rRNAseq revealed a distinct microbial profile, including a decline in pathways linked to short-chain-fatty-acid production in aged male compared to aged female mice (F, G). Finally, FMTs of aged male stool induced higher IL-17A levels in CNS γδ T cells in young male recipient mice post-stroke compared to aged female stool transfers, highlighting the modulation by the microbiota (H).
Conclusion
IL-17 neutralization improves 90d long-term outcomes in aged male but not aged female mice, which is associated with a microbial profile strengthening the IL-17A axis in aged males. These findings highlight the potential of IL-17A-centered stroke therapies and underscore the importance of considering sex differences in future stroke studies.
Sex differences in the IL-17A axis determine post-stroke outcome in aged mice
- A Infarct volume 3d,
- B Survival and
- C Neuroscore 90d post-tMCAO of IL-17Ab or IgG treated aged male or female mice.
- D % IL-17A+ CNS γδ T cells 3d post-tMCAO.
- E Brain neutrophil counts/mm2 of IL-17Ab or IgG treated aged male or female mice 1d post-tMCAO.
- F PCA and
- G Pathway analysis of 16s rRNAseq of stool content of aged male or female mice.
- H % IL-17A+ CNS γδ T cells of young male mice after FMT of aged male, female or young male stool 3d post-tMCAO. // P<0.05/0.01/0.001,
- A, D, E unpaired student’s t-Test, B χ2-test, C Mann-Whitney U-test, H one-way ANOVA (Sidak‘s multiple comparison).
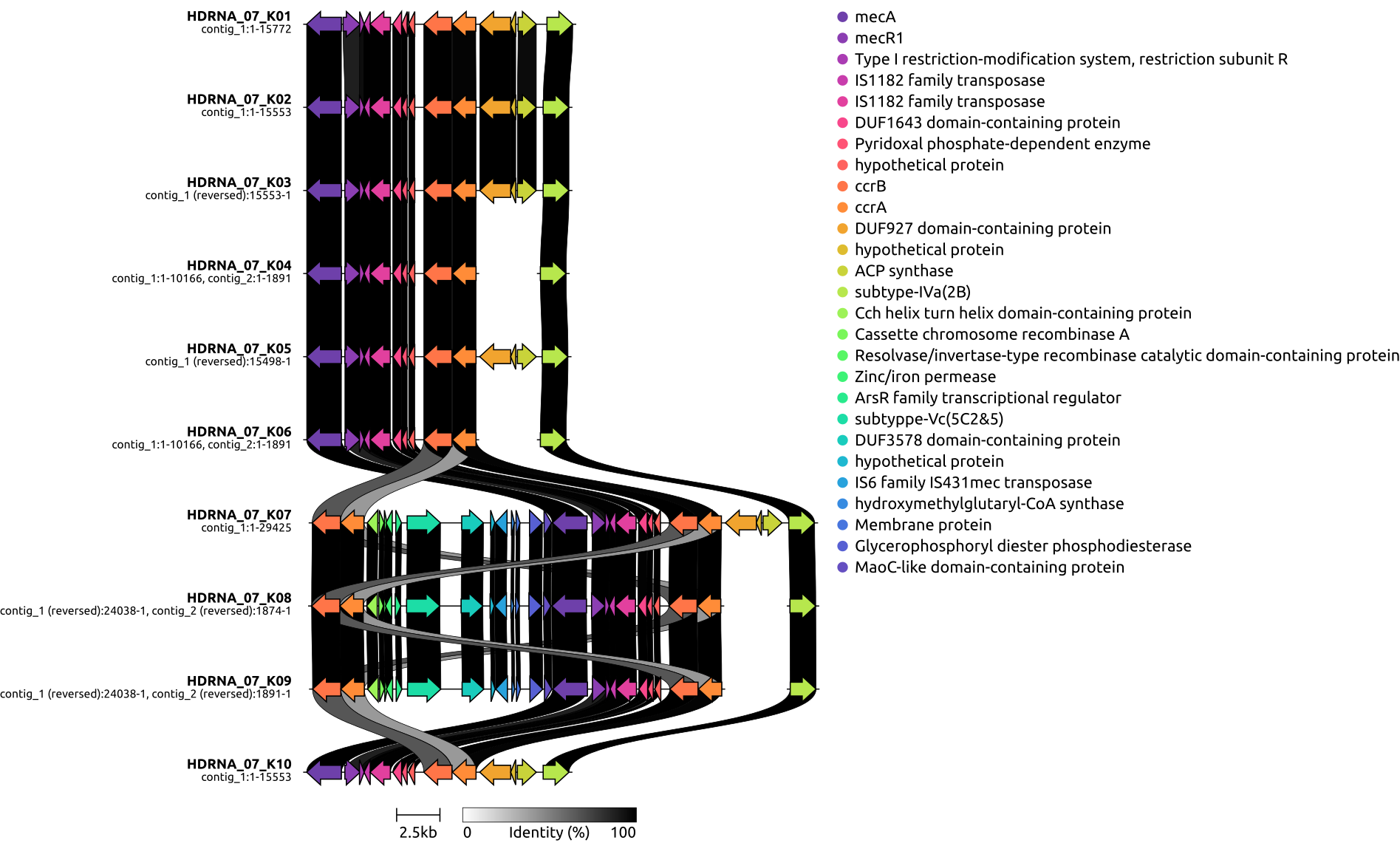
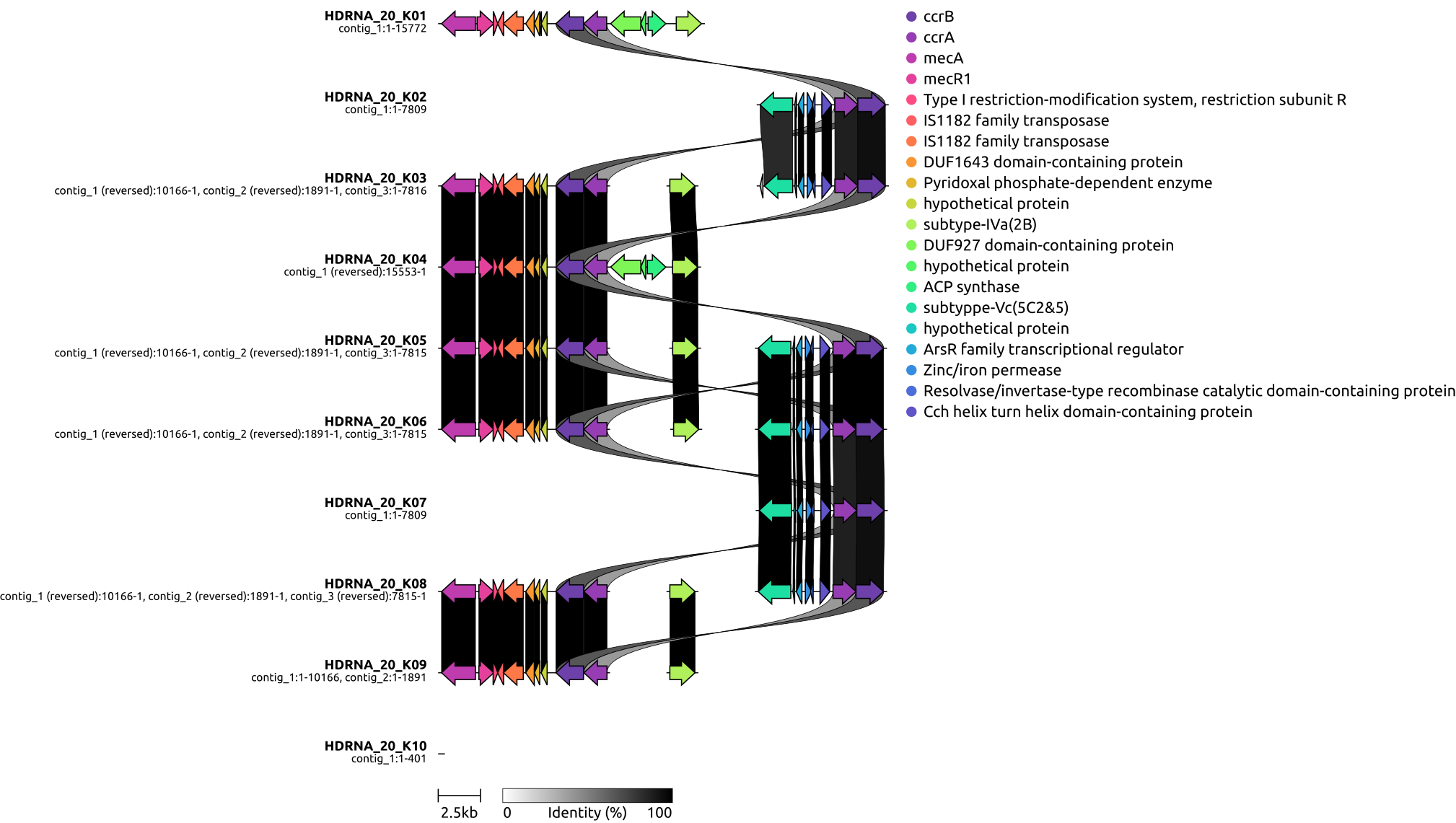
draw graphics on local genetic environments of SCCmec using clinker
TODO: read some papers about recombination and horizontal gene transfer of SCCmec
Identification and characterization of SCCmec typing with psm-mec positivity in staphylococci from patients with coagulase-negative staphylococci peritoneal dialysis-related peritonitis
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10517493/
SCCmec in staphylococci: genes on the move
1, SCCmec background
SCCmec is a 21 to 60 kb long genetic element that confers broad-spectrum β-lactam resistance to MRSA.[2] Moreover, additional genetic elements like Tn554, pT181, and pUB110 can be found in SCCmec, which have the capability to render resistance to various non-β-lactam drugs.
The mec complex is divided further into five types (I through V) based on the arrangement of regulatory genetic features such as mecR1, an inducer.[7] The mec gene complex in SCCmec, comprising mec gene, its regulators (mecR1, mecI), and insertion sequences (IS), is categorized into five classes (A to E).
- Class A includes mecA, full mecR1, mecI, and IS431.
- Class B has IS1272, mecA, partial mecR1, and IS431.
- Class C, with two versions (C1, C2), contains mecA, partial mecR1, IS431, differing in IS431 orientation.
- Class D includes IS431, mecA, partial mecR1;
- Class E consists of blaZ, mecC, mecR1, mecI.[7][8][9]2, prepare files in directory gbs (cd ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/gbs/)
cp /home/jhuang/Tools/bacto/db/CP133676.gb_converted.fna ./
#cp ../snippy_CP133676/shovill/HDRNA_01_K01/contigs.fa HDRNA_01_K01_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K02/contigs.fa HDRNA_01_K02_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K03/contigs.fa HDRNA_01_K03_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K04/contigs.fa HDRNA_01_K04_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K05/contigs.fa HDRNA_01_K05_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K06/contigs.fa HDRNA_01_K06_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K07/contigs.fa HDRNA_01_K07_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K08/contigs.fa HDRNA_01_K08_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K09/contigs.fa HDRNA_01_K09_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K10/contigs.fa HDRNA_01_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133677.gb_converted.fna ./
#cp ../snippy_CP133677/shovill/HDRNA_03_K01/contigs.fa HDRNA_03_K01_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K02/contigs.fa HDRNA_03_K02_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K03/contigs.fa HDRNA_03_K03_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K04/contigs.fa HDRNA_03_K04_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K05/contigs.fa HDRNA_03_K05_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K06/contigs.fa HDRNA_03_K06_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K07/contigs.fa HDRNA_03_K07_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K08/contigs.fa HDRNA_03_K08_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K09/contigs.fa HDRNA_03_K09_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K10/contigs.fa HDRNA_03_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133678.gb_converted.fna ./
#cp ../snippy_CP133678/shovill/HDRNA_06_K01/contigs.fa HDRNA_06_K01_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K02/contigs.fa HDRNA_06_K02_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K03/contigs.fa HDRNA_06_K03_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K04/contigs.fa HDRNA_06_K04_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K05/contigs.fa HDRNA_06_K05_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K06/contigs.fa HDRNA_06_K06_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K07/contigs.fa HDRNA_06_K07_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K08/contigs.fa HDRNA_06_K08_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K09/contigs.fa HDRNA_06_K09_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K10/contigs.fa HDRNA_06_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133680.gb_converted.fna ./
#cp ../snippy_CP133680/shovill/HDRNA_07_K01/contigs.fa HDRNA_07_K01_contigs.fa
#cp ../snippy_CP133680/shovill/HDRNA_07_K01-BB28/contigs.fa HDRNA_07_K01-BB28_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K02/contigs.fa HDRNA_07_K02_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K03/contigs.fa HDRNA_07_K03_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K04/contigs.fa HDRNA_07_K04_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K05/contigs.fa HDRNA_07_K05_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K06/contigs.fa HDRNA_07_K06_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K07/contigs.fa HDRNA_07_K07_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K08/contigs.fa HDRNA_07_K08_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K09/contigs.fa HDRNA_07_K09_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K10/contigs.fa HDRNA_07_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133682.gb_converted.fna ./
#cp ../snippy_CP133682/shovill/HDRNA_08_K01/contigs.fa HDRNA_08_K01_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K02/contigs.fa HDRNA_08_K02_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K03/contigs.fa HDRNA_08_K03_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K04/contigs.fa HDRNA_08_K04_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K05/contigs.fa HDRNA_08_K05_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K06/contigs.fa HDRNA_08_K06_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K07/contigs.fa HDRNA_08_K07_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K08/contigs.fa HDRNA_08_K08_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K09/contigs.fa HDRNA_08_K09_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K10/contigs.fa HDRNA_08_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133684.gb_converted.fna ./
#cp ../snippy_CP133684/shovill/HDRNA_12_K01/contigs.fa HDRNA_12_K01_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K02/contigs.fa HDRNA_12_K02_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K03/contigs.fa HDRNA_12_K03_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K04/contigs.fa HDRNA_12_K04_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K05/contigs.fa HDRNA_12_K05_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K06/contigs.fa HDRNA_12_K06_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K07/contigs.fa HDRNA_12_K07_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K08/contigs.fa HDRNA_12_K08_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K09/contigs.fa HDRNA_12_K09_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K10/contigs.fa HDRNA_12_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133688.gb_converted.fna ./
#cp ../snippy_CP133688/shovill/HDRNA_16_K01/contigs.fa HDRNA_16_K01_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K02/contigs.fa HDRNA_16_K02_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K03/contigs.fa HDRNA_16_K03_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K04/contigs.fa HDRNA_16_K04_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K05/contigs.fa HDRNA_16_K05_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K06/contigs.fa HDRNA_16_K06_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K07/contigs.fa HDRNA_16_K07_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K08/contigs.fa HDRNA_16_K08_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K09/contigs.fa HDRNA_16_K09_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K10/contigs.fa HDRNA_16_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133693.gb_converted.fna ./
#cp ../snippy_CP133693/shovill/HDRNA_17_K01/contigs.fa HDRNA_17_K01_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K02/contigs.fa HDRNA_17_K02_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K03/contigs.fa HDRNA_17_K03_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K04/contigs.fa HDRNA_17_K04_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K05/contigs.fa HDRNA_17_K05_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K06/contigs.fa HDRNA_17_K06_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K07/contigs.fa HDRNA_17_K07_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K08/contigs.fa HDRNA_17_K08_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K09/contigs.fa HDRNA_17_K09_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K10/contigs.fa HDRNA_17_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133696.gb_converted.fna ./
#cp ../snippy_CP133696/shovill/HDRNA_19_K01/contigs.fa HDRNA_19_K01_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K02/contigs.fa HDRNA_19_K02_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K03/contigs.fa HDRNA_19_K03_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K04/contigs.fa HDRNA_19_K04_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K05/contigs.fa HDRNA_19_K05_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K06/contigs.fa HDRNA_19_K06_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K07/contigs.fa HDRNA_19_K07_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K08/contigs.fa HDRNA_19_K08_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K09/contigs.fa HDRNA_19_K09_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K10/contigs.fa HDRNA_19_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133700.gb_converted.fna ./
#cp ../snippy_CP133700/shovill/HDRNA_20_K01/contigs.fa HDRNA_20_K01_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K02/contigs.fa HDRNA_20_K02_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K03/contigs.fa HDRNA_20_K03_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K04/contigs.fa HDRNA_20_K04_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K05/contigs.fa HDRNA_20_K05_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K06/contigs.fa HDRNA_20_K06_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K07/contigs.fa HDRNA_20_K07_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K08/contigs.fa HDRNA_20_K08_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K09/contigs.fa HDRNA_20_K09_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K10/contigs.fa HDRNA_20_K10_contigs.fa3, draw graphics on local genetic environments of SCCmec using clinker
# - draw a graphics for the gene organization of SCCmec regions for 10 groups, using phylogenetic tree for the 100 isolates.
# - regenerate a SNP Indels table for each groups, marked the SNPs and Indels in the regions of SCCmec yellow.
#in snippy_CP133676
cp /home/jhuang/Tools/bacto/db/CP133676.gb_converted.fna ./
#cp ../snippy_CP133676/shovill/HDRNA_01_K01/contigs.fa HDRNA_01_K01_contigs.fa #SCCmec_type_IVa(2B) --> 40444-55810 --> chr:30444-65810
cp ../snippy_CP133676/shovill/HDRNA_01_K02/contigs.fa HDRNA_01_K02_contigs.fa #SCCmec_type_IVa(2B) --> contig00005+contig00011
cp ../snippy_CP133676/shovill/HDRNA_01_K03/contigs.fa HDRNA_01_K03_contigs.fa #SCCmec_type_IVa(2B) --> contig00006+contig00004
cp ../snippy_CP133676/shovill/HDRNA_01_K04/contigs.fa HDRNA_01_K04_contigs.fa #SCCmec_type_IVa(2B) --> contig00002
cp ../snippy_CP133676/shovill/HDRNA_01_K05/contigs.fa HDRNA_01_K05_contigs.fa #SCCmec_type_IVa(2B) --> contig00006+contig00004
cp ../snippy_CP133676/shovill/HDRNA_01_K06/contigs.fa HDRNA_01_K06_contigs.fa #SCCmec_type_IVa(2B) --> contig00002
cp ../snippy_CP133676/shovill/HDRNA_01_K07/contigs.fa HDRNA_01_K07_contigs.fa #SCCmec_type_IVa(2B) --> contig00002
cp ../snippy_CP133676/shovill/HDRNA_01_K08/contigs.fa HDRNA_01_K08_contigs.fa #SCCmec_type_IVa(2B) --> contig00021+contig00011
cp ../snippy_CP133676/shovill/HDRNA_01_K09/contigs.fa HDRNA_01_K09_contigs.fa #SCCmec_type_IVa(2B) --> contig00006+contig00004
cp ../snippy_CP133676/shovill/HDRNA_01_K10/contigs.fa HDRNA_01_K10_contigs.fa #SCCmec_type_IVa(2B) --> contig00008+contig00002
#samtools faidx CP133676.gb_converted.fna CP133676:30444-65810 > CP133676_30444-65810.fasta
#samtools faidx HDRNA_01_K02_contigs.fa contig00005 > HDRNA_01_K02_selected.fasta
##samtools faidx HDRNA_01_K02_contigs.fa contig00011 >> HDRNA_01_K02_selected.fasta
#samtools faidx HDRNA_01_K03_contigs.fa contig00006 > HDRNA_01_K03_selected.fasta
##samtools faidx HDRNA_01_K03_contigs.fa contig00004 >> HDRNA_01_K03_selected.fasta
#samtools faidx HDRNA_01_K04_contigs.fa contig00002 > HDRNA_01_K04_selected.fasta
#samtools faidx HDRNA_01_K05_contigs.fa contig00006 > HDRNA_01_K05_selected.fasta
##samtools faidx HDRNA_01_K05_contigs.fa contig00004 >> HDRNA_01_K05_selected.fasta
#samtools faidx HDRNA_01_K06_contigs.fa contig00002 > HDRNA_01_K06_selected.fasta
#samtools faidx HDRNA_01_K07_contigs.fa contig00002 > HDRNA_01_K07_selected.fasta
#samtools faidx HDRNA_01_K08_contigs.fa contig00021 > HDRNA_01_K08_selected.fasta
##samtools faidx HDRNA_01_K08_contigs.fa contig00011 >> HDRNA_01_K08_selected.fasta
#samtools faidx HDRNA_01_K09_contigs.fa contig00006 > HDRNA_01_K09_selected.fasta
##samtools faidx HDRNA_01_K09_contigs.fa contig00004 >> HDRNA_01_K09_selected.fasta
#samtools faidx HDRNA_01_K10_contigs.fa contig00008 > HDRNA_01_K10_selected.fasta
##samtools faidx HDRNA_01_K10_contigs.fa contig00002 >> HDRNA_01_K10_selected.fasta
# -- install bakta --
conda create --name bakta
conda activate bakta
mamba install -c conda-forge -c bioconda bakta
bakta_db list
bakta_db download --output /mnt/nvme0n1p1/REFs --type full
mv /mnt/nvme0n1p1/REFs/db /mnt/nvme0n1p1/REFs/bakta_db
#Run Bakta using '--db /mnt/nvme0n1p1/REFs/bakta_db' or set a BAKTA_DB environment variable: 'export BAKTA_DB=/mnt/nvme0n1p1/REFs/db'
# -- HDRNA_01_K01=CP133676
#mecA:12:AB505628 100.00 2010/2010 CP133676 40444..42453
#dmecR1:1:AB033763 100.00 987/987 CP133676 42550..43536
#IS1272:3:AM292304 100.00 1843/1843 CP133676 43525..45367
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133676 47209..48858
#ccrA2:7:81108:AB096217 99.63 1350/1350 CP133676 48859..50203
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133676 54320..55810 #subtype-IVa(2B) is always located in another contig --> ignoring this entity!
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133676.gb_converted.fna
#ADAPT the gene_name and positions following the info below in CP133676_adapted.gb, e.g. adding /gene="mecA" and correct the positions.
python3 ~/Scripts/extract_subregion.py CP133676.gb_converted.gbff 40244 56010 CP133676_sub.gbff
#50303-52324 contains DUF927 domain-containing protein
#54320-40244=14076
#14076..15566
#/product="AAA family ATPase"
# -- HDRNA_01_K02
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00005 3825..5169
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00005 5170..6819
#IS1272:3:AM292304 100.00 1843/1843 contig00005 8661..10503
#dmecR1:1:AB033763 100.00 987/987 contig00005 10492..11478
#mecA:12:AB505628 100.00 2010/2010 contig00005 11575..13584
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00011 86471..87961
#python3 extract_gb_from_gbk.py snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.gbk contig00005 HDRNA_01_K02_contig00005.gb
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.fna contig00005:3625-13784 > HDRNA_01_K02_sub.fna
#+4117=92278
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.fna contig00011:86271-88161 >> HDRNA_01_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K02_sub.fna
#python3 ~/Scripts/extract_subregion.py HDRNA_01_K02_contig00005.gbff 3725 13684 HDRNA_01_K02_contig00005_sub.gb
# -- HDRNA_01_K03
#dmecR1:1:AB033763 100.00 987/987 contig00006 10492..11478
#mecA:12:AB505628 100.00 2010/2010 contig00006 11575..13584
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00006 3825..5169
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00006 5170..6819
#IS1272:3:AM292304 100.00 1843/1843 contig00006 8661..10503
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00004 211325..212815
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K03/HDRNA_01_K03.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K03/HDRNA_01_K03.fna contig00006:3625-13784 > HDRNA_01_K03_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K03/HDRNA_01_K03.fna contig00004:211125-213015 >> HDRNA_01_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K03_sub.fna
# -- HDRNA_01_K04
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00002 389566..390910
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 390911..392560
#IS1272:3:AM292304 100.00 1843/1843 contig00002 394402..396244
#dmecR1:1:AB033763 100.00 987/987 contig00002 396233..397219
#mecA:12:AB505628 100.00 2010/2010 contig00002 397316..399325
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 384124..385614
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K04/HDRNA_01_K04.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K04/HDRNA_01_K04.fna contig00002:383924-399525 > HDRNA_01_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K04_sub.fna
# -- HDRNA_01_K05
#dmecR1:1:AB033763 100.00 987/987 contig00006 10657..11643
#mecA:12:AB505628 100.00 2010/2010 contig00006 11740..13749
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00006 3990..5334
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00006 5335..6984
#IS1272:3:AM292304 100.00 1843/1843 contig00006 8826..10668
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00004 212178..213668
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K05/HDRNA_01_K05.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K05/HDRNA_01_K05.fna contig00006:3790-13949 > HDRNA_01_K05_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K05/HDRNA_01_K05.fna contig00004:211978-213868 >> HDRNA_01_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K05_sub.fna
# -- HDRNA_01_K06
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00002 389999..391343
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 391344..392993
#IS1272:3:AM292304 100.00 1843/1843 contig00002 394835..396677
#dmecR1:1:AB033763 100.00 987/987 contig00002 396666..397652
#mecA:12:AB505628 100.00 2010/2010 contig00002 397749..399758
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 384557..386047
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K06/HDRNA_01_K06.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K06/HDRNA_01_K06.fna contig00002:384357-399858 > HDRNA_01_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K06_sub.fna
# -- HDRNA_01_K07
#mecA:12:AB505628 100.00 2010/2010 contig00002 172913..174922
#dmecR1:1:AB033763 100.00 987/987 contig00002 175019..176005
#IS1272:3:AM292304 100.00 1843/1843 contig00002 175994..177836
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 179678..181327
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00002 181328..182672
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 186624..188114
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K07/HDRNA_01_K07.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K07/HDRNA_01_K07.fna contig00002:172813-188314 > HDRNA_01_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K07_sub.fna
# -- HDRNA_01_K08
#dmecR1:1:AB033763 100.00 987/987 contig00021 10445..11431
#mecA:3:AB037671 100.00 2004/2007 contig00021 11535..13538
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00021 3778..5122
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00021 5123..6772
#IS1272:3:AM292304 100.00 1843/1843 contig00021 8614..10456
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00011 72699..74189
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K08/HDRNA_01_K08.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K08/HDRNA_01_K08.fna contig00021:3578-13738 > HDRNA_01_K08_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K08/HDRNA_01_K08.fna contig00011:72499-74389 >> HDRNA_01_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K08_sub.fna
# -- HDRNA_01_K09
#dmecR1:1:AB033763 100.00 987/987 contig00006 10492..11478
#mecA:12:AB505628 100.00 2010/2010 contig00006 11575..13584
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00006 3825..5169
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00006 5170..6819
#IS1272:3:AM292304 100.00 1843/1843 contig00006 8661..10503
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00004 211303..212793
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K09/HDRNA_01_K09.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K09/HDRNA_01_K09.fna contig00006:3625-13784 > HDRNA_01_K09_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K09/HDRNA_01_K09.fna contig00004:211103-212993 >> HDRNA_01_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K09_sub.fna
# -- HDRNA_01_K10
#dmecR1:1:AB033763 100.00 987/987 contig00008 10417..11403
#mecA:12:AB505628 100.00 2010/2010 contig00008 11500..13509
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00008 3750..5094
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00008 5095..6744
#IS1272:3:AM292304 100.00 1843/1843 contig00008 8586..10428
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 382889..384379
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K10/HDRNA_01_K10.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K10/HDRNA_01_K10.fna contig00008:3550-13709 > HDRNA_01_K10_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K10/HDRNA_01_K10.fna contig00002:382689-384579 >> HDRNA_01_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K10_sub.fna
# -- CP133677
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133677 2112527..2114017
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133677 2117969..2119318
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133677 2119319..2120968
#IS1272:3:AM292304 99.95 1844/1843 CP133677 2122810..2124653
#dmecR1:1:AB033763 100.00 987/987 CP133677 2124642..2125628
#mecA:12:AB505628 100.00 2010/2010 CP133677 2125725..2127734
#bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133677.gb_converted.fna
#python3 ~/Scripts/extract_subregion.py CP133677.gb_converted.gbff 2112327 2127934 HDRNA_03_K01.gbff
revseq ~/Tools/bacto/db/CP133677.gb_converted.fna
#Re-submit cp133677.rev to https://cge.food.dtu.dk/services/SCCmecFinder-1.2/
#mecA:12:AB505628 100.00 2010/2010 CP133677 462542..464551
#dmecR1:1:AB033763 100.00 987/987 CP133677 464648..465634
#IS1272:3:AM292304 99.95 1844/1843 CP133677 465623..467466
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133677 469308..470957
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133677 470958..472307
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133677 476259..477749
bakta --db /mnt/nvme0n1p1/REFs/bakta_db cp133677.rev
python3 ~/Scripts/extract_subregion.py cp133677.gbff 462342 477949 HDRNA_03_K01.gbff
# -- HDRNA_03_K02
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00026 13584..14933
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00026 14934..16583
#IS1272:3:AM292304 99.95 1844/1843 contig00026 18425..20268
#dmecR1:1:AB033763 100.00 987/987 contig00026 20257..21243
#mecA:12:AB505628 100.00 2010/2010 contig00026 21340..23349
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00026 8252..9742
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K02/HDRNA_03_K02.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K02/HDRNA_03_K02.fna contig00026:8052-23549 > HDRNA_03_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K02_sub.fna
# -- HDRNA_03_K03
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00029 13619..14968
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 14969..16618
#IS1272:3:AM292304 99.95 1844/1843 contig00029 18460..20303
#dmecR1:1:AB033763 100.00 987/987 contig00029 20292..21278
#mecA:12:AB505628 100.00 2010/2010 contig00029 21375..23384
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 8232..9722
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K03/HDRNA_03_K03.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K03/HDRNA_03_K03.fna contig00029:8032-23584 > HDRNA_03_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K03_sub.fna
# -- HDRNA_03_K04
#dmecR1:1:AB033763 100.00 987/987 contig00032 10498..11484
#mecA:12:AB505628 100.00 2010/2010 contig00032 11581..13590
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00032 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00032 5175..6824
#IS1272:3:AM292304 99.95 1844/1843 contig00032 8666..10509
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00036 8277..9767
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K04/HDRNA_03_K04.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K04/HDRNA_03_K04.fna contig00032:3625-13790 > HDRNA_03_K04_sub.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K04/HDRNA_03_K04.fna contig00036:8077-9967 >> HDRNA_03_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K04_sub.fna
# -- HDRNA_03_K05
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00029 13774..15123
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 15124..16773
#IS1272:3:AM292304 99.95 1844/1843 contig00029 18615..20458
#dmecR1:1:AB033763 100.00 987/987 contig00029 20447..21433
#mecA:12:AB505628 100.00 2010/2010 contig00029 21530..23539
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 8387..9877
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K05/HDRNA_03_K05.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K05/HDRNA_03_K05.fna contig00029:8187-23739 > HDRNA_03_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K05_sub.fna
# -- HDRNA_03_K06
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00030 13739..15088
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00030 15089..16738
#IS1272:3:AM292304 99.95 1844/1843 contig00030 18580..20423
#dmecR1:1:AB033763 100.00 987/987 contig00030 20412..21398
#mecA:12:AB505628 100.00 2010/2010 contig00030 21495..23504
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00030 8352..9842
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K06/HDRNA_03_K06.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K06/HDRNA_03_K06.fna contig00030:8152-23704 > HDRNA_03_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K06_sub.fna
# -- HDRNA_03_K07
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00027 11087..12436
#mecA:12:AB505628 100.00 2010/2010 contig00027 2671..4680
#dmecR1:1:AB033763 100.00 987/987 contig00027 4777..5763
#IS1272:3:AM292304 99.95 1844/1843 contig00027 5752..7595
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 9437..11086
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00027 16333..17823
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K07/HDRNA_03_K07.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K07/HDRNA_03_K07.fna contig00027:2471-18023 > HDRNA_03_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K07_sub.fna
# -- HDRNA_03_K08
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00029 13631..14980
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 14981..16630
#IS1272:3:AM292304 99.95 1844/1843 contig00029 18472..20315
#dmecR1:1:AB033763 100.00 987/987 contig00029 20304..21290
#mecA:12:AB505628 100.00 2010/2010 contig00029 21387..23396
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 8244..9734
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K08/HDRNA_03_K08.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K08/HDRNA_03_K08.fna contig00029:8044-23596 > HDRNA_03_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K08_sub.fna
# -- HDRNA_03_K09
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00025 25311..26660
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00025 26661..28310
#IS1272:3:AM292304 99.95 1844/1843 contig00025 30152..31995
#dmecR1:1:AB033763 100.00 987/987 contig00025 31984..32970
#mecA:12:AB505628 100.00 2010/2010 contig00025 33067..35076
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00025 19924..21414
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K09/HDRNA_03_K09.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K09/HDRNA_03_K09.fna contig00025:19724-35276 > HDRNA_03_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K09_sub.fna
# -- HDRNA_03_K10
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00027 13754..15103
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 15104..16753
#IS1272:3:AM292304 99.95 1844/1843 contig00027 18595..20438
#dmecR1:1:AB033763 100.00 987/987 contig00027 20427..21413
#mecA:12:AB505628 100.00 2010/2010 contig00027 21510..23519
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00027 8367..9857
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K10/HDRNA_03_K10.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K10/HDRNA_03_K10.fna contig00027:8167-23719 > HDRNA_03_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K10_sub.fna
#CP133678
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 CP133678 42063..43217
#ccrA2:7:81108:AB096217 99.93 1350/1350 CP133678 47234..48583
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 CP133678 48584..50233
#IS1272:2:AB033763 100.00 1585/1585 CP133678 52075..53659
#dmecR1:1:AB033763 100.00 987/987 CP133678 55466..56452
#mecA:12:AB505628 100.00 2010/2010 CP133678 56549..58558
#bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133678.gb_converted.fna
#python3 ~/Scripts/extract_subregion.py CP133678.gb_converted.gbff 41863 58758 HDRNA_06_K01.gbff
revseq ~/Tools/bacto/db/CP133678.gb_converted.fna
#Re-submit cp133678.rev to https://cge.food.dtu.dk/services/SCCmecFinder-1.2/
#mecA:12:AB505628 100.00 2010/2010 CP133678 2406703..2408712
#dmecR1:1:AB033763 100.00 987/987 CP133678 2408809..2409795
#IS1272:2:AB033763 100.00 1585/1585 CP133678 2411602..2413186
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 CP133678 2415028..2416677
#ccrA2:7:81108:AB096217 99.93 1350/1350 CP133678 2416678..2418027
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 CP133678 2422044..2423198
bakta --db /mnt/nvme0n1p1/REFs/bakta_db cp133678.rev
python3 ~/Scripts/extract_subregion.py cp133678.gbff 2406503 2423398 HDRNA_06_K01.gbff
#HDRNA_06_K02
##IS1272:2:AB033763 91.00 1523/1585 contig00036 86..1608
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00031 1464..2618
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K02/HDRNA_06_K02.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K02/HDRNA_06_K02.fna contig00031:1264-2818 > HDRNA_06_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K02_sub.fna
#HDRNA_06_K03
##IS1272:2:AB033763 91.00 1523/1585 contig00038 1..1523
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00033 4048..5202
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K03/HDRNA_06_K03.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K03/HDRNA_06_K03.fna contig00033:3848-5402 > HDRNA_06_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K03_sub.fna
#HDRNA_06_K04
##IS1272:2:AB033763 91.00 1523/1585 contig00043 1..1523
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00039 1444..2598
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K04/HDRNA_06_K04.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K04/HDRNA_06_K04.fna contig00039:1244-2798 > HDRNA_06_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K04_sub.fna
#HDRNA_06_K05
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00025 13961..15310
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00025 15311..16960
#IS1272:2:AB033763 100.00 1585/1585 contig00025 18802..20386
#dmecR1:1:AB033763 100.00 987/987 contig00033 357..1343
#mecA:12:AB505628 100.00 2010/2010 contig00033 1440..3449
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00025 8790..9944
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K05/HDRNA_06_K05.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K05/HDRNA_06_K05.fna contig00025:8590-20586 > HDRNA_06_K05_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K05/HDRNA_06_K05.fna contig00033:157-3649 >> HDRNA_06_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K05_sub.fna
#HDRNA_06_K06
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00027 11981..13330
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00027 13331..14980
#IS1272:2:AB033763 100.00 1585/1585 contig00027 16822..18406
#mecA:12:AB505628 100.00 2010/2010 contig00032 1634..3643
#dmecR1:1:AB033763 100.00 987/987 contig00032 3740..4726
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00027 6810..7964
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K06/HDRNA_06_K06.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K06/HDRNA_06_K06.fna contig00027:6610-18606 > HDRNA_06_K06_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K06/HDRNA_06_K06.fna contig00032:1434-4926 >> HDRNA_06_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K06_sub.fna
#HDRNA_06_K07
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00024 13961..15310
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00024 15311..16960
#IS1272:2:AB033763 100.00 1585/1585 contig00024 18802..20386
#mecA:12:AB505628 100.00 2010/2010 contig00032 1633..3642
#dmecR1:1:AB033763 100.00 987/987 contig00032 3739..4725
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00024 8790..9944
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K07/HDRNA_06_K07.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K07/HDRNA_06_K07.fna contig00024:8590-20586 > HDRNA_06_K07_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K07/HDRNA_06_K07.fna contig00032:1433-4925 >> HDRNA_06_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K07_sub.fna
#HDRNA_06_K08
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00026 6635..7984
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00026 7985..9634
#IS1272:2:AB033763 100.00 1585/1585 contig00026 11476..13060
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00026 1464..2618
#dmecR1:1:AB033763 100.00 987/987 contig00029 357..1343
#mecA:12:AB505628 100.00 2010/2010 contig00029 1440..3449
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K08/HDRNA_06_K08.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K08/HDRNA_06_K08.fna contig00026:1264-13260 > HDRNA_06_K08_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K08/HDRNA_06_K08.fna contig00029:157-3649 >> HDRNA_06_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K08_sub.fna
#HDRNA_06_K09
##IS1272:2:AB033763 91.00 1523/1585 contig00045 1..1523
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00039 4048..5202
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K09/HDRNA_06_K09.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K09/HDRNA_06_K09.fna contig00039:3848-5402 > HDRNA_06_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K09_sub.fna
#HDRNA_06_K10
#dmecR1:1:AB033763 100.00 987/987 contig00031 357..1343
#mecA:12:AB505628 100.00 2010/2010 contig00031 1440..3449
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00026 6635..7984
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00026 7985..9634
#IS1272:2:AB033763 100.00 1585/1585 contig00026 11476..13060
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00026 1464..2618
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K10/HDRNA_06_K10.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K10/HDRNA_06_K10.fna contig00026:1264-13260 > HDRNA_06_K10_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K10/HDRNA_06_K10.fna contig00031:157-3649 >> HDRNA_06_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K10_sub.fna
#CP133680
#mecA:12:AB505628 100.00 2010/2010 CP133680 37611..39620
#dmecR1:1:AB033763 100.00 987/987 CP133680 39717..40703
#IS1272:3:AM292304 99.95 1844/1843 CP133680 40692..42535
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133680 44377..46026
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133680 46027..47376
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133680 51493..52983
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133680.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133680.gb_converted.gbff 37411 53183 CP133680_sub.gbff
#HDRNA_07_K02
#mecA:12:AB505628 100.00 2010/2010 contig00003 137822..139831
#dmecR1:1:AB033763 100.00 987/987 contig00003 139928..140914
#IS1272:3:AM292304 99.95 1844/1843 contig00003 140903..142746
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 144588..146237
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 146238..147587
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 151484..152974
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K02/HDRNA_07_K02.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K02/HDRNA_07_K02.fna contig00003:137622-153174 > HDRNA_07_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K02_sub.fna
#HDRNA_07_K03
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 52228..53718
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 57615..58964
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 58965..60614
#IS1272:3:AM292304 99.95 1844/1843 contig00003 62456..64299
#dmecR1:1:AB033763 100.00 987/987 contig00003 64288..65274
#mecA:12:AB505628 100.00 2010/2010 contig00003 65371..67380
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K03/HDRNA_07_K03.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K03/HDRNA_07_K03.fna contig00003:52028-67580 > HDRNA_07_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K03_sub.fna
#HDRNA_07_K04
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 11087..12436
#mecA:12:AB505628 100.00 2010/2010 contig00027 2671..4680
#dmecR1:1:AB033763 100.00 987/987 contig00027 4777..5763
#IS1272:3:AM292304 99.95 1844/1843 contig00027 5752..7595
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 9437..11086
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 231..1721
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K04/HDRNA_07_K04.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K04/HDRNA_07_K04.fna contig00027:2471-12636 > HDRNA_07_K04_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K04/HDRNA_07_K04.fna contig00016:31-1921 >> HDRNA_07_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K04_sub.fna
#HDRNA_07_K05
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 52208..53698
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00015 57540..58889
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00015 58890..60539
#IS1272:3:AM292304 99.95 1844/1843 contig00015 62381..64224
#dmecR1:1:AB033763 100.00 987/987 contig00015 64213..65199
#mecA:12:AB505628 100.00 2010/2010 contig00015 65296..67305
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K05/HDRNA_07_K05.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K05/HDRNA_07_K05.fna contig00015:52008-67505 > HDRNA_07_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K05_sub.fna
#HDRNA_07_K06
#mecA:12:AB505628 100.00 2010/2010 contig00005 137910..139919
#dmecR1:1:AB033763 100.00 987/987 contig00005 140016..141002
#IS1272:3:AM292304 99.95 1844/1843 contig00005 140991..142834
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00005 144676..146325
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00005 146326..147675
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00017 211..1701
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K06/HDRNA_07_K06.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K06/HDRNA_07_K06.fna contig00005:137710-147875 > HDRNA_07_K06_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K06/HDRNA_07_K06.fna contig00017:11-1901 >> HDRNA_07_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K06_sub.fna
#TODO_1: DESCRIBE it as the fact below, one is ccrB4+ccrA4+subtyppe-Vc(5C2&5), another is ccrB2+ccrA2+subtype-IVa(2B)!
#TODO_2: CONSIDERING whether "#IS1272:2:AB033763 91.00" should be added to the plot?
#HDRNA_07_K07
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00002 142538..144172
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00002 144169..145530
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00002 148018..149952
#mecA:12:AB505628 100.00 2010/2010 contig00002 156410..158419
#dmecR1:1:AB033763 100.00 987/987 contig00002 158516..159502
#IS1272:3:AM292304 99.95 1844/1843 contig00002 159491..161334
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 163176..164825
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00002 164826..166175
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 170072..171562
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K07/HDRNA_07_K07.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K07/HDRNA_07_K07.fna contig00002:142338-171762 > HDRNA_07_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K07_sub.fna
#mecR1
#IS1182 family transposase
#Cation-transporting P-type ATPase --> subtyppe-Vc(5C2&5)
#AAA family ATPase --> subtype-IVa(2B)
#HDRNA_07_K08
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5175..6824
#IS1272:3:AM292304 99.95 1844/1843 contig00004 8666..10509
#dmecR1:1:AB033763 100.00 987/987 contig00004 10498..11484
#mecA:12:AB505628 100.00 2010/2010 contig00004 11581..13590
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 20048..21982
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 24470..25831
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 25828..27462
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 50618..52108
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K08/HDRNA_07_K08.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K08/HDRNA_07_K08.fna contig00004:3625-27662 > HDRNA_07_K08_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K08/HDRNA_07_K08.fna contig00016:50418-52308 >> HDRNA_07_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K08_sub.fna
#HDRNA_07_K09
#dmecR1:1:AB033763 100.00 987/987 contig00004 10423..11409
#mecA:12:AB505628 100.00 2010/2010 contig00004 11506..13515
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 19973..21907
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 24395..25756
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 25753..27387
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5100..6749
#IS1272:3:AM292304 99.95 1844/1843 contig00004 8591..10434
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 52208..53698
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K09/HDRNA_07_K09.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K09/HDRNA_07_K09.fna contig00004:3550-27587 > HDRNA_07_K09_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K09/HDRNA_07_K09.fna contig00016:52008-53898 >> HDRNA_07_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K09_sub.fna
#HDRNA_07_K10
#mecA:12:AB505628 100.00 2010/2010 contig00003 137894..139903
#dmecR1:1:AB033763 100.00 987/987 contig00003 140000..140986
#IS1272:3:AM292304 99.95 1844/1843 contig00003 140975..142818
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 144660..146309
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 146310..147659
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 151556..153046
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K10/HDRNA_07_K10.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K10/HDRNA_07_K10.fna contig00003:137694-153246 > HDRNA_07_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K10_sub.fna
#CP133682
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 CP133682 47094..48722
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 CP133682 55568..57439
#mecA:12:AB505628 100.00 2010/2010 CP133682 64626..66635
#mecR1:1:D86934 100.00 1758/1758 CP133682 66732..68489
#mecI:1:D86934 100.00 372/372 CP133682 68489..68860
##IS1272:2:AB033763 91.06 1577/1585 CP133682 865163..866739
##IS1272:2:AB033763 91.06 1577/1585 CP133682 968562..970138
##IS1272:2:AB033763 91.06 1577/1585 CP133682 1659758..1661334
##IS1272:2:AB033763 91.06 1577/1585 CP133682 2357991..2359567
##IS1272:2:AB033763 91.06 1577/1585 CP133682 293604..295180
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133682.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133682.gb_converted.gbff 46894 69060 CP133682_sub.gbff
#Zinc-ribbon domain-containing protein-->
#HDRNA_08_K02
#mecI:1:D86934 100.00 372/372 contig00005 34179..34550
#mecR1:1:D86934 100.00 1758/1758 contig00005 34550..36307
#mecA:12:AB505628 100.00 2010/2010 contig00005 36404..38413
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 45600..47471
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 54317..55945
##IS1272:2:AB033763 91.01 1524/1585 contig00020 66..1589
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K02/HDRNA_08_K02.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K02/HDRNA_08_K02.fna contig00005:33979-56145 > HDRNA_08_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K02_sub.fna
#HDRNA_08_K03
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 111644..113272
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 120118..121989
#mecA:12:AB505628 100.00 2010/2010 contig00005 129176..131185
#mecR1:1:D86934 100.00 1758/1758 contig00005 131282..133039
#mecI:1:D86934 100.00 372/372 contig00005 133039..133410
##IS1272:2:AB033763 91.01 1524/1585 contig00020 66..1589
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K03/HDRNA_08_K03.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K03/HDRNA_08_K03.fna contig00005:111444-133610 > HDRNA_08_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K03_sub.fna
#HDRNA_08_K04
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 111072..112700
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 119546..121417
#mecA:12:AB505628 100.00 2010/2010 contig00005 128604..130613
#mecR1:1:D86934 100.00 1758/1758 contig00005 130710..132467
#mecI:1:D86934 100.00 372/372 contig00005 132467..132838
##IS1272:2:AB033763 91.06 1577/1585 contig00019 7..1583
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K04/HDRNA_08_K04.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K04/HDRNA_08_K04.fna contig00005:110872-133038 > HDRNA_08_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K04_sub.fna
#HDRNA_08_K05
##IS1272:2:AB033763 91.01 1524/1585 contig00021 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 34179..34550
#mecR1:1:D86934 100.00 1758/1758 contig00006 34550..36307
#mecA:12:AB505628 100.00 2010/2010 contig00006 36404..38413
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45600..47471
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 54317..55945
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K05/HDRNA_08_K05.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K05/HDRNA_08_K05.fna contig00006:33979-56145 > HDRNA_08_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K05_sub.fna
#HDRNA_08_K06
##IS1272:2:AB033763 91.01 1524/1585 contig00020 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 34179..34550
#mecR1:1:D86934 100.00 1758/1758 contig00006 34550..36307
#mecA:12:AB505628 100.00 2010/2010 contig00006 36404..38413
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45600..47471
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 54317..55945
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K06/HDRNA_08_K06.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K06/HDRNA_08_K06.fna contig00006:33979-56145 > HDRNA_08_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K06_sub.fna
#HDRNA_08_K07
#mecI:1:D86934 100.00 372/372 contig00005 32299..32670
#mecR1:1:D86934 100.00 1758/1758 contig00005 32670..34427
#mecA:12:AB505628 100.00 2010/2010 contig00005 34524..36533
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 43720..45591
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 52437..54065
##IS1272:2:AB033763 91.01 1524/1585 contig00025 66..1589
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K07/HDRNA_08_K07.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K07/HDRNA_08_K07.fna contig00005:32099-54265 > HDRNA_08_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K07_sub.fna
#HDRNA_08_K08
##IS1272:2:AB033763 91.01 1524/1585 contig00022 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 33701..34072
#mecR1:1:D86934 100.00 1758/1758 contig00006 34072..35829
#mecA:12:AB505628 100.00 2010/2010 contig00006 35926..37935
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45122..46993
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 53839..55467
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K08/HDRNA_08_K08.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K08/HDRNA_08_K08.fna contig00006:33501-55667 > HDRNA_08_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K08_sub.fna
#HDRNA_08_K09
##IS1272:2:AB033763 91.01 1524/1585 contig00021 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 34178..34549
#mecR1:1:D86934 100.00 1758/1758 contig00006 34549..36306
#mecA:12:AB505628 100.00 2010/2010 contig00006 36403..38412
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45599..47470
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 54316..55944
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K09/HDRNA_08_K09.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K09/HDRNA_08_K09.fna contig00006:33978-56144 > HDRNA_08_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K09_sub.fna
#HDRNA_08_K10
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 111072..112700
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 119546..121417
#mecA:12:AB505628 100.00 2010/2010 contig00005 128604..130613
#mecR1:1:D86934 100.00 1758/1758 contig00005 130710..132467
#mecI:1:D86934 100.00 372/372 contig00005 132467..132838
##IS1272:2:AB033763 91.06 1577/1585 contig00018 7..1583
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K10/HDRNA_08_K10.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K10/HDRNA_08_K10.fna contig00005:110872-133038 > HDRNA_08_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K10_sub.fna
#CP133684
#mecA:12:AB505628 100.00 2010/2010 CP133684 37596..39605
#dmecR1:1:AB033763 100.00 987/987 CP133684 39702..40688
#IS1272:3:AM292304 99.95 1844/1843 CP133684 40677..42520
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133684 44362..46011
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133684 46012..47361
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133684 51478..52968
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133684.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133684.gb_converted.gbff 37396 53168 CP133684_sub.gbff
#HDRNA_12_K02
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00012 52208..53698
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00012 57540..58889
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00012 58890..60539
#IS1272:3:AM292304 99.95 1844/1843 contig00012 62381..64224
#dmecR1:1:AB033763 100.00 987/987 contig00012 64213..65199
#mecA:12:AB505628 100.00 2010/2010 contig00012 65296..67305
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K02/HDRNA_12_K02.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K02/HDRNA_12_K02.fna contig00012:52008-67505 > HDRNA_12_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K02_sub.fna
#HDRNA_12_K03
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00029 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00029 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00029 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00029 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 211..1701
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K03/HDRNA_12_K03.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K03/HDRNA_12_K03.fna contig00029:2451-12616 > HDRNA_12_K03_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K03/HDRNA_12_K03.fna contig00015:11-1901 >> HDRNA_12_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K03_sub.fna
#HDRNA_12_K04
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00031 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00031 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00031 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00031 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00031 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 184..1674
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K04/HDRNA_12_K04.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K04/HDRNA_12_K04.fna contig00031:2451-12616 > HDRNA_12_K04_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K04/HDRNA_12_K04.fna contig00014:84-1874 >> HDRNA_12_K04_sub.fna
bakta --skip-crispr --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K04_sub.fna
#HDRNA_12_K05
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00030 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00030 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00030 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00030 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00030 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 211..1701
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K05/HDRNA_12_K05.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K05/HDRNA_12_K05.fna contig00030:2451-12616 > HDRNA_12_K05_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K05/HDRNA_12_K05.fna contig00016:11-1901 >> HDRNA_12_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K05_sub.fna
#HDRNA_12_K06
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 51961..53451
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00014 57293..58642
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00014 58643..60292
#IS1272:3:AM292304 99.95 1844/1843 contig00014 62134..63977
#dmecR1:1:AB033763 100.00 987/987 contig00014 63966..64952
#mecA:12:AB505628 100.00 2010/2010 contig00014 65049..67058
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K06/HDRNA_12_K06.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K06/HDRNA_12_K06.fna contig00014:51761-67258 > HDRNA_12_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K06_sub.fna
#HDRNA_12_K07
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 40711..42201
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00014 46153..47502
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00014 47503..49152
#IS1272:3:AM292304 99.95 1844/1843 contig00014 50994..52837
#dmecR1:1:AB033763 100.00 987/987 contig00014 52826..53812
#mecA:12:AB505628 100.00 2010/2010 contig00014 53909..55918
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K07/HDRNA_12_K07.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K07/HDRNA_12_K07.fna contig00014:40511-56118 > HDRNA_12_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K07_sub.fna
#HDRNA_12_K08
#dmecR1:1:AB033763 100.00 987/987 contig00027 10441..11427
#mecA:12:AB505628 100.00 2010/2010 contig00027 11524..13533
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3768..5117
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5118..6767
#IS1272:3:AM292304 99.95 1844/1843 contig00027 8609..10452
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00005 137459..138949
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K08/HDRNA_12_K08.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K08/HDRNA_12_K08.fna contig00027:3568-13733 > HDRNA_12_K08_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K08/HDRNA_12_K08.fna contig00005:137259-139149 >> HDRNA_12_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K08_sub.fna
#HDRNA_12_K09
#dmecR1:1:AB033763 100.00 987/987 contig00028 10368..11354
#mecA:12:AB505628 100.00 2010/2010 contig00028 11451..13460
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 3695..5044
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 5045..6694
#IS1272:3:AM292304 99.95 1844/1843 contig00028 8536..10379
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 52207..53697
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K09/HDRNA_12_K09.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K09/HDRNA_12_K09.fna contig00028:3495-13660 > HDRNA_12_K09_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K09/HDRNA_12_K09.fna contig00014:52007-53897 >> HDRNA_12_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K09_sub.fna
#HDRNA_12_K10
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00028 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00028 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00028 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 211..1701
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K10/HDRNA_12_K10.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K10/HDRNA_12_K10.fna contig00028:2451-12616 > HDRNA_12_K10_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K10/HDRNA_12_K10.fna contig00014:11-1901 >> HDRNA_12_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K10_sub.fna
#CP133688
#mecA:12:AB505628 100.00 2010/2010 CP133688 37716..39725
#dmecR1:1:AB033763 100.00 987/987 CP133688 39822..40808
#IS1272:3:AM292304 100.00 1843/1843 CP133688 40797..42639
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133688 44481..46130
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133688 46131..47480
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133688 51377..52867
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133688.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133688.gb_converted.gbff 37516 53067 CP133688_sub.gbff
#HDRNA_16_K02
#dmecR1:1:AB033763 100.00 987/987 contig00012 10440..11426
#mecA:12:AB505628 100.00 2010/2010 contig00012 11523..13532
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00012 3768..5117
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00012 5118..6767
#IS1272:3:AM292304 100.00 1843/1843 contig00012 8609..10451
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 71060..72550
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K02/HDRNA_16_K02.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K02/HDRNA_16_K02.fna contig00012:3568-13732 > HDRNA_16_K02_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K02/HDRNA_16_K02.fna contig00013:70860-72750 >> HDRNA_16_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K02_sub.fna
#HDRNA_16_K03
#dmecR1:1:AB033763 100.00 987/987 contig00014 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00014 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00014 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00014 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00014 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 56677..58167
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K03/HDRNA_16_K03.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K03/HDRNA_16_K03.fna contig00014:3550-13714 > HDRNA_16_K03_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K03/HDRNA_16_K03.fna contig00015:56477-58367 >> HDRNA_16_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K03_sub.fna
#HDRNA_16_K04
#dmecR1:1:AB033763 100.00 987/987 contig00017 10367..11353
#mecA:12:AB505628 100.00 2010/2010 contig00017 11450..13459
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00017 3695..5044
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00017 5045..6694
#IS1272:3:AM292304 100.00 1843/1843 contig00017 8536..10378
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00012 56677..58167
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K04/HDRNA_16_K04.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K04/HDRNA_16_K04.fna contig00017:3495-13659 > HDRNA_16_K04_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K04/HDRNA_16_K04.fna contig00012:56477-58367 >> HDRNA_16_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K04_sub.fna
#HDRNA_16_K05
#mecA:12:AB505628 100.00 2010/2010 contig00013 64162..66171
#dmecR1:1:AB033763 100.00 987/987 contig00013 66268..67254
#IS1272:3:AM292304 100.00 1843/1843 contig00013 67243..69085
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00013 70927..72576
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00013 72577..73926
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 67300..68790
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K05/HDRNA_16_K05.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K05/HDRNA_16_K05.fna contig00013:63962-74126 > HDRNA_16_K05_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K05/HDRNA_16_K05.fna contig00014:67100-68990 >> HDRNA_16_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K05_sub.fna
#HDRNA_16_K06
#dmecR1:1:AB033763 100.00 987/987 contig00004 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00004 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00004 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 67555..69045
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K06/HDRNA_16_K06.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K06/HDRNA_16_K06.fna contig00004:3550-13714 > HDRNA_16_K06_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K06/HDRNA_16_K06.fna contig00013:67355-69245 >> HDRNA_16_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K06_sub.fna
#HDRNA_16_K07
#mecA:12:AB505628 100.00 2010/2010 contig00003 156854..158863
#dmecR1:1:AB033763 100.00 987/987 contig00003 158960..159946
#IS1272:3:AM292304 100.00 1843/1843 contig00003 159935..161777
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 163619..165268
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 165269..166618
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 170515..172005
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K07/HDRNA_16_K07.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K07/HDRNA_16_K07.fna contig00003:156654-172205 > HDRNA_16_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K07_sub.fna
#HDRNA_16_K08
#dmecR1:1:AB033763 100.00 987/987 contig00011 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00011 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00011 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00011 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00011 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 67300..68790
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K08/HDRNA_16_K08.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K08/HDRNA_16_K08.fna contig00011:3550-13714 > HDRNA_16_K08_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K08/HDRNA_16_K08.fna contig00013:67100-68990 >> HDRNA_16_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K08_sub.fna
#HDRNA_16_K09
#dmecR1:1:AB033763 100.00 987/987 contig00017 10497..11483
#mecA:12:AB505628 100.00 2010/2010 contig00017 11580..13589
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00017 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00017 5175..6824
#IS1272:3:AM292304 100.00 1843/1843 contig00017 8666..10508
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00012 52313..53803
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K09/HDRNA_16_K09.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K09/HDRNA_16_K09.fna contig00017:3625-13789 > HDRNA_16_K09_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K09/HDRNA_16_K09.fna contig00012:52113-54003 >> HDRNA_16_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K09_sub.fna
#HDRNA_16_K10
#mecA:12:AB505628 100.00 2010/2010 contig00005 156908..158917
#dmecR1:1:AB033763 100.00 987/987 contig00005 159014..160000
#IS1272:3:AM292304 100.00 1843/1843 contig00005 159989..161831
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00005 163673..165322
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00005 165323..166672
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 211..1701
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K10/HDRNA_16_K10.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K10/HDRNA_16_K10.fna contig00005:156708-166872 > HDRNA_16_K10_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K10/HDRNA_16_K10.fna contig00013:11-1901 >> HDRNA_16_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K10_sub.fna
#CP133693
#mecA:12:AB505628 100.00 2010/2010 CP133693 37128..39137
#dmecR1:1:AB033763 100.00 987/987 CP133693 39234..40220
#IS1272:3:AM292304 100.00 1843/1843 CP133693 40209..42051
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133693 43893..45542
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133693 45543..46892
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133693 51009..52499
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133693.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133693.gb_converted.gbff 36928 52699 CP133693_sub.gbff
#HDRNA_17_K02
#dmecR1:1:AB033763 100.00 987/987 contig00028 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00028 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00028 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00031 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K02/HDRNA_17_K02.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K02/HDRNA_17_K02.fna contig00028:3550-13714 > HDRNA_17_K02_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K02/HDRNA_17_K02.fna contig00031:4666-6556 >> HDRNA_17_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K02_sub.fna
#HDRNA_17_K03
#dmecR1:1:AB033763 100.00 987/987 contig00025 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00025 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00025 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00025 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00025 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00028 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K03/HDRNA_17_K03.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K03/HDRNA_17_K03.fna contig00025:3550-13714 > HDRNA_17_K03_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K03/HDRNA_17_K03.fna contig00028:4666-6556 >> HDRNA_17_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K03_sub.fna
#HDRNA_17_K04
#dmecR1:1:AB033763 100.00 987/987 contig00027 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00027 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00027 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00030 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K04/HDRNA_17_K04.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K04/HDRNA_17_K04.fna contig00027:3550-13714 > HDRNA_17_K04_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K04/HDRNA_17_K04.fna contig00030:4666-6556 >> HDRNA_17_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K04_sub.fna
#HDRNA_17_K05
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00024 11321..12670
#mecA:12:AB505628 100.00 2010/2010 contig00024 2906..4915
#dmecR1:1:AB033763 100.00 987/987 contig00024 5012..5998
#IS1272:3:AM292304 100.00 1843/1843 contig00024 5987..7829
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00024 9671..11320
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00024 16567..18057
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K05/HDRNA_17_K05.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K05/HDRNA_17_K05.fna contig00024:2706-18257 > HDRNA_17_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K05_sub.fna
#HDRNA_17_K06
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 11301..12650
#mecA:12:AB505628 100.00 2010/2010 contig00027 2886..4895
#dmecR1:1:AB033763 100.00 987/987 contig00027 4992..5978
#IS1272:3:AM292304 100.00 1843/1843 contig00027 5967..7809
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 9651..11300
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 211..1701
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K06/HDRNA_17_K06.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K06/HDRNA_17_K06.fna contig00027:2686-12850 > HDRNA_17_K06_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K06/HDRNA_17_K06.fna contig00029:11-1901 >> HDRNA_17_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K06_sub.fna
#HDRNA_17_K07
#dmecR1:1:AB033763 100.00 987/987 contig00026 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00026 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00026 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00026 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00026 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00030 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K07/HDRNA_17_K07.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K07/HDRNA_17_K07.fna contig00026:3550-13714 > HDRNA_17_K07_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K07/HDRNA_17_K07.fna contig00030:4666-6556 >> HDRNA_17_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K07_sub.fna
#HDRNA_17_K08
#dmecR1:1:AB033763 100.00 987/987 contig00027 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00027 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00027 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00031 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K08/HDRNA_17_K08.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K08/HDRNA_17_K08.fna contig00027:3550-13714 > HDRNA_17_K08_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K08/HDRNA_17_K08.fna contig00031:4666-6556 >> HDRNA_17_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K08_sub.fna
#HDRNA_17_K09
#dmecR1:1:AB033763 100.00 987/987 contig00025 10497..11483
#mecA:12:AB505628 100.00 2010/2010 contig00025 11580..13589
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00025 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00025 5175..6824
#IS1272:3:AM292304 100.00 1843/1843 contig00025 8666..10508
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00028 4886..6376
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K09/HDRNA_17_K09.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K09/HDRNA_17_K09.fna contig00025:3625-13789 > HDRNA_17_K09_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K09/HDRNA_17_K09.fna contig00028:4686-6576 >> HDRNA_17_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K09_sub.fna
#HDRNA_17_K10
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00024 10273..11622
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00024 11623..13272
#IS1272:3:AM292304 100.00 1843/1843 contig00024 15114..16956
#dmecR1:1:AB033763 100.00 987/987 contig00024 16945..17931
#mecA:12:AB505628 100.00 2010/2010 contig00024 18028..20037
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00024 4886..6376
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K10/HDRNA_17_K10.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K10/HDRNA_17_K10.fna contig00024:4686-20237 > HDRNA_17_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K10_sub.fna
#CP133696
#ccrB2:1:N315:D86934 98.96 1629/1629 CP133696 42277..43905
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 CP133696 43927..45276
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133696.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133696.gb_converted.gbff 42077 45476 CP133696_sub.gbff
#HDRNA_19_K02
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K02/HDRNA_19_K02.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K02/HDRNA_19_K02.fna contig00030:2360-5759 > HDRNA_19_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K02_sub.fna
#HDRNA_19_K03
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K03/HDRNA_19_K03.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K03/HDRNA_19_K03.fna contig00031:2360-5759 > HDRNA_19_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K03_sub.fna
#HDRNA_19_K04
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K04/HDRNA_19_K04.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K04/HDRNA_19_K04.fna contig00031:2360-5759 > HDRNA_19_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K04_sub.fna
#HDRNA_19_K05
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K05/HDRNA_19_K05.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K05/HDRNA_19_K05.fna contig00031:2360-5759 > HDRNA_19_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K05_sub.fna
#HDRNA_19_K06
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2580..3929
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3951..5579
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K06/HDRNA_19_K06.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K06/HDRNA_19_K06.fna contig00031:2380-5779 > HDRNA_19_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K06_sub.fna
#HDRNA_19_K07
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2580..3929
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3951..5579
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K07/HDRNA_19_K07.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K07/HDRNA_19_K07.fna contig00031:2380-5779 > HDRNA_19_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K07_sub.fna
#HDRNA_19_K08
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2577..3926
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3948..5576
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K08/HDRNA_19_K08.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K08/HDRNA_19_K08.fna contig00030:2377-5776 > HDRNA_19_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K08_sub.fna
#HDRNA_19_K09
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K09/HDRNA_19_K09.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K09/HDRNA_19_K09.fna contig00030:2360-5759 > HDRNA_19_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K09_sub.fna
#HDRNA_19_K10
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K10/HDRNA_19_K10.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K10/HDRNA_19_K10.fna contig00030:2360-5759 > HDRNA_19_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K10_sub.fna
#CP133700
#mecA:12:AB505628 100.00 2010/2010 CP133700 37713..39722
#dmecR1:1:AB033763 100.00 987/987 CP133700 39819..40805
#IS1272:3:AM292304 99.95 1844/1843 CP133700 40794..42637
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133700 44479..46128
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133700 46129..47478
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133700 51595..53085
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133700.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133700.gb_converted.gbff 37513 53285 CP133700_sub.gbff
#HDRNA_20_K02
#subtyppe-Vc(5C2&5):10:AB505629 99.90 1935/1935 contig00009 2050..3984
#ccrA4:2:BK20781:FJ670542 100.00 1362/1362 contig00009 6472..7833
#ccrB4:2:BK20781:FJ670542 100.00 1629/1629 contig00009 7830..9458
##IS1272:2:AB033763 91.06 1577/1585 contig00030 7..1583
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K02/HDRNA_20_K02.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K02/HDRNA_20_K02.fna contig00009:1850-9658 > HDRNA_20_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K02_sub.fna
#HDRNA_20_K03
#dmecR1:1:AB033763 100.00 987/987 contig00028 10498..11484
#mecA:12:AB505628 100.00 2010/2010 contig00028 11581..13590
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 5175..6824
#IS1272:3:AM292304 99.95 1844/1843 contig00028 8666..10509
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 51961..53451
#subtyppe-Vc(5C2&5):10:AB505629 99.79 1936/1935 contig00004 4168..6103
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 8591..9952
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 9949..11583
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna contig00028:3625-13790 > HDRNA_20_K03_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna contig00015:51761-53651 >> HDRNA_20_K03_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna contig00004:3968-11783 >> HDRNA_20_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K03_sub.fna
#HDRNA_20_K04
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 51961..53451
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00013 57348..58697
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00013 58698..60347
#IS1272:3:AM292304 99.95 1844/1843 contig00013 62189..64032
#dmecR1:1:AB033763 100.00 987/987 contig00013 64021..65007
#mecA:12:AB505628 100.00 2010/2010 contig00013 65104..67113
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K04/HDRNA_20_K04.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K04/HDRNA_20_K04.fna contig00013:51761-67313 > HDRNA_20_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K04_sub.fna
#HDRNA_20_K05
#dmecR1:1:AB033763 100.00 987/987 contig00032 10423..11409
#mecA:12:AB505628 100.00 2010/2010 contig00032 11506..13515
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00032 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00032 5100..6749
#IS1272:3:AM292304 99.95 1844/1843 contig00032 8591..10434
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 51407..52897
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 3319..5253
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 7741..9102
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 9099..10733
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna contig00032:3550-13715 > HDRNA_20_K05_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna contig00016:51207-53097 >> HDRNA_20_K05_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna contig00004:3119-10933 >> HDRNA_20_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K05_sub.fna
#HDRNA_20_K06
#dmecR1:1:AB033763 100.00 987/987 contig00027 10441..11427
#mecA:12:AB505628 100.00 2010/2010 contig00027 11524..13533
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3768..5117
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5118..6767
#IS1272:3:AM292304 99.95 1844/1843 contig00027 8609..10452
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00006 159812..161302
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 14074..16008
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 18496..19857
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 19854..21488
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna contig00027:3568-13733 > HDRNA_20_K06_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna contig00006:159612-161502 >> HDRNA_20_K06_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna contig00004:13874-21688 >> HDRNA_20_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K06_sub.fna
#HDRNA_20_K07
##IS1272:2:AB033763 91.01 1524/1585 contig00032 1..1524
#subtyppe-Vc(5C2&5):10:AB505629 99.90 1935/1935 contig00021 2048..3982
#ccrA4:2:BK20781:FJ670542 100.00 1362/1362 contig00021 6470..7831
#ccrB4:2:BK20781:FJ670542 100.00 1629/1629 contig00021 7828..9456
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K07/HDRNA_20_K07.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K07/HDRNA_20_K07.fna contig00021:1848-9656 > HDRNA_20_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K07_sub.fna
#HDRNA_20_K08
#dmecR1:1:AB033763 100.00 987/987 contig00034 10423..11409
#mecA:12:AB505628 100.00 2010/2010 contig00034 11506..13515
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00034 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00034 5100..6749
#IS1272:3:AM292304 99.95 1844/1843 contig00034 8591..10434
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 51407..52897
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 142407..144041
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 144038..145399
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 147887..149821
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna contig00034:3550-13715 > HDRNA_20_K08_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna contig00016:51207-53097 >> HDRNA_20_K08_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna contig00004:142207-150021 >> HDRNA_20_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K08_sub.fna
cp HDRNA_20_K08_sub.gbff gbff_HDRNA_20/HDRNA_20_K08.gbff
#HDRNA_20_K09
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00029 11177..12526
#mecA:12:AB505628 100.00 2010/2010 contig00029 2761..4770
#dmecR1:1:AB033763 100.00 987/987 contig00029 4867..5853
#IS1272:3:AM292304 99.95 1844/1843 contig00029 5842..7685
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 9527..11176
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 211..1701
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K09/HDRNA_20_K09.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K09/HDRNA_20_K09.fna contig00029:2561-12726 > HDRNA_20_K09_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K09/HDRNA_20_K09.fna contig00015:11-1901 >> HDRNA_20_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K09_sub.fna
#TODO_3: HOW to draw an empty unit?
#HDRNA_20_K10
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K10/HDRNA_20_K10.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K10/HDRNA_20_K10.fna contig00001:1-401 > HDRNA_20_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K10_sub.fna
#write in the result-mail: In total, we have 4 different subtype
# - subtype-IVa(2B):1:CA05:AB063172
- HDRNA_01_K01 subtype-IVa(2B)
- HDRNA_03_K01 subtype-IVa(2B)
- HDRNA_12_K01 subtype-IVa(2B)
- HDRNA_16_K01 subtype-IVa(2B)
- HDRNA_17_K01 subtype-IVa(2B)
# - subtype-IVc(2B):3:81108:AB096217
- HDRNA_06_K01
# - subtype-IVd(2B):4:JCSC4469:AB097677
- HDRNA_08_K01
# - subtyppe-Vc(5C2&5):10:AB505629 + subtype-IVa(2B):1:CA05:AB063172
- HDRNA_07_K01
- HDRNA_20_K01
mv *_sub.gbff gbff_all
cd gbff_all
mv CP133676_sub.gbff HDRNA_01_K01.gbff
mv CP133677_sub.gbff HDRNA_03_K01.gbff
mv CP133678_sub.gbff HDRNA_06_K01.gbff
mv CP133680_sub.gbff HDRNA_07_K01.gbff
mv CP133682_sub.gbff HDRNA_08_K01.gbff
mv CP133684_sub.gbff HDRNA_12_K01.gbff
mv CP133688_sub.gbff HDRNA_16_K01.gbff
mv CP133693_sub.gbff HDRNA_17_K01.gbff
mv CP133696_sub.gbff HDRNA_19_K01.gbff
mv CP133700_sub.gbff HDRNA_20_K01.gbff
for f in *_sub.gbff; do mv "$f" "${f/_sub.gbff/.gbff}"; done
#mkdir ../subtype-IVa_2B ../subtype-IVc_2B ../subtype-IVd_2B ../subtype-Vc_5C2and5_subtype-IVa_2B
mkdir gbff_HDRNA_01 gbff_HDRNA_03 gbff_HDRNA_06 gbff_HDRNA_07 gbff_HDRNA_08 gbff_HDRNA_12 gbff_HDRNA_16 gbff_HDRNA_17 gbff_HDRNA_19 gbff_HDRNA_20
# copy or move the corresponding gbff to its directory
rm *.json
clinker *_sub.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_01
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_03
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_06
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_07
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_08
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
...
cd gbff_HDRNA_20
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cp ./gbff_HDRNA_01/clinker.png HDRNA_01_clinker.png
cp ./gbff_HDRNA_03/clinker.png HDRNA_03_clinker.png
cp ./gbff_HDRNA_06/clinker.png HDRNA_06_clinker.png
cp ./gbff_HDRNA_07/clinker.png HDRNA_07_clinker.png
cp ./gbff_HDRNA_08/clinker.png HDRNA_08_clinker.png
cp ./gbff_HDRNA_12/clinker.png HDRNA_12_clinker.png
cp ./gbff_HDRNA_16/clinker.png HDRNA_16_clinker.png
cp ./gbff_HDRNA_17/clinker.png HDRNA_17_clinker.png
cp ./gbff_HDRNA_19/clinker.png HDRNA_19_clinker.png
cp ./gbff_HDRNA_20/clinker.png HDRNA_20_clinker.png
#TODO_4: rearrange the Reihenfolge of Isolate in the alignments zu den Reihenfolge in the phylogenetic tree! --> B-region.
# generate a C-region for ACME (similar as 1-s2.0-S1368764622001108-gr1.jpg)
# !!!!!! or generate a new plot for ACME region with the phylogenetic tree !!!!!!
# Evolution of RND efflux pumps in the development of a successful pathogen
# #change the SNP-tables by controlling the REF and isolate1 identical, change the REF--> isolate1, delete the isolate1 column!
# 先自己calculate ACME!-
code of extract_gb_from_gbk.py
import argparse from Bio import SeqIO def extract_contig(genbank_file, contig_id, output_file): with open(genbank_file, "r") as input_handle: # Iterate over each record (contig) in the GenBank file for record in SeqIO.parse(input_handle, "genbank"): if record.id == contig_id: # Write the specific contig to a new file with open(output_file, "w") as output_handle: SeqIO.write(record, output_handle, "genbank") print(f"Contig {contig_id} has been extracted to {output_file}") return print(f"Contig {contig_id} not found in {genbank_file}") def main(): parser = argparse.ArgumentParser(description="Extract a specific contig from a GenBank file.") parser.add_argument("genbank_file", help="The input GenBank file.") parser.add_argument("contig_id", help="The ID of the contig to extract.") parser.add_argument("output_file", help="The output file to save the extracted contig.") args = parser.parse_args() extract_contig(args.genbank_file, args.contig_id, args.output_file) if __name__ == "__main__": main() #python3 extract_gb_from_gbk.py snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.gbk contig00005 HDRNA_01_K02_contig00005.gb -
code of extract_subregion.py
#!/usr/bin/env python3 import sys from Bio import SeqIO def extract_subregion(genbank_path, start, end, output_file): for record in SeqIO.parse(genbank_path, "genbank"): subregion = record[start:end] SeqIO.write(subregion, output_file, "genbank") if __name__ == "__main__": if len(sys.argv) < 5: print("Usage: python extract_subregion.py <GenBank file> <start> <end> <output file>") sys.exit(1) genbank_file = sys.argv[1] start = int(sys.argv[2]) end = int(sys.argv[3]) output_file = sys.argv[4] extract_subregion(genbank_file, start, end, output_file) -
(optional) merge images
from Bio import Phylo import matplotlib.pyplot as plt from PIL import Image, ImageDraw def generate_tree_image(newick, tree_image_path): # Parse the Newick format tree tree = Phylo.read(newick, "newick") # Create a matplotlib figure fig = plt.figure(figsize=(10, 10)) ax = fig.add_subplot(1, 1, 1) # Draw the tree Phylo.draw(tree, do_show=False, axes=ax) # Save the figure plt.savefig(tree_image_path) plt.close() def merge_images(tree_image_path, sccmec_image_path, output_image_path): # Load the images tree_image = Image.open(tree_image_path) sccmec_image = Image.open(sccmec_image_path) # Resize images to have the same height tree_width, tree_height = tree_image.size sccmec_width, sccmec_height = sccmec_image.size new_height = max(tree_height, sccmec_height) new_tree_width = int(tree_width * (new_height / tree_height)) new_sccmec_width = int(sccmec_width * (new_height / sccmec_height)) tree_image = tree_image.resize((new_tree_width, new_height), Image.ANTIALIAS) sccmec_image = sccmec_image.resize((new_sccmec_width, new_height), Image.ANTIALIAS) # Create a new image with a white background total_width = new_tree_width + new_sccmec_width combined_image = Image.new('RGB', (total_width, new_height), (255, 255, 255)) # Paste the images into the combined image combined_image.paste(tree_image, (0, 0)) combined_image.paste(sccmec_image, (new_tree_width, 0)) # Optionally, draw connecting lines (example with a single line for demonstration) draw = ImageDraw.Draw(combined_image) draw.line((new_tree_width, 100, new_tree_width + new_sccmec_width, 100), fill="black", width=2) # Save the combined image combined_image.save(output_image_path) if __name__ == "__main__": # Example Newick format tree (replace with your own tree) newick = "((A:0.1,B:0.2,(C:0.3,D:0.4):0.5):0.6,E:0.7);" tree_image_path = 'tree_image.png' sccmec_image_path = 'sccmec_image.png' # Replace with your actual SCCmec region image path output_image_path = 'combined_image.png' # Generate the tree image generate_tree_image(newick, tree_image_path) # Merge images merge_images(tree_image_path, sccmec_image_path, output_image_path) print(f"Combined image saved to {output_image_path}") #pip3 install biopython matplotlib pillow #python3 generate_and_merge_images.py
Analysis of SNPs, InDels, transposons, and IS elements in 5 A. baumannii strains
1, call variant calling using snippy
mkdir snippy_CP059040;
for sample in snippy_CP059040; do
cd ${sample};
# -- hard-copy --
#git clone https://github.com/huang/bacto
#mv bacto/* ./
#rm -rf bacto
# -- soft-link --
ln -s ~/Tools/bacto/db/ .;
ln -s ~/Tools/bacto/envs/ .;
ln -s ~/Tools/bacto/local/ .;
cp ~/Tools/bacto/Snakefile .;
cp ~/Tools/bacto/bacto-0.1.json .;
cp ~/Tools/bacto/cluster.json .;
cd ..;
done
mkdir raw_data; cd raw_data;
ln -s ../../01.RawData/Tig1/Tig1_1.fq.gz Tig1_R1.fastq.gz
ln -s ../../01.RawData/Tig1/Tig1_2.fq.gz Tig1_R2.fastq.gz
ln -s ../../01.RawData/Tig2/Tig2_1.fq.gz Tig2_R1.fastq.gz
ln -s ../../01.RawData/Tig2/Tig2_2.fq.gz Tig2_R2.fastq.gz
ln -s ../../01.RawData/W/W_1.fq.gz W_R1.fastq.gz
ln -s ../../01.RawData/W/W_2.fq.gz W_R2.fastq.gz
ln -s ../../01.RawData/Y/Y_1.fq.gz Y_R1.fastq.gz
ln -s ../../01.RawData/Y/Y_2.fq.gz Y_R2.fastq.gz
ln -s ../../01.RawData/△adeIJ/△adeIJ_1.fq.gz _adeIJ_R1.fastq.gz
ln -s ../../01.RawData/△adeIJ/△adeIJ_2.fq.gz _adeIJ_R2.fastq.gz
#download CP059040.gb from GenBank
mv ~/Downloads/sequence\(1\).gb db/CP059040.gb
#setting the following in bacto-0.1.json
"genus": "Acinetobacter",
"kingdom": "Bacteria",
"species": "baumannii",
"reference": "db/CP059040.gb"
conda activate bengal3_ac3
(bengal3_ac3) cd snippy_CP059040
(bengal3_ac3) /home/jhuang/miniconda3/envs/snakemake_4_3_1/bin/snakemake --printshellcmds2, using spandx calling variants (almost the same results to the one from viral-ngs!)
mkdir ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/CP059040
cp CP059040.gb ~/miniconda3/envs/spandx/share/snpeff-5.1-2/data/CP059040/genes.gbk
vim ~/miniconda3/envs/spandx/share/snpeff-5.1-2/snpEff.config
/home/jhuang/miniconda3/envs/spandx/bin/snpEff build CP059040 #-d
~/Scripts/genbank2fasta.py CP059040.gb
mv CP059040.gb_converted.fna CP059040.fasta #rename "CP059040.1 xxxxx" to "CP059040" in the fasta-file
ln -s /home/jhuang/Tools/spandx/ spandx
(spandx) nextflow run spandx/main.nf --fastq "snippy_CP059040/trimmed/*_P_{1,2}.fastq" --ref CP059040.fasta --annotation --database CP059040 -resume3, summarize all SNPs and Indels from the snippy result directory.
#Output: snippy_CP059040/snippy/summary_snps_indels.csv
# adapt the array isolates = ["Tig1", "Tig2", "Y", "W", "_adeIJ"]
python3 ~/Scripts/summarize_snippy_res.py snippy_CP059040/snippy
grep -v "None,,,,,,None,None" summary_snps_indels.csv > summary_snps_indels_.csv
grep -v "/" All_SNPs_indels_annotated.txt > All_SNPs_indels_annotated_.txt4, merge the two files summary_snpsindels.csv and All_SNPs_indelsannotated.txt by merging the results from two variant calling methods snippy and spandx
cut -d$'\t' -f2 All_SNPs_indels_annotated_.txt > ../../id1
cut -d',' -f2 summary_snps_indels_.csv > ../../id2
diff id1 id25, (optional, since it should not happed) filter rows without change (between REF and the isolates) in snippy_CP059040/snippy/summary_snpsindels.csv (94)
awk -F, '
NR == 1 {print; next} # Print the header line
{
ref = $3; # Reference base (assuming REF is in the 3rd column)
same = 1; # Flag to check if all bases are the same as reference
samples = ""; # Initialize variable to hold sample data
for (i = 6; i <= NF - 8; i++) { # Loop through all sample columns, adjusting for the number of fixed columns
samples = samples $i " "; # Collect sample data
if ($i != ref) {
same = 0;
}
}
if (!same) {
print $0; # Print the entire line if not all bases are the same as the reference
#print "Samples: " samples; # Print all sample data for checking
}
}
' merged_variants_CP133676.csv > merged_variants_CP133676_.csv
#Explanation:
# -F, sets the field separator to a comma.
# NR == 1 {print; next} prints the header line and skips to the next line.
# ref = $3 sets the reference base (assumed to be in the 3rd column).
# same = 1 initializes a flag to check if all sample bases are the same as the reference.
# samples = ""; initializes a string to collect sample data.
# for (i = 6; i <= NF - 8; i++) loops through the sample columns. This assumes the first 5 columns are fixed (CHROM, POS, REF, ALT, TYPE), and the last 8 columns are fixed (Effect, Impact, Functional_Class, Codon_change, Protein_and_nucleotide_change, Amino_Acid_Length, Gene_name, Biotype).
# samples = samples $i " "; collects sample data.
# if ($i != ref) { same = 0; } checks if any sample base is different from the reference base. If found, it sets same to 0.
# if (!same) { print $0; print "Samples: " samples; } prints the entire line and the sample data if not all sample bases are the same as the reference.6, improve the header
sed -i '1s/_trimmed_P//g' merged_variants_CP059040.csv7, draw local genetic environments of △adeIJ
mkdir gbff_files
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/Tig1/contigs.fa --prefix Tig1
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/Tig2/contigs.fa --prefix Tig2
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/Y/contigs.fa --prefix Y
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/W/contigs.fa --prefix W
(bakta) bakta --db /mnt/nvme0n1p1/REFs/bakta_db snippy_CP059040/shovill/_adeIJ/contigs.fa --prefix _adeIJ
# -- find the gene positions in the gff3-file --
#tetR: 204019
#adeJ: complement(208370..211546)
#adeI: complement(211559..212809)
#DUF3298 domain-containing protein 218792..219580
#python3 ~/Scripts/extract_subregion.py Tig1.gbff 203819 219780 adeIJ_sub/Tig1.gbff
samtools faidx Tig1.fna
samtools faidx Tig1.fna contig_7:203819-219780 > Tig1_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db Tig1_sub.fna
mv Tig1_sub.gbff gbff_files/Tig1.gbff
samtools faidx Tig2.fna
samtools faidx Tig2.fna contig_7:206569-222530 > Tig2_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db Tig2_sub.fna
mv Tig2_sub.gbff gbff_files/Tig2.gbff
samtools faidx Y.fna
samtools faidx Y.fna contig_8:203819-215344 > Y_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db Y_sub.fna
mv Y_sub.gbff gbff_files/Y.gbff
samtools faidx W.fna
samtools faidx W.fna contig_7:37321-48846 > W_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db W_sub.fna
mv W_sub.gbff gbff_files/W.gbff
samtools faidx _adeIJ.fna
samtools faidx _adeIJ.fna contig_7:203819-215344 > _adeIJ_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db _adeIJ_sub.fna
mv _adeIJ_sub.gbff gbff_files/delta_adeIJ.gbff
makeblastdb -in ~/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna -dbtype 'nucl' -out CP059040.gb_converted.fna.db
blastn -db CP059040.gb_converted.fna.db -query Tig1_sub.fna -out adeKJI_on_CP059040.blastn -evalue 1e-10 -num_threads 15 -outfmt 6
samtools faidx ~/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna
samtools faidx ~/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna CP059040:732682-748643 > CP059040_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db CP059040_sub.fna
mv CP059040_sub.gbff gbff_files/CP059040.gbff
cd gbff_files
rm *.json
clinker *.gbff -p plot.html --dont_set_origin -s session.json -o alignments.csv -dl "," -dc 4
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig1_.gff3 Tig2_.gff3 Y_.gff3 W_.gff3 _adeIJ_.gff3 -d$'\t' -o gff3.xls8, show difference between the genome using BRIG
This is an issue with java version. I was running Java 1.8 version and had same problem as the OP's question. Then I followed the answers and finally changing the java version worked for me.
I ran the following steps:
1) Go to link: https://www.oracle.com/java/technologies/javase-java-archive-javase6-downloads.html
2) mkdir Java1.6
3) Download "jdk-6u45-linux-x64.bin" and save in Java1.6
3) cd Java1.6 && chmod +x jdk-6u45-linux-x64.bin && ./jdk-6u45-linux-x64.bin
4) You should see java file in Java1.6/jdk1.6.0_45/bin
5) Java1.6/jdk1.6.0_45/bin/java -version - should give "1.6.0_45"
6) (base) conda install bioconda::blast-legacy # install blastall 2.2.26
7) #set lastLocation="/home/jhuang/miniconda3/bin/" in default-BRIG.xml --> /home/jhuang/miniconda3/bin/blastall is used for blast!
8) Open BRIG using Java1.6 as follows: ~/Tools/Java1.6/jdk1.6.0_45/bin/java -Xmx15000M -jar BRIG.jar
Note: I did not uninstall my original Java1.8 rather used the downloded java1.6 execulatable file using path. That way I retain both versions without any problem.
This gave me all the rings without any issue!! Hope this helps someone!
Users who wish to run BRIG from the command-line need to:
* Navigate to the unpacked BRIG folder in a command-line interface (terminal, console, command prompt).
* Run 'java -Xmx1500M -jar BRIG.jar'. Where -Xmx specifies the amount of memory allocated to BRIG.
Note:
* BLAST legacy comes as a compressed package, which will unzip the BLAST binaries where ever the package is. We advise users to first create a BLAST directory (in either the home or applications directory), copy the downloaded BLAST package to that directory and unzip the package.
* BRIG supports both BLAST+ & BLAST Legacy. If BRIG cannot find BLAST it will prompt users at runtime. Users can specify the location of their BLAST installation in the BRIG options menu which is: Main window > Preferences > BRIG options.
* N.B: If BOTH BLAST+ and legacy versions are in the same location, BRIG will prefer BLAST+
convert CP059040.png -crop 4000x2780+3840+0 CP059040_.png9, try to use bactopia
# -- ERROR --> Aborted during installation! --
mamba create -y -n bactopia -c conda-forge -c bioconda bactopia
conda activate bactopia
bactopia datasets
# Paired-end
bactopia --R1 R1.fastq.gz --R2 R2.fastq.gz --sample SAMPLE_NAME \
--datasets datasets/ --outdir OUTDIR
# Single-End
bactopia --SE SAMPLE.fastq.gz --sample SAMPLE --datasets datasets/ --outdir OUTDIR
# Multiple Samples
bactopia prepare MY-FASTQS/ > fastqs.txt
bactopia --fastqs fastqs.txt --datasets datasets --outdir OUTDIR
# Single ENA/SRA Experiment
bactopia --accession SRX000000 --datasets datasets --outdir OUTDIR
# Multiple ENA/SRA Experiments
bactopia search "staphylococcus aureus" > accessions.txt
bactopia --accessions accessions.txt --dataset datasets --outdir ${OUTDIR}10, IS elments using ISEScan
(base) isescan.py --seqfile Tig1.fna --output Tig1_isescan_out --nthread 20
isescan.py --seqfile Tig2.fna --output Tig2_isescan_out --nthread 20
isescan.py --seqfile Y.fna --output Y_isescan_out --nthread 20
isescan.py --seqfile W.fna --output W_isescan_out --nthread 20
isescan.py --seqfile _adeIJ.fna --output deltaAdeIJ_isescan_out --nthread 20
~/Tools/csv2xls-0.4/csv_to_xls.py ./Tig1_isescan_out/Tig1.fna.csv ./Tig2_isescan_out/Tig2.fna.csv ./Y_isescan_out/Y.fna.csv ./W_isescan_out/W.fna.csv ./deltaAdeIJ_isescan_out/_adeIJ.fna.csv -d',' -o ISEScan_res.xls
#extracted sequence segments from the two isolates, specifically:
# ATCC19606: 930469 to 951674 — segment1
# ATCC17978: 2,934,384 to 3,000,721 — segment2
#Then, I compared the two segments and found that positions 1-11055 of segment1 mapped to 66338-55284 of segment2, and positions 11049-21206 of segment1 mapped to 10158-23 of segment2. This means the sequence from 10159-55283 of segment2 (about 45 kb nt) is not mapped. I then extracted the 45 kb sequence (see the attached fasta file). I attempted to detect IS elements using the tool ISEScan (https://academic.oup.com/bioinformatics/article/33/21/3340/3930124). Four ISs were detected (see 45kb.fasta.xlsx; for more detailed results, see 45kb.fasta.zip).
#samtools faidx Acinetobacter_baumannii_ATCC19606.gbk_converted.fna CP059040.1:930469-951674 > ../ATCC19606_segment.fasta
vim ./gbks/A.baumannii_ATCC17978.gbk
#LOCUS CP000521 3976747 bp DNA circular BCT 31-JAN-2014
#DEFINITION Acinetobacter baumannii ATCC 17978, complete genome.
I used the following commands extracted a 45kb fasta. Then using a tools get IS elements.
samtools faidx A.baumannii_ATCC17978.gbk_converted.fna CP000521.1:2934384-3000721 > ../ATCC17978_segment.fasta
makeblastdb -in ATCC17978_segment.fasta -dbtype nucl
blastn -db ATCC17978_segment.fasta -query ATCC19606_segment.fasta -num_threads 15 -outfmt 6 -strand both -evalue 0.1 > ATCC19606_segment_on_ATCC17978_segment.blastn
samtools faidx ATCC17978_segment.fasta CP000521.1_2934384_3000721:10159-55283 > 45kb.fasta
please update the following tables in which all positons referred to the 45kb sequence to the complete genome in ATCC17978.
#seqID: sequence identifier
#family: family name of IS element
#cluster: Tpase cluster
#isBegin and isEnd: genome coordinates of the predicted IS element
#isLen: length of the predicted IS element
#ncopy4is: number of predicted IS copies including full-length and partial IS copies
#start1, end1, start2, end2: genome coordinates of the IRs
#score: score of the IRs
#irId: number of identical matches in pairwise alignment of left and righ hand invered repeats
#irLen, length of inverted repeats
#nGaps: number of gaps in IRs
#orfBegin, orfEnd: genome coordinates of the predicted Tpase ORF
#strand: strand where the Tpase is
#orfLen: length of predicted Tpase ORF
#E-value: the best E-value among all IS copies for the same IS element, the smaller the better
#E-value4copy: the E-value of the reported IS copy, the smaller the better
#type: type of IS element copy, 'c' for complete IS element and 'p' for partial IS element
#ov: ov number returned by hmmer search
#tir: terminal inverted repeat sequences
seqID family cluster isBegin isEnd isLen ncopy4is start1 end1 start2 end2 score irId irLen nGaps orfBegin orfEnd strand orfLen E-value E-value4copy type ov tir
CP000521.1_2934384_3000721:10159-55283 IS5 IS5_222 5818 8737 2920 1 5818 5842 8713 8737 18 17 25 0 5931 8822 + 2892 3.3E-74 3.3E-74 c 1 TGATTAAACTTTGCGGATTAAATTG:TGATTAAATCTAATGTGTTGAATTG
CP000521.1_2934384_3000721:10159-55283 IS3 IS3_176 8745 9849 1105 1 8745 8761 9833 9849 26 15 17 0 8916 9775 - 860 9E-38 9E-38 p 1 ATTGATGATAGCCAAAA:ATTGATCCTAGCCAAAA
CP000521.1_2934384_3000721:10159-55283 IS5 IS5_226 9983 10411 429 1 9983 9996 10398 10411 20 12 14 0 9850 10364 + 515 7.2E-28 7.2E-28 p 1 TATCATTCATTATA:TATCATTCAGCATA
CP000521.1_2934384_3000721:10159-55283 IS5 IS5_302 23918 24796 879 1 23918 23953 24762 24796 54 33 36 1 23947 24699 - 753 3E-82 3E-82 c 1 AAAATCAAAATAATGCTTAGGGCGTGTCCTCATTTG:AAAATCAAAATGATGC-TAGGGCGTGTCTTCATTTG11, transposons
Insertion sequences (IS) and transposon elements are both types of mobile genetic elements, but they are distinct from each other.
Insertion Sequences (IS):
Insertion sequences are the simplest type of transposable element.
They typically consist of a short DNA sequence that encodes only the proteins necessary for their own transposition.
An IS element usually includes a transposase gene flanked by inverted repeats, which are short, repeated sequences that are necessary for the insertion process.
IS elements do not carry additional genes other than those required for their own mobility, such as antibiotic resistance genes.
Transposons:
Transposons, or transposable elements, are more complex and can be divided into two main categories: insertion sequences and composite transposons.
Composite transposons consist of two IS elements flanking a central region that often contains additional genes, such as those conferring antibiotic resistance or other functional traits.
Transposons generally include the genes required for their movement (transposase) and can carry additional genes unrelated to transposition.
Key Differences:
Insertion Sequences are the simplest type of transposon and consist of only the basic components necessary for transposition.
Transposons can include IS elements as part of their structure, but they also often contain additional genes.
So, to answer your question: Insertion Sequences do not necessarily "contain" transposon elements, but they are a type of transposon themselves. More complex transposons might include IS elements as part of their structure, along with additional genes.12, transposon detection tools
There are several tools designed to identify and analyze transposons in genomic sequences, similar to how ISEScan is used for identifying insertion sequences (IS). Here are some popular tools for transposon detection:
* RepeatMasker (https://github.com/rmhubley/RepeatMasker/blob/master/RepeatMasker):
- Description: RepeatMasker is a widely used tool for identifying and masking repetitive elements in genomic sequences. It can detect various types of transposable elements, including both Class I (retrotransposons) and Class II (DNA transposons) elements.
- Website: RepeatMasker
* Transposome:
- Description: Transposome is a tool designed to detect and annotate transposable elements in genomic sequences. It integrates with various databases and can provide detailed information about the transposable elements found.
- Website: Transposome
* TEscan:
- Description: TEScan is a tool specifically designed for the identification and annotation of transposable elements in genomic sequences. It uses a database of transposable elements to scan the genome.
- Website: TEscan
* RepeatModeler:
- Description: RepeatModeler is used for de novo repeat identification and annotation. It helps in building custom repeat libraries and can identify transposable elements not present in pre-existing databases.
- Website: RepeatModeler
* DIGGER:
- Description: DIGGER is a tool for the detection and characterization of transposable elements in eukaryotic genomes. It uses a combination of sequence similarity searches and structural analysis.
- Website: DIGGER
* TEannotator:
- Description: TEannotator provides comprehensive annotation of transposable elements by combining different prediction methods and databases.
- Website: TEannotator
* MITE-Hunter:
- Description: MITE-Hunter is specialized for identifying miniature inverted-repeat transposable elements (MITEs), which are a subset of transposable elements.
- Website: MITE-HunterEach of these tools has its own strengths and may be suited to different types of analyses or types of genomes. The choice of tool might depend on the specific needs of your analysis, such as the type of transposable elements you are interested in, the complexity of your genome, and the level of detail required.
conda create --name transposons #python=3.9
conda activate transposons
# TODO: send the results and fasta files of the 5 strains so that he can do some own analysis if necessary.
1. https://github.com/Dfam-consortium/TETools
Dfam TE Tools includes RepeatMasker, RepeatModeler, and coseg. This container is an easy way to get a minimal yet fully functional installation of RepeatMasker and RepeatModeler and is additionally useful for testing or reproducibility purposes.
1. RepeatMasker
# Install RepeatMasker
mamba install -c bioconda repeatmasker
# Run RepeatMasker
RepeatMasker -species "species_name" input_file.fasta
# Replace "species_name" with the target species and input_file.fasta with your genomic sequence file.
RepeatMasker -species "baumannii" Tig1.fna --output Tig1_repeatmasker_out
#https://www.biostars.org/p/215531/
#https://www.repeatmasker.org/RepeatMasker/
#https://github.com/Dfam-consortium/FamDB
#https://www.dfam.org/releases/Dfam_3.8/families/
#https://www.dfam.org/releases/Dfam_3.8/families/FamDB/README.txt
Acinetobacter baumannii
To find the transposons in Acinetobacter baumannii using FamDB, you should download the partition that includes bacterial data. Based on the partition information provided:
Partition 0 [dfam38_full.0.h5]: This partition includes Bacteria.
Steps to Download and Use Partition 0
Download Partition 0: wget https://dfam.org/releases/Dfam_3.8/famdb/dfam38_full.0.h5
Move the downloaded file to the correct directory: mv dfam38_full.0.h5 /home/jhuang/Tools/RepeatMasker/Libraries/famdb
Verify the file is in the correct location: ls /home/jhuang/Tools/RepeatMasker/Libraries/famdb
Run the famdb.py script using the downloaded partition:
./famdb.py --path /home/jhuang/Tools/RepeatMasker/Libraries/famdb/dfam38_full.0.h5
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ names baumanni
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ info
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ info
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ lineage -ad --format totals 470
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ lineage -ad --format totals "Acinetobacter baumannii"
1342 entries in ancestors; 52 lineage-specific entries; found in partitions: 0;
wget https://dfam.org/releases/Dfam_3.8/famdb/dfam38_full.0.h5 -P /home/jhuang/Tools/RepeatMasker/Libraries/famdb/
wget https://dfam.org/releases/Dfam_3.8/famdb/dfam38_full.0.h5 -P /home/jhuang/Tools/RepeatMasker/Libraries/famdb/
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ names "bacteria"
Thus, Acinetobacter baumannii does not belong to any of the partitions listed (which include Bacteria, Bacilli, Bacillaeota, Firmicutes, and Terrabacteria group). Instead, it belongs to the partition containing the Proteobacteria phylum.
./famdb.py -i /home/jhuang/Tools/RepeatMasker/Libraries/famdb/ names "Acinetobacter"
Verify the Installation and Usage
Ensure the path provided to famdb.py points to the exact location of dfam38_full.0.h5. Adjust the script command if needed, and confirm the file permissions to avoid access issues.
By following these steps, you should be able to use the FamDB database partition that contains bacterial transposons data to analyze Acinetobacter baumannii.
#https://bioinformatics.stackexchange.com/questions/2373/is-it-wise-to-use-repeatmasker-on-prokaryotes
# align genome against itself
nucmer --maxmatch --nosimplify genome.fasta genome.fasta
# select repeats and convert the corrdinates to bed format
show-coords -r -T -H out.delta | awk '{if ($1 != $3 && $2 != $4) print $0}' | awk '{print $8"\t"$1"\t"$2}' > repeats.bed
# mask those bases with bedtools
bedtools maskfasta -fi genome.fasta -bed repeats.bed -fo masked.fasta
https://bioinformatics.stackexchange.com/questions/2373/is-it-wise-to-use-repeatmasker-on-prokaryotes
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-015-0654-5
Detection and Characterization of Transposons in Bacteria
https://pubmed.ncbi.nlm.nih.gov/31584155/
2. TnCentral: a Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis
https://journals.asm.org/doi/10.1128/mbio.02060-21
https://tncentral.ncc.unesp.br/tnfinder/
usage: Tn3+TA_finder.py [-h] [-v] -f Sequences.fasta [Sequences.fasta ...] [-o Directory] [-g]
[-t cores] [-p percentage] [-c percentage] [-d base pairs] [-m]
[-e base pairs]
~/Tools/tncomp_finder/TnComp_finder.py
usage: TnComp_finder.py [-h] [-v] -f sequences.fasta [sequences.fasta ...] [-o directory]
[-p threads] [-g] [-i %] [-c %] [-d bp] [-e bp] [-k] [-s | -t]
TnComp_finder.py: error: the following arguments are required: -f/--files
#Tn3 transposon/toxin finder
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f Tig1.fna -t 100 -o Tig1_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f Tig2.fna -t 100 -o Tig2_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f Y.fna -t 100 -o Y_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f W.fna -t 100 -o W_Tn3_out
~/Tools/tn3-ta_finder/Tn3+TA_finder.py -f _adeIJ.fna -t 30 -o deltaAdeIJ_Tn3_out
#composite transposon finder
~/Tools/tncomp_finder/TnComp_finder.py -f Tig1.fna -p 100 -o Tig1_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f Tig2.fna -p 100 -o Tig2_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f Y.fna -p 100 -o Y_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f W.fna -p 100 -o W_TnComp_out
~/Tools/tncomp_finder/TnComp_finder.py -f _adeIJ.fna -p 100 -o deltaAdeIJ_TnComp_out
find . -type f -empty -delete
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig1_contig13.tblastn Tig1_contig19.tblastn Tig1_contig2.tblastn Tig1_contig6.tblastn Tig1_contig17.tblastn _Tig1_contig1.tblastn Tig1_contig3.tblastn Tig1_contig7.tblastn -d$'\t' -o Tig1_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig2_contig11.tblastn Tig2_contig16.tblastn _Tig2_contig1.tblastn Tig2_contig6.tblastn Tig2_contig12.tblastn Tig2_contig17.tblastn Tig2_contig2.tblastn Tig2_contig7.tblastn Tig2_contig13.tblastn Tig2_contig18.tblastn Tig2_contig3.tblastn -d$'\t' -o Tig2_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py Y_contig12.tblastn Y_contig14.tblastn Y_contig18.tblastn _Y_contig1.tblastn Y_contig3.tblastn Y_contig8.tblastn Y_contig13.tblastn Y_contig17.tblastn Y_contig19.tblastn Y_contig2.tblastn Y_contig7.tblastn -d$'\t' -o Y_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py W_contig13.tblastn _W_contig1.tblastn W_contig3.tblastn W_contig7.tblastn W_contig18.tblastn W_contig20.tblastn W_contig6.tblastn -d$'\t' -o W_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py _adeIJ_contig13.tblastn _adeIJ_contig19.tblastn _adeIJ_contig3.tblastn _adeIJ_contig7.tblastn _adeIJ_contig17.tblastn __adeIJ_contig1.tblastn _adeIJ_contig6.tblastn -d$'\t' -o deltaAdeIJ_Tn3_finder_res.xls
#~/Tools/csv2xls-0.4/csv_to_xls.py Tig1_TnComp_out/blastn/Tig1.blastn Tig2_TnComp_out/blastn/Tig2.blastn Y_TnComp_out/blastn/Y.blastn W_TnComp_out/blastn/W.blastn deltaAdeIJ_TnComp_out/blastn/_adeIJ.blastn -d$'\t' -o TnComp_finder_res.xls
#shorten the sheet names of the generated xls-file.
3. Transposome
# Clone the Transposome repository
git clone https://github.com/sestaton/Transposome.git
cd Transposome
# Install
sudo apt-get install -y build-essential lib32z1 git ncbi-blast+ curl
#The command curl -L cpanmin.us | perl - --installdeps . is used to install the Perl module dependencies specified in the current directory (typically a Makefile.PL or Build.PL file). By default, cpanm (App::cpanminus) installs Perl modules in the system Perl library directories. The exact location depends on the system and how Perl is configured. Common locations include: /usr/local/lib/perl5/, /usr/lib/perl5/, or ~/perl5/lib/perl5/
#/home/jhuang/miniconda3/envs/transposons/bin/perl
#The 70 perl libraries are installed under ~/miniconda3/envs/transposons/lib/perl5/site_perl
curl -L cpanmin.us | perl - --installdeps .
perl Makefile.PL
make
make test
make install
# Run Transposome
transposome run --config config_file.yml
# Create a configuration file config_file.yml with appropriate parameters and input files.
4. TEscan
# Install from Bioconda
conda install -c bioconda teannot
# Run TEannot with TEScan
TEannot -i Tig1.fna -o Tig1_TEScan_out
# Replace input_file.fasta with your genomic sequence file and specify an output_directory.
5. RepeatModeler
# Install RepeatModeler
conda install -c bioconda repeatmodeler
# Build a database for the input genome
BuildDatabase -name genomeDB input_file.fasta
# Run RepeatModeler
RepeatModeler -database genomeDB -pa 4
# Replace input_file.fasta with your genomic sequence file and 4 with the number of CPUs to use.
6. DIGGER
# Clone the DIGGER repository
git clone https://github.com/rosenb/digger.git
cd digger
# Install dependencies using conda
conda create -n digger -c bioconda biopython blast
conda activate digger
# Run DIGGER
python digger.py -i input_file.fasta -d database_file -o output_directory
# Replace input_file.fasta with your genomic sequence file, database_file with a suitable transposon database, and specify an output_directory.
7. TEannotator
# Clone the TEannotator repository
git clone https://github.com/teannotator/TEannotator.git
cd TEannotator
# Install dependencies using conda
conda create -n teannotator -c bioconda biopython blast
conda activate teannotator
# Run TEannotator
python TEannotator.py -i input_file.fasta -d database_file -o output_directory
# Replace input_file.fasta with your genomic sequence file, database_file with a suitable transposon database, and specify an output_directory.
8. MITE-Hunter
# Download MITE-Hunter
wget http://target.iplantcollaborative.org/ufloader/MITE_Hunter.tar.gz
tar -xzf MITE_Hunter.tar.gz
cd MITE_Hunter
# Install dependencies using conda
conda create -n mitehunter -c bioconda biopython
conda activate mitehunter
# Run MITE-Hunter
perl MITE_Hunter_manager.pl -i input_file.fasta -g genome_size -c 10
# Replace input_file.fasta with your genomic sequence file and genome_size with the estimated genome size.13, draw the chromosome comparisons using BRIG, java version causing errors
<?xml version="1.0" encoding="UTF-8"?>
<BRIG blastOptions="" legendPosition="upper-right" queryFile="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna" outputFolder="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out" blastPlus="yes" outputFile="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/xxxxxx" title="CP059040" imageFormat="svg" queryFastaFile="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/db/CP059040.gb_converted.fna" cgXML="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/CP059040.gb_converted.fna.xml">
<cgview_settings arrowheadLength="medium" backboneColor="black" backboneRadius="700" backboneThickness="medium" backgroundColor="white" borderColor="black" featureSlotSpacing="medium" featureThickness="30" giveFeaturePositions="false" globalLabel="true" height="4500" isLinear="false" labelFont="SansSerif,plain,30" labelLineLength="medium" labelLineThickness="medium" labelPlacementQuality="best" labelsToKeep="1000" longTickColor="black" minimumFeatureLength="medium" moveInnerLabelsToOuter="true" origin="12" rulerFont="SansSerif,plain,35" rulerFontColor="black" rulerPadding="40" rulerUnits="bases" shortTickColor="black" shortTickThickness="medium" showBorder="true" showShading="true" showWarning="false" tickDensity="0.2333" tickThickness="medium" titleFont="SansSerif,plain,55" titleFontColor="black" useColoredLabelBackgrounds="false" useInnerLabels="true" warningFont="Default,plain,35" warningFontColor="black" width="4500" zeroTickColor="black" />
<brig_settings Ring1="172,14,225" Ring2="222,149,220" Ring3="161,221,231" Ring4="49,34,221" Ring5="116,152,226" Ring6="224,206,38" Ring7="40,191,140" Ring8="158,223,139" Ring9="226,38,122" Ring10="211,41,77" defaultUpper="70" defaultLower="50" defaultMinimum="50" genbankFiles="gbk,gb,genbank" fastaFiles="fna,faa,fas,fasta,fa" emblFiles="embl" blastLocation="" divider="3" multiplier="3" memory="1500" defaultSpacer="0" />
<special value="GC Content" />
<special value="GC Skew" />
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig1">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig1/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig2">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig2/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Y">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Y/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/W">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/W/contigs.fa" />
</refDir>
<refDir location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/_adeIJ">
<refFile location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/_adeIJ/contigs.fa" />
</refDir>
<ring position="0" colour="172,14,225" name="Tig1" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig1/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="172,14,225" name="Tig2" position="1" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Tig2/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="222,149,220" name="Y" position="2" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/Y/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="161,221,231" name="W" position="3" upperInt="70" lowerInt="50" legend="yes" size="30" labels="no" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/W/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="49,34,221" name="△adeIJ" position="4" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="/home/jhuang/DATA/Data_Tam_variant_calling/snippy_CP059040/shovill/_adeIJ/contigs.fa" blastResults="/home/jhuang/DATA/Data_Tam_variant_calling/brig_out/scratch/contigs.faVsCP059040.gb_converted.fna.tab" />
</ring>
<ring colour="225,0,0" name="GC Content" position="5" upperInt="70" lowerInt="50" legend="yes" size="30" labels="no" blastType="blastn">
<sequence location="GC Content" />
</ring>
<ring colour="225,0,0" name="GC Skew" position="6" upperInt="70" lowerInt="50" legend="yes" size="30" labels="yes" blastType="blastn">
<sequence location="GC Skew" />
</ring>
</BRIG>14, (optional) MANUALLY REMOVE the column f6 in filtered_merged_variants_CP133676.csv, and rename CHROM to HDRNA_01_K01 in the header, summarize chr and plasmids SNPs of a sample together to a single list, save as an Excel-file.
#TOMORROW_TODO_FROM_HERE, generate local genetic environments of adeIJ (Figure_1); make the variant_calling.xls; search all SNP, InDel,transposons, IS elements using http://xgenes.com/article/article-content/316/insertion-sequence-is-element-detection/, however, we have to align the found positions of Treffer; generate a BRIG using Tig1 as the reference, Tig2, delta_adeIJ, Y, W to look if 99% are idential (Figure_2).
# I am not sure what do you mean about transposons. Actually I now processed a project of the Transposons (search for the manuscript of Patricia). What do you mean? They have some specifial expected transposons so that I can check the positions of pre-defined transponsons by comparing the genomes (see the slide of Patrick)?
#TODO: Using the method using for Patricia, I can compare the five genomes to REF to look for the positions newly inserted.15, code of summarize_snippy_res.py
import pandas as pd
import glob
import argparse
import os
#python3 summarize_snps_indels.py snippy_HDRNA_01/snippy
#The following record for 2365295 is wrong, since I am sure in the HDRNA_01_K010, it should be a 'G', since in HDRNA_01_K010.csv
#CP133676,2365295,snp,A,G,G:178 A:0
#
#The current output is as follows:
#CP133676,2365295,A,snp,A,A,A,A,A,A,A,A,A,A,None,,,,,,None,None
#CP133676,2365295,A,snp,A,A,A,A,A,A,A,A,A,A,nan,,,,,,nan,nan
#grep -v "None,,,,,,None,None" summary_snps_indels.csv > summary_snps_indels_.csv
#BUG: CP133676,2365295,A,snp,A,A,A,A,A,A,A,A,A,A,nan,,,,,,nan,nan
import pandas as pd
import glob
import argparse
import os
def main(base_directory):
# List of isolate identifiers
#isolates = [f"HDRNA_01_K0{i}" for i in range(1, 11)]
isolates = ["Tig1", "Tig2", "Y", "W", "_adeIJ"]
expected_columns = ["CHROM", "POS", "REF", "ALT", "TYPE", "EFFECT", "LOCUS_TAG", "GENE", "PRODUCT"]
# Find all CSV files in the directory and its subdirectories
csv_files = glob.glob(os.path.join(base_directory, '**', '*.csv'), recursive=True)
# Create an empty DataFrame to store the summary
summary_df = pd.DataFrame()
# Iterate over each CSV file
for file_path in csv_files:
# Extract isolate identifier from the file name
isolate = os.path.basename(file_path).replace('.csv', '')
df = pd.read_csv(file_path)
# Ensure all expected columns are present, adding missing ones as empty columns
for col in expected_columns:
if col not in df.columns:
df[col] = None
# Extract relevant columns
df = df[expected_columns]
# Ensure consistent data types
df = df.astype({"CHROM": str, "POS": int, "REF": str, "ALT": str, "TYPE": str, "EFFECT": str, "LOCUS_TAG": str, "GENE": str, "PRODUCT": str})
# Add the isolate column with the ALT allele
df[isolate] = df["ALT"]
# Drop columns that are not needed for the summary
df = df.drop(["ALT"], axis=1)
if summary_df.empty:
summary_df = df
else:
summary_df = pd.merge(summary_df, df, on=["CHROM", "POS", "REF", "TYPE", "EFFECT", "LOCUS_TAG", "GENE", "PRODUCT"], how="outer")
# Fill missing values with the REF allele for each isolate column
for isolate in isolates:
if isolate in summary_df.columns:
summary_df[isolate] = summary_df[isolate].fillna(summary_df["REF"])
else:
summary_df[isolate] = summary_df["REF"]
# Rename columns to match the required format
summary_df = summary_df.rename(columns={
"CHROM": "CHROM",
"POS": "POS",
"REF": "REF",
"TYPE": "TYPE",
"EFFECT": "Effect",
"LOCUS_TAG": "Gene_name",
"GENE": "Biotype",
"PRODUCT": "Product"
})
# Replace any remaining None or NaN values in the non-isolate columns with empty strings
summary_df = summary_df.fillna("")
# Add empty columns for Impact, Functional_Class, Codon_change, Protein_and_nucleotide_change, Amino_Acid_Length
summary_df["Impact"] = ""
summary_df["Functional_Class"] = ""
summary_df["Codon_change"] = ""
summary_df["Protein_and_nucleotide_change"] = ""
summary_df["Amino_Acid_Length"] = ""
# Reorder columns
cols = ["CHROM", "POS", "REF", "TYPE"] + isolates + ["Effect", "Impact", "Functional_Class", "Codon_change", "Protein_and_nucleotide_change", "Amino_Acid_Length", "Gene_name", "Biotype"]
summary_df = summary_df.reindex(columns=cols)
# Remove duplicate rows
summary_df = summary_df.drop_duplicates()
# Save the summary DataFrame to a CSV file
output_file = os.path.join(base_directory, "summary_snps_indels.csv")
summary_df.to_csv(output_file, index=False)
print("Summary CSV file created successfully at:", output_file)
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="Summarize SNPs and Indels from CSV files.")
parser.add_argument("directory", type=str, help="Base directory containing the CSV files in subdirectories.")
args = parser.parse_args()
main(args.directory)How to List All Methods for an Object in R
In R, objects can belong to different classes (S3 or S4), and the methods applicable to these objects depend on their class. Below are the steps to list all methods available for an object in both the S3 and S4 systems.
-
Determine the Class of the Object
Before listing methods, it is important to determine the class of the object. You can use the class() function to identify the class.
# Check the class of the object class_name <- class(dat) print(class_name) -
List Methods for S3 Objects
If the object is an S3 object, you can list all methods that are associated with its class using the methods() function:
# List all S3 methods applicable to the object's class methods(class = class_name)For specific generic functions related to an S3 object, such as “plot”, you can see which methods are available:
# List S3 methods for a specific generic function methods("plot")To get the function definition of a specific S3 method:
# Get the definition of a specific S3 method getS3method("plot", class_name) -
List Methods for S4 Objects
If the object is an S4 object, you need to use showMethods() to list all methods that are defined for the object’s class:
# List all S4 methods applicable to the object's class showMethods(classes = class_name)To list methods for a specific S4 function related to the object:
# Show all methods for a specific S4 function showMethods("functionName")To get more details about a specific method for an S4 object, use getMethod():
# Get details about a specific S4 method getMethod("plot", "VoltRon") -
List All Functions in a Specific Package
If you are working with a specific package (e.g., “VoltRon”) and want to list all functions (including methods) available in that package, you can use:
# List all functions available in the "VoltRon" package ls("package:VoltRon")Or, if you want to see the structure of functions (i.e., only the function names):
# List all functions (methods) associated with the "VoltRon" package lsf.str("package:VoltRon")
Summary
- S3 Methods: Use methods(class = class_name) to list methods applicable to an S3 object’s class.
- S4 Methods: Use showMethods(classes = class_name) to list methods for an S4 object.
- Package Functions: Use ls(“package:PackageName”) or lsf.str(“package:PackageName”) to list all functions in a specific package.
- Use getS3method() or getMethod() to retrieve specific method definitions.
By following these steps, you can effectively list and explore all methods available for an object in R, regardless of whether it is an S3 or S4 object.Phyloseq + MicrobiotaProcess + PICRUSt2
-
Phyloseq.Rmd
author: "" date: '`r format(Sys.time(), "%d %m %Y")`' header-includes: - \usepackage{color, fancyvrb} output: rmdformats::readthedown: highlight: kate number_sections : yes pdf_document: toc: yes toc_depth: 2 number_sections : yes --- ```{r, echo=FALSE, warning=FALSE} ## Global options # TODO: reproduce the html with the additional figure/SVN-files for editing. # IMPORTANT NOTE: needs before "mkdir figures" #rmarkdown::render('Phyloseq.Rmd',output_file='Phyloseq.html') ``` ```{r load-packages, include=FALSE} library(knitr) library(rmdformats) library(readxl) library(dplyr) library(kableExtra) options(max.print="75") knitr::opts_chunk$set(fig.width=8, fig.height=6, eval=TRUE, cache=TRUE, echo=TRUE, prompt=FALSE, tidy=TRUE, comment=NA, message=FALSE, warning=FALSE) opts_knit$set(width=85) # Phyloseq R library #* Phyloseq web site : https://joey711.github.io/phyloseq/index.html #* See in particular tutorials for # - importing data: https://joey711.github.io/phyloseq/import-data.html # - heat maps: https://joey711.github.io/phyloseq/plot_heatmap-examples.html ``` # Data Import raw data and assign sample key: ```{r, echo=TRUE, warning=FALSE} #extend map_corrected.txt with Diet and Flora #setwd("~/DATA/Data_Laura_16S_2/core_diversity_e4753") map_corrected <- read.csv("map_corrected.txt", sep="\t", row.names=1) knitr::kable(map_corrected) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) ``` # Prerequisites to be installed * R : https://pbil.univ-lyon1.fr/CRAN/ * R studio : https://www.rstudio.com/products/rstudio/download/#download ```R install.packages("dplyr") # To manipulate dataframes install.packages("readxl") # To read Excel files into R install.packages("ggplot2") # for high quality graphics install.packages("heatmaply") source("https://bioconductor.org/biocLite.R") biocLite("phyloseq") ``` ```{r libraries, echo=TRUE, message=FALSE} library("readxl") # necessary to import the data from Excel file library("ggplot2") # graphics library("picante") library("microbiome") # data analysis and visualisation library("phyloseq") # also the basis of data object. Data analysis and visualisation library("ggpubr") # publication quality figures, based on ggplot2 library("dplyr") # data handling, filter and reformat data frames library("RColorBrewer") # nice color options library("heatmaply") library(vegan) library(gplots) ``` # Read the data and create phyloseq objects Three tables are needed * OTU * Taxonomy * Samples ```{r, echo=TRUE, warning=FALSE} #Change your working directory to where the files are located ps.ng.tax <- import_biom("./table_even42369.biom", "./clustering/rep_set.tre") sample <- read.csv("./map_corrected.txt", sep="\t", row.names=1) SAM = sample_data(sample, errorIfNULL = T) rownames(SAM) <- c("1","2","3","5","6","7","8","9","10","12","13","14","15","16","17","18","19","20","21","22","23","24","25","26","27","28","29","30","31","32","33","34","35","36","37","38","39","40","41","42","43","44","46","47","48","49","50","51","52","53","55") ps.ng.tax <- merge_phyloseq(ps.ng.tax, SAM) print(ps.ng.tax) colnames(tax_table(ps.ng.tax)) <- c("Domain","Phylum","Class","Order","Family","Genus","Species") saveRDS(ps.ng.tax, "./ps.ng.tax.rds") ``` Visualize data ```{r, echo=TRUE, warning=FALSE} sample_names(ps.ng.tax) rank_names(ps.ng.tax) sample_variables(ps.ng.tax) ``` Normalize number of reads in each sample using median sequencing depth. ```{r, echo=TRUE, warning=FALSE} # RAREFACTION #set.seed(9242) # This will help in reproducing the filtering and nomalisation. #ps.ng.tax <- rarefy_even_depth(ps.ng.tax, sample.size = 42369) #total <- 42369 # NORMALIZE number of reads in each sample using median sequencing depth. total = median(sample_sums(ps.ng.tax)) #> total #[1] 42369 standf = function(x, t=total) round(t * (x / sum(x))) ps.ng.tax = transform_sample_counts(ps.ng.tax, standf) ps.ng.tax_rel <- microbiome::transform(ps.ng.tax, "compositional") saveRDS(ps.ng.tax, "./ps.ng.tax.rds") hmp.meta <- meta(ps.ng.tax) hmp.meta$sam_name <- rownames(hmp.meta) ``` # Heatmaps ```{r, echo=TRUE, warning=FALSE} #MOVE_FROM_ABOVE: The number of reads used for normalization is **`r sprintf("%.0f", total)`**. #A basic heatmap using the default parameters. # plot_heatmap(ps.ng.tax, method = "NMDS", distance = "bray") #NOTE that giving the correct OTU numbers in the text (1%, 0.5%, ...)!!! ``` We consider the most abundant OTUs for heatmaps. For example one can only take OTUs that represent at least 1% of reads in at least one sample. Remember we normalized all the sampples to median number of reads (total). We are left with only 168 OTUS which makes the reading much more easy. ```{r, echo=TRUE, warning=FALSE} # Custom function to plot a heatmap with the specified sample order #plot_heatmap_custom <- function(ps, sample_order, method = "NMDS", distance = "bray") { ps.ng.tax_abund <- phyloseq::filter_taxa(ps.ng.tax, function(x) sum(x > total*0.01) > 0, TRUE) kable(otu_table(ps.ng.tax_abund)) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) # Calculate the relative abundance for each sample ps.ng.tax_abund_rel <- transform_sample_counts(ps.ng.tax_abund, function(x) x / sum(x)) datamat_ = as.data.frame(otu_table(ps.ng.tax_abund)) #datamat <- datamat_[c("1","2","5","6","7", "8","9","10","12","13","14", "15","16","17","18","19","20", "21","22","23","24","25","26","27","28", "29","30","31","32", "33","34","35","36","37","38","39","51", "40","41","42","43","44","46", "47","48","49","50","52","53","55")] datamat <- datamat_[c("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")] hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.08) mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN"); mycol = mycol[as.vector(mycl)] sampleCols <- rep('GREY',ncol(datamat)) #names(sampleCols) <- c("Group1", "Group1", "Group1", "Group1", "Group1", "Group2", "Group2", "Group2", "Group2", "Group2","Group2", "Group3", "Group3", "Group3", "Group3", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1", "Group1") #sampleCols[colnames(datamat)=='1'] <- '#a6cee3' #sampleCols[colnames(datamat)=='2'] <- '#a6cee3' #sampleCols[colnames(datamat)=='5'] <- '#a6cee3' #sampleCols[colnames(datamat)=='6'] <- '#a6cee3' #sampleCols[colnames(datamat)=='7'] <- '#a6cee3' sampleCols[colnames(datamat)=='8'] <- '#1f78b4' sampleCols[colnames(datamat)=='9'] <- '#1f78b4' sampleCols[colnames(datamat)=='10'] <- '#1f78b4' sampleCols[colnames(datamat)=='12'] <- '#1f78b4' sampleCols[colnames(datamat)=='13'] <- '#1f78b4' sampleCols[colnames(datamat)=='14'] <- '#1f78b4' #sampleCols[colnames(datamat)=='15'] <- '#b2df8a' #sampleCols[colnames(datamat)=='16'] <- '#b2df8a' #sampleCols[colnames(datamat)=='17'] <- '#b2df8a' #sampleCols[colnames(datamat)=='18'] <- '#b2df8a' #sampleCols[colnames(datamat)=='19'] <- '#b2df8a' #sampleCols[colnames(datamat)=='20'] <- '#b2df8a' sampleCols[colnames(datamat)=='21'] <- '#33a02c' sampleCols[colnames(datamat)=='22'] <- '#33a02c' sampleCols[colnames(datamat)=='23'] <- '#33a02c' sampleCols[colnames(datamat)=='24'] <- '#33a02c' sampleCols[colnames(datamat)=='25'] <- '#33a02c' sampleCols[colnames(datamat)=='26'] <- '#33a02c' sampleCols[colnames(datamat)=='27'] <- '#33a02c' sampleCols[colnames(datamat)=='28'] <- '#33a02c' #sampleCols[colnames(datamat)=='29'] <- '#fb9a99' #sampleCols[colnames(datamat)=='30'] <- '#fb9a99' #sampleCols[colnames(datamat)=='31'] <- '#fb9a99' #sampleCols[colnames(datamat)=='32'] <- '#fb9a99' sampleCols[colnames(datamat)=='33'] <- '#e31a1c' sampleCols[colnames(datamat)=='34'] <- '#e31a1c' sampleCols[colnames(datamat)=='35'] <- '#e31a1c' sampleCols[colnames(datamat)=='36'] <- '#e31a1c' sampleCols[colnames(datamat)=='37'] <- '#e31a1c' sampleCols[colnames(datamat)=='38'] <- '#e31a1c' sampleCols[colnames(datamat)=='39'] <- '#e31a1c' sampleCols[colnames(datamat)=='51'] <- '#e31a1c' #sampleCols[colnames(datamat)=='40'] <- '#cab2d6' #sampleCols[colnames(datamat)=='41'] <- '#cab2d6' #sampleCols[colnames(datamat)=='42'] <- '#cab2d6' #sampleCols[colnames(datamat)=='43'] <- '#cab2d6' #sampleCols[colnames(datamat)=='44'] <- '#cab2d6' #sampleCols[colnames(datamat)=='46'] <- '#cab2d6' sampleCols[colnames(datamat)=='47'] <- '#6a3d9a' sampleCols[colnames(datamat)=='48'] <- '#6a3d9a' sampleCols[colnames(datamat)=='49'] <- '#6a3d9a' sampleCols[colnames(datamat)=='50'] <- '#6a3d9a' sampleCols[colnames(datamat)=='52'] <- '#6a3d9a' sampleCols[colnames(datamat)=='53'] <- '#6a3d9a' sampleCols[colnames(datamat)=='55'] <- '#6a3d9a' #bluered(75) #color_pattern <- colorRampPalette(c("blue", "white", "red"))(100) library(RColorBrewer) custom_palette <- colorRampPalette(brewer.pal(9, "Blues")) heatmap_colors <- custom_palette(100) #colors <- heatmap_color_default(100) png("figures/heatmap.png", width=1200, height=2400) #par(mar=c(2, 2, 2, 2)) , lwid=1 lhei=c(0.7, 10)) # Adjust height of color keys keysize=0.3, heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row', scale='row',trace='none',col=heatmap_colors, cexRow=1.2, cexCol=1.5, RowSideColors = mycol, ColSideColors = sampleCols, srtCol=15, labRow=row.names(datamat), key=TRUE, margins=c(10, 15), lhei=c(0.7, 15), lwid=c(1,8)) dev.off() ``` ```{r, echo=TRUE, warning=FALSE, fig.cap="Heatmap", out.width = '100%', fig.align= "center"} knitr::include_graphics("./figures/heatmap.png") ``` ```{r, echo=FALSE, warning=FALSE} #It is possible to use different distances and different multivaraite methods. Many different built-in distances can be used. #dist_methods <- unlist(distanceMethodList) #print(dist_methods) ``` \pagebreak # Taxonomic summary ## Bar plots in phylum level ```{r, echo=FALSE, warning=FALSE} #Make the bargraph nicer by removing OTUs boundaries. This is done by adding ggplot2 modifier. # 1: uniform color. Color is for the border, fill is for the inside #ggplot(mtcars, aes(x=as.factor(cyl) )) + # geom_bar(color="blue", fill=rgb(0.1,0.4,0.5,0.7) ) # 2: Using Hue #ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) + # geom_bar( ) + # scale_fill_hue(c = 40) + # theme(legend.position="none") # 3: Using RColorBrewer #ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) + # geom_bar( ) + # scale_fill_brewer(palette = "Set1") + # theme(legend.position="none") # 4: Using greyscale: #ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) + # geom_bar( ) + # scale_fill_grey(start = 0.25, end = 0.75) + # theme(legend.position="none") # 5: Set manualy #ggplot(mtcars, aes(x=as.factor(cyl), fill=as.factor(cyl) )) + # geom_bar( ) + # scale_fill_manual(values = c("red", "green", "blue") ) + # theme(legend.position="none") #NOT SUCCESSFUL! #allGroupsColors<- c( # "grey0", "grey50", "dodgerblu", "deepskyblue", # "red", "darkred", "green", "green4") # plot_bar(ps.ng.tax_rel, fill="Phylum") + # geom_bar(stat="identity", position="stack") + scale_color_manual(values = allGroupsColors) #, fill=Phylum + scale_fill_brewer(palette = "Set1") # ##### Keep only the most abundant phyla and #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Actinobacteria")) #1.57 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Bacteroidetes")) #27.27436 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Cyanobacteria")) #0.02244249 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Epsilonbacteraeota")) #0.01309145 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Euryarchaeota")) #0.1210024 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Firmicutes")) #32.50589 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Lentisphaerae")) #0.0001870208 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Patescibacteria")) #0.008789976 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Planctomycetes")) #0.01365252 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Proteobacteria")) #6.769216 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Synergistetes")) #0.005049561 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Tenericutes")) #0.0005610623 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Verrucomicrobia")) #2.076304 #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c(NA)) #sum(otu_table(ps.ng.tax_most)) = 2.619413 ``` ```{r, echo=TRUE, warning=FALSE} library(ggplot2) geom.text.size = 6 theme.size = 8 #(14/5) * geom.text.size #ps.ng.tax_most <- subset_taxa(ps.ng.tax_rel, Phylum %in% c("D_1__Actinobacteria", "D_1__Bacteroidetes", "D_1__Firmicutes", "D_1__Proteobacteria", "D_1__Verrucomicrobia", NA)) ps.ng.tax_most = phyloseq::filter_taxa(ps.ng.tax_rel, function(x) mean(x) > 0.001, TRUE) #CONSOLE(OPTIONAL): for sampleid in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73; do #echo "otu_table(ps.ng.tax_most)[,${sampleid}]=otu_table(ps.ng.tax_most)[,${sampleid}]/sum(otu_table(ps.ng.tax_most)[,${sampleid}])" done #OR ps.ng.tax_most_ = transform_sample_counts(ps.ng.tax_most, function(x) x / sum(x)) ``` ```{r, echo=FALSE, warning=FALSE} ##--Creating 100% stacked bar plots with less abundant taxa in a sub-category #901-- ##https://github.com/joey711/phyloseq/issues/901 ##ps.ng.tax_most_df <- psmelt(ps.ng.tax_most_) #5986x19 #glom <- tax_glom(ps.ng.tax_most_, taxrank = 'Phylum') #tax_table(glom) # should list # taxa as # phyla #data <- psmelt(glom) # create dataframe from phyloseq object #data$Phylum <- as.character(data$Phylum) #convert to character ##simple way to rename phyla with < 1% abundance #data$Phylum[data$Abundance < 0.001] <- "< 0.1% abund." # #library(plyr) #medians <- ddply(data, ~Phylum, function(x) c(median=median(x$Abundance))) #remainder <- medians[medians$median <= 0.001,]$Phylum #data[data$Phylum %in% remainder,]$Phylum <- "Phyla < 0.1% abund." #data$Phylum[data$Abundance < 0.001] <- "Phyla < 0.1% abund." ##--> data are not used! # ##in class level #glom <- tax_glom(ps.ng.tax_most_, taxrank = 'Class') #tax_table(glom) # should list # taxa as # phyla #data <- psmelt(glom) # create dataframe from phyloseq object #data$Class <- as.character(data$Class) #convert to character # ##simple way to rename phyla with < 1% abundance #data$Class[data$Abundance < 0.001] <- "< 0.1% abund." #Count = length(unique(data$Class)) # ##unique(data$Class) ##data$Class <- factor(data$Class, levels = c("Bacilli", "Bacteroidia", "Verrucomicrobiae", "Clostridia", "Gammaproteobacteria", "Alphaproteobacteria", "Actinobacteria", "Negativicutes", "Erysipelotrichia", "Methanobacteria", "< 0.1% abund.")) ##------- Creating 100% stacked bar plots END -------- library(stringr) #FITTING1: # tax_table(ps.ng.tax_most_)[1,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Domain"], "__")[[1]][2] # ... ... # tax_table(ps.ng.tax_most_)[167,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Species"], "__")[[1]][2] #ps.ng.tax_most_ #in total [ 89 taxa and 55 samples ] #otu_table() OTU Table: [ 166 taxa and 54 samples ] #otu_table() OTU Table: [ 168 taxa and 50 samples ] #for id in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166; do #echo "tax_table(ps.ng.tax_most_)[${id},\"Domain\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Domain\"], \"__\")[[1]][2]" #echo "tax_table(ps.ng.tax_most_)[${id},\"Phylum\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Phylum\"], \"__\")[[1]][2]" #echo "tax_table(ps.ng.tax_most_)[${id},\"Class\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Class\"], \"__\")[[1]][2]" #echo "tax_table(ps.ng.tax_most_)[${id},\"Order\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Order\"], \"__\")[[1]][2]" #echo "tax_table(ps.ng.tax_most_)[${id},\"Family\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Family\"], \"__\")[[1]][2]" #echo "tax_table(ps.ng.tax_most_)[${id},\"Genus\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Genus\"], \"__\")[[1]][2]" #echo "tax_table(ps.ng.tax_most_)[${id},\"Species\"] <- str_split(tax_table(ps.ng.tax_most_)[${id},\"Species\"], \"__\")[[1]][2]" #done tax_table(ps.ng.tax_most_)[1,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Domain"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[1,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Phylum"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[1,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Class"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[1,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Order"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[1,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Family"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[1,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Genus"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[1,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[1,"Species"], "__")[[1]][2] #... ... tax_table(ps.ng.tax_most_)[167,"Domain"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Domain"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[167,"Phylum"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Phylum"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[167,"Class"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Class"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[167,"Order"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Order"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[167,"Family"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Family"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[167,"Genus"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Genus"], "__")[[1]][2] tax_table(ps.ng.tax_most_)[167,"Species"] <- str_split(tax_table(ps.ng.tax_most_)[167,"Species"], "__")[[1]][2] ``` ```{r, echo=TRUE, warning=FALSE} #aes(color="Phylum", fill="Phylum") --> aes() #ggplot(data=data, aes(x=Sample, y=Abundance, fill=Phylum)) plot_bar(ps.ng.tax_most_, fill="Phylum") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=2)) #6 instead of theme.size ``` ```{r, echo=FALSE, warning=FALSE} #png("abc.png") #knitr::include_graphics("./Phyloseq_files/figure-html/unnamed-chunk-7-1.png") #dev.off() ``` \pagebreak Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth. ```{r, echo=TRUE, warning=FALSE} ps.ng.tax_most_pre_post_stroke <- merge_samples(ps.ng.tax_most_, "pre_post_stroke") ps.ng.tax_most_pre_post_stroke_ = transform_sample_counts(ps.ng.tax_most_pre_post_stroke, function(x) x / sum(x)) #plot_bar(ps.ng.tax_most_SampleType_, fill = "Phylum") + geom_bar(aes(color=Phylum, fill=Phylum), stat="identity", position="stack") plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Phylum") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = theme.size, colour="black")) ``` \pagebreak Use color according to phylum. Do separate panels Stroke and Sex_age. ```{r, echo=TRUE, warning=FALSE} ps.ng.tax_most_copied <- data.table::copy(ps.ng.tax_most_) #FITTING6: regulate the bar height if it has replicates: 5+6+6+8+4+8+6+7=25+25=50 otu_table(ps.ng.tax_most_)[,c("1")] <- otu_table(ps.ng.tax_most_)[,c("1")]/5 otu_table(ps.ng.tax_most_)[,c("2")] <- otu_table(ps.ng.tax_most_)[,c("2")]/5 otu_table(ps.ng.tax_most_)[,c("5")] <- otu_table(ps.ng.tax_most_)[,c("5")]/5 otu_table(ps.ng.tax_most_)[,c("6")] <- otu_table(ps.ng.tax_most_)[,c("6")]/5 otu_table(ps.ng.tax_most_)[,c("7")] <- otu_table(ps.ng.tax_most_)[,c("7")]/5 otu_table(ps.ng.tax_most_)[,c("8")] <- otu_table(ps.ng.tax_most_)[,c("8")]/6 otu_table(ps.ng.tax_most_)[,c("9")] <- otu_table(ps.ng.tax_most_)[,c("9")]/6 otu_table(ps.ng.tax_most_)[,c("10")] <- otu_table(ps.ng.tax_most_)[,c("10")]/6 otu_table(ps.ng.tax_most_)[,c("12")] <- otu_table(ps.ng.tax_most_)[,c("12")]/6 otu_table(ps.ng.tax_most_)[,c("13")] <- otu_table(ps.ng.tax_most_)[,c("13")]/6 otu_table(ps.ng.tax_most_)[,c("14")] <- otu_table(ps.ng.tax_most_)[,c("14")]/6 otu_table(ps.ng.tax_most_)[,c("15")] <- otu_table(ps.ng.tax_most_)[,c("15")]/6 otu_table(ps.ng.tax_most_)[,c("16")] <- otu_table(ps.ng.tax_most_)[,c("16")]/6 otu_table(ps.ng.tax_most_)[,c("17")] <- otu_table(ps.ng.tax_most_)[,c("17")]/6 otu_table(ps.ng.tax_most_)[,c("18")] <- otu_table(ps.ng.tax_most_)[,c("18")]/6 otu_table(ps.ng.tax_most_)[,c("19")] <- otu_table(ps.ng.tax_most_)[,c("19")]/6 otu_table(ps.ng.tax_most_)[,c("20")] <- otu_table(ps.ng.tax_most_)[,c("20")]/6 otu_table(ps.ng.tax_most_)[,c("21")] <- otu_table(ps.ng.tax_most_)[,c("21")]/8 otu_table(ps.ng.tax_most_)[,c("22")] <- otu_table(ps.ng.tax_most_)[,c("22")]/8 otu_table(ps.ng.tax_most_)[,c("23")] <- otu_table(ps.ng.tax_most_)[,c("23")]/8 otu_table(ps.ng.tax_most_)[,c("24")] <- otu_table(ps.ng.tax_most_)[,c("24")]/8 otu_table(ps.ng.tax_most_)[,c("25")] <- otu_table(ps.ng.tax_most_)[,c("25")]/8 otu_table(ps.ng.tax_most_)[,c("26")] <- otu_table(ps.ng.tax_most_)[,c("26")]/8 otu_table(ps.ng.tax_most_)[,c("27")] <- otu_table(ps.ng.tax_most_)[,c("27")]/8 otu_table(ps.ng.tax_most_)[,c("28")] <- otu_table(ps.ng.tax_most_)[,c("28")]/8 otu_table(ps.ng.tax_most_)[,c("29")] <- otu_table(ps.ng.tax_most_)[,c("29")]/4 otu_table(ps.ng.tax_most_)[,c("30")] <- otu_table(ps.ng.tax_most_)[,c("30")]/4 otu_table(ps.ng.tax_most_)[,c("31")] <- otu_table(ps.ng.tax_most_)[,c("31")]/4 otu_table(ps.ng.tax_most_)[,c("32")] <- otu_table(ps.ng.tax_most_)[,c("32")]/4 otu_table(ps.ng.tax_most_)[,c("33")] <- otu_table(ps.ng.tax_most_)[,c("33")]/8 otu_table(ps.ng.tax_most_)[,c("34")] <- otu_table(ps.ng.tax_most_)[,c("34")]/8 otu_table(ps.ng.tax_most_)[,c("35")] <- otu_table(ps.ng.tax_most_)[,c("35")]/8 otu_table(ps.ng.tax_most_)[,c("36")] <- otu_table(ps.ng.tax_most_)[,c("36")]/8 otu_table(ps.ng.tax_most_)[,c("37")] <- otu_table(ps.ng.tax_most_)[,c("37")]/8 otu_table(ps.ng.tax_most_)[,c("38")] <- otu_table(ps.ng.tax_most_)[,c("38")]/8 otu_table(ps.ng.tax_most_)[,c("39")] <- otu_table(ps.ng.tax_most_)[,c("39")]/8 otu_table(ps.ng.tax_most_)[,c("51")] <- otu_table(ps.ng.tax_most_)[,c("51")]/8 otu_table(ps.ng.tax_most_)[,c("40")] <- otu_table(ps.ng.tax_most_)[,c("40")]/6 otu_table(ps.ng.tax_most_)[,c("41")] <- otu_table(ps.ng.tax_most_)[,c("41")]/6 otu_table(ps.ng.tax_most_)[,c("42")] <- otu_table(ps.ng.tax_most_)[,c("42")]/6 otu_table(ps.ng.tax_most_)[,c("43")] <- otu_table(ps.ng.tax_most_)[,c("43")]/6 otu_table(ps.ng.tax_most_)[,c("44")] <- otu_table(ps.ng.tax_most_)[,c("44")]/6 otu_table(ps.ng.tax_most_)[,c("46")] <- otu_table(ps.ng.tax_most_)[,c("46")]/6 otu_table(ps.ng.tax_most_)[,c("47")] <- otu_table(ps.ng.tax_most_)[,c("47")]/7 otu_table(ps.ng.tax_most_)[,c("48")] <- otu_table(ps.ng.tax_most_)[,c("48")]/7 otu_table(ps.ng.tax_most_)[,c("49")] <- otu_table(ps.ng.tax_most_)[,c("49")]/7 otu_table(ps.ng.tax_most_)[,c("50")] <- otu_table(ps.ng.tax_most_)[,c("50")]/7 otu_table(ps.ng.tax_most_)[,c("52")] <- otu_table(ps.ng.tax_most_)[,c("52")]/7 otu_table(ps.ng.tax_most_)[,c("53")] <- otu_table(ps.ng.tax_most_)[,c("53")]/7 otu_table(ps.ng.tax_most_)[,c("55")] <- otu_table(ps.ng.tax_most_)[,c("55")]/7 #plot_bar(ps.ng.tax_most_swab_, x="Phylum", fill = "Phylum", facet_grid = Patient~RoundDay) + geom_bar(aes(color=Phylum, fill=Phylum), stat="identity", position="stack") + theme(axis.text = element_text(size = theme.size, colour="black")) plot_bar(ps.ng.tax_most_, x="Phylum", fill="Phylum", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=2)) ``` ```{r, echo=FALSE, warning=FALSE} #knitr::include_graphics("./Phyloseq_files/figure-html/unnamed-chunk-10-1.png") #> tax_table(carbom) #Taxonomy Table: [205 taxa by 7 taxonomic ranks]: # Domain Supergroup Division Class #Otu001 "Eukaryota" "Archaeplastida" "Chlorophyta" "Mamiellophyceae" # Order Family Genus #Otu001 "Mamiellales" "Bathycoccaceae" "Ostreococcus" #sample_data(ps.ng.tax) ``` ## Bar plots in class level ```{r, echo=TRUE, warning=FALSE} plot_bar(ps.ng.tax_most_copied, fill="Class") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=3)) ``` Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth. ```{r, echo=TRUE, warning=FALSE} plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Class") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = theme.size, colour="black")) ``` \pagebreak Use color according to class. Do separate panels Stroke and Sex_age. ```{r, echo=TRUE, warning=FALSE} #-- If existing replicates, to be processed as follows -- plot_bar(ps.ng.tax_most_, x="Class", fill="Class", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=3)) ``` ## Bar plots in order level ```{r, echo=TRUE, warning=FALSE} plot_bar(ps.ng.tax_most_copied, fill="Order") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=4)) ``` Regroup together pre vs post stroke and normalize number of reads in each group using median sequencing depth. ```{r, echo=TRUE, warning=FALSE} plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Order") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = theme.size, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=4)) ``` \pagebreak Use color according to order. Do separate panels Stroke and Sex_age. ```{r, echo=TRUE, warning=FALSE} #FITTING7: regulate the bar height if it has replicates plot_bar(ps.ng.tax_most_, x="Order", fill="Order", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("darkblue", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "lightskyblue", "darkgreen", "deeppink", "khaki2", "firebrick", "brown1", "darkorange1", "cyan1", "royalblue4", "darksalmon", "darkblue","royalblue4", "dodgerblue3", "steelblue1", "lightskyblue", "darkseagreen", "darkgoldenrod1", "darkseagreen", "darkorchid", "darkolivegreen1", "brown1", "darkorange1", "cyan1", "darkgrey")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=4)) ``` ## Bar plots in family level ```{r, echo=TRUE, warning=FALSE} plot_bar(ps.ng.tax_most_copied, fill="Family") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = 5, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=8)) ``` Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth. ```{r, echo=TRUE, warning=FALSE} plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Family") + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = theme.size, colour="black")) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=8)) ``` \pagebreak Use color according to family. Do separate panels Stroke and Sex_age. ```{r, echo=TRUE, warning=FALSE} #-- If existing replicates, to be processed as follows -- plot_bar(ps.ng.tax_most_, x="Family", fill="Family", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") + scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = 5, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=8)) ``` ```{r, echo=FALSE, warning=FALSE} #MOVE_FROM_ABOVE: ## Bar plots in genus level #MOVE_FROM_ABOVE: Regroup together pre vs post stroke samples and normalize number of reads in each group using median sequencing depth. #plot_bar(ps.ng.tax_most_pre_post_stroke_, fill="Genus") + geom_bar(aes(), stat="identity", position="stack") + #scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = theme.size, colour="black")) + theme(legend.position="bottom") ``` \pagebreak ```{r, echo=FALSE, warning=FALSE} #MOVE_FROM_ABOVE: Use color according to genus. Do separate panels Stroke and Sex_age. ##-- If existing replicates, to be processed as follows -- #plot_bar(ps.ng.tax_most_, x="Genus", fill="Genus", facet_grid = pre_post_stroke~Sex_age) + geom_bar(aes(), stat="identity", position="stack") + #scale_fill_manual(values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#FFFFFF", "#FFFF00", "#00FFFF", "#FFA500", "#00FF00", "#808080", "#FF00FF", "#800080", "#FDD017", "#0000A0", "#3BB9FF", "#008000", "#800000", "#ADD8E6", "#F778A1", "#800517", "#736F6E", "#F52887", "#C11B17", "#5CB3FF", "#A52A2A", "#FF8040", "#2B60DE", "#736AFF", "#1589FF", "#98AFC7", "#8D38C9", "#307D7E", "#F6358A", "#151B54", "#6D7B8D", "#FDEEF4", "#FF0080", "#F88017", "#2554C7", "#FFF8C6", "#D4A017", "#306EFF", "#151B8D", "#9E7BFF", "#EAC117", "#E0FFFF", "#15317E", "#6C2DC7", "#FBB917", "#FCDFFF", "#15317E", "#254117", "#FAAFBE", "#357EC7")) + theme(axis.text = element_text(size = 6, colour="black"), axis.text.x=element_blank(), axis.ticks=element_blank()) + theme(legend.position="bottom") + guides(fill=guide_legend(nrow=18)) ``` \pagebreak # Alpha diversity Plot Chao1 richness estimator, Observed OTUs, Shannon index, and Phylogenetic diversity. Regroup together samples from the same group. ```{r, echo=FALSE, warning=FALSE} # using rarefied data #FITTING2: CONSOLE: #gunzip table_even4753.biom.gz #alpha_diversity.py -i table_even42369.biom --metrics chao1,observed_otus,shannon,PD_whole_tree -o adiv_even.txt -t ../clustering/rep_set.tre #gunzip table_even4753.biom.gz #alpha_diversity.py -i table_even4753.biom --metrics chao1,observed_otus,shannon,PD_whole_tree -o adiv_even.txt -t ../clustering_stool/rep_set.tre #gunzip table_even4753.biom.gz #alpha_diversity.py -i table_even4753.biom --metrics chao1,observed_otus,shannon,PD_whole_tree -o adiv_even.txt -t ../clustering_swab/rep_set.tre ``` ```{r, echo=TRUE, warning=FALSE} hmp.div_qiime <- read.csv("adiv_even.txt", sep="\t") colnames(hmp.div_qiime) <- c("sam_name", "chao1", "observed_otus", "shannon", "PD_whole_tree") row.names(hmp.div_qiime) <- hmp.div_qiime$sam_name div.df <- merge(hmp.div_qiime, hmp.meta, by = "sam_name") div.df2 <- div.df[, c("Group", "chao1", "shannon", "observed_otus", "PD_whole_tree")] colnames(div.df2) <- c("Group", "Chao-1", "Shannon", "OTU", "Phylogenetic Diversity") #colnames(div.df2) options(max.print=999999) #27 H47 830.5000 5.008482 319 10.60177 #FITTING4: if occuring "Computation failed in `stat_signif()`:not enough 'y' observations" #means: the patient H47 contains only one sample, it should be removed for the statistical p-values calculations. #delete H47(1) #div.df2 <- div.df2[-c(3), ] #div.df2 <- div.df2[-c(55,54, 45,40,39,27,26,25,1), ] knitr::kable(div.df2) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) #https://uc-r.github.io/t_test #We can perform the test with t.test and transform our data and we can also perform the nonparametric test with the wilcox.test function. stat.test.Shannon <- compare_means( Shannon ~ Group, data = div.df2, method = "t.test" ) knitr::kable(stat.test.Shannon) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) div_df_melt <- reshape2::melt(div.df2) #head(div_df_melt) #https://plot.ly/r/box-plots/#horizontal-boxplot #http://www.sthda.com/english/wiki/print.php?id=177 #https://rpkgs.datanovia.com/ggpubr/reference/as_ggplot.html #http://www.sthda.com/english/articles/24-ggpubr-publication-ready-plots/82-ggplot2-easy-way-to-change-graphical-parameters/ #https://plot.ly/r/box-plots/#horizontal-boxplot #library("gridExtra") #par(mfrow=c(4,1)) p <- ggboxplot(div_df_melt, x = "Group", y = "value", facet.by = "variable", scales = "free", width = 0.5, fill = "gray", legend= "right") #ggpar(p, xlab = FALSE, ylab = FALSE) lev <- levels(factor(div_df_melt$Group)) # get the variables #FITTING4: delete H47(1) in lev #lev <- lev[-c(3)] # make a pairwise list that we want to compare. #my_stat_compare_means #https://stackoverflow.com/questions/47839988/indicating-significance-with-ggplot2-in-a-boxplot-with-multiple-groups L.pairs <- combn(seq_along(lev), 2, simplify = FALSE, FUN = function(i) lev[i]) #%>% filter(p.signif != "ns") my_stat_compare_means <- function (mapping = NULL, data = NULL, method = NULL, paired = FALSE, method.args = list(), ref.group = NULL, comparisons = NULL, hide.ns = FALSE, label.sep = ", ", label = NULL, label.x.npc = "left", label.y.npc = "top", label.x = NULL, label.y = NULL, tip.length = 0.03, symnum.args = list(), geom = "text", position = "identity", na.rm = FALSE, show.legend = NA, inherit.aes = TRUE, ...) { if (!is.null(comparisons)) { method.info <- ggpubr:::.method_info(method) method <- method.info$method method.args <- ggpubr:::.add_item(method.args, paired = paired) if (method == "wilcox.test") method.args$exact <- FALSE pms <- list(...) size <- ifelse(is.null(pms$size), 0.3, pms$size) color <- ifelse(is.null(pms$color), "black", pms$color) map_signif_level <- FALSE if (is.null(label)) label <- "p.format" if (ggpubr:::.is_p.signif_in_mapping(mapping) | (label %in% "p.signif")) { if (ggpubr:::.is_empty(symnum.args)) { map_signif_level <- c(`****` = 1e-04, `***` = 0.001, `**` = 0.01, `*` = 0.05, ns = 1) } else { map_signif_level <- symnum.args } if (hide.ns) names(map_signif_level)[5] <- " " } step_increase <- ifelse(is.null(label.y), 0.12, 0) ggsignif::geom_signif(comparisons = comparisons, y_position = label.y, test = method, test.args = method.args, step_increase = step_increase, size = size, color = color, map_signif_level = map_signif_level, tip_length = tip.length, data = data) } else { mapping <- ggpubr:::.update_mapping(mapping, label) layer(stat = StatCompareMeans, data = data, mapping = mapping, geom = geom, position = position, show.legend = show.legend, inherit.aes = inherit.aes, params = list(label.x.npc = label.x.npc, label.y.npc = label.y.npc, label.x = label.x, label.y = label.y, label.sep = label.sep, method = method, method.args = method.args, paired = paired, ref.group = ref.group, symnum.args = symnum.args, hide.ns = hide.ns, na.rm = na.rm, ...)) } } p2 <- p + stat_compare_means( method="t.test", comparisons = list(c("Group1", "Group2"), c("Group1", "Group3"), c("Group1", "Group4"), c("Group1", "Group6"), c("Group1", "Group8"), c("Group2", "Group5"),c("Group4", "Group5"),c("Group4", "Group6"),c("Group4", "Group7"),c("Group6", "Group7")), label = "p.signif", symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")) ) #comparisons = L.pairs, #symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05), symbols = c("****", "***", "**", "*")), #stat_pvalue_manual #print(p2) #https://stackoverflow.com/questions/20500706/saving-multiple-ggplots-from-ls-into-one-and-separate-files-in-r #FITTING3: mkdir figures ggsave("./figures/alpha_diversity_Group.png", device="png", height = 10, width = 12) ggsave("./figures/alpha_diversity_Group.svg", device="svg", height = 10, width = 12) p3 <- p + stat_compare_means( method="t.test", comparisons = list(c("Group2", "Group4"), c("Group2", "Group6"), c("Group4", "Group8"), c("Group6", "Group8")), label = "p.signif", symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")), ) #symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05), symbols = c("****", "***", "**", "*")), #stat_pvalue_manual #print(p2) #https://stackoverflow.com/questions/20500706/saving-multiple-ggplots-from-ls-into-one-and-separate-files-in-r #FITTING3: mkdir figures ggsave("./figures/alpha_diversity_Group2.png", device="png", height = 10, width = 12) ggsave("./figures/alpha_diversity_Group2.svg", device="svg", height = 10, width = 12) ``` # Selected alpha diversity ```{r, echo=TRUE, warning=FALSE, fig.cap="Alpha diversity", out.width = '100%', fig.align= "center"} knitr::include_graphics("./figures/alpha_diversity_Group2.png") selected_alpha_diversities<-read.csv("selected_alpha_diversities.txt",sep="\t") knitr::kable(selected_alpha_diversities) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) ``` # Beta diversity ```{r, echo=TRUE, warning=FALSE, fig.cap="Beta diversity", out.width = '100%', fig.align= "center"} #file:///home/jhuang/DATA/Data_Marius_16S/core_diversity_e42369/bdiv_even42369_Group/unweighted_unifrac_boxplots/Group_Stats.txt beta_diversity_group_stats<-read.csv("unweighted_unifrac_boxplots_Group_Stats.txt",sep="\t") knitr::kable(beta_diversity_group_stats) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) #NOTE: Run this Phyloseq0.Rmd, then run the code of MicrobiotaProcess.R to manually generate PCoA.png, then run this Phyloseq.Rmd! #NOTE: AT_FIRST_DEACTIVATE_THIS_LINE: knitr::include_graphics("./figures/PCoA.png") ``` # Differential abundance analysis Differential abundance analysis aims to find the differences in the abundance of each taxa between two groups of samples, assigning a significance value to each comparison. ## Group2 vs Group4 ```{r, echo=TRUE, warning=FALSE} library("DESeq2") #ALTERNATIVE using ps.ng.tax_most_copied: ps.ng.tax (40594) vs. ps.ng.tax_most_copied (166) ps.ng.tax_sel <- ps.ng.tax #FITTING5: correct the id of the group members, see FITTING6 otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("8","9","10","12","13","14", "21","22","23","24","25","26","27","28")] diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group) diagdds$Group <- relevel(diagdds$Group, "Group4") diagdds = DESeq(diagdds, test="Wald", fitType="parametric") resultsNames(diagdds) res = results(diagdds, cooksCutoff = FALSE) alpha = 0.05 sigtab = res[which(res$padj < alpha), ] sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix")) sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ] kable(sigtab) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) #rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)) #subv %in% v ### returns a vector TRUE FALSE #is.element(subv, v) ### returns a vector TRUE FALSE library("ggplot2") theme_set(theme_bw()) scale_fill_discrete <- function(palname = "Set1", ...) { scale_fill_brewer(palette = palname, ...) } x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x)) x = sort(x) sigtab$Order = factor(as.character(sigtab$Order), levels=names(x)) x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x)) x = sort(x) sigtab$Family = factor(as.character(sigtab$Family), levels=names(x)) ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+ theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5)) #Error in checkForExperimentalReplicates(object, modelMatrix) : # The design matrix has the same number of samples and coefficients to fit, # so estimation of dispersion is not possible. Treating samples # as replicates was deprecated in v1.20 and no longer supported since v1.22. ``` ## Group2 vs Group6 ```{r, echo=TRUE, warning=FALSE} ps.ng.tax_sel <- ps.ng.tax otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("8","9","10","12","13","14", "33","34","35","36","37","38","39","51")] diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group) diagdds$Group <- relevel(diagdds$Group, "Group6") diagdds = DESeq(diagdds, test="Wald", fitType="parametric") resultsNames(diagdds) res = results(diagdds, cooksCutoff = FALSE) alpha = 0.05 sigtab = res[which(res$padj < alpha), ] sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix")) sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ] kable(sigtab) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) library("ggplot2") theme_set(theme_bw()) scale_fill_discrete <- function(palname = "Set1", ...) { scale_fill_brewer(palette = palname, ...) } x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x)) x = sort(x) sigtab$Order = factor(as.character(sigtab$Order), levels=names(x)) x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x)) x = sort(x) sigtab$Family = factor(as.character(sigtab$Family), levels=names(x)) ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+ theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5)) ``` ## Group4 vs Group8 ```{r, echo=TRUE, warning=FALSE} ps.ng.tax_sel <- ps.ng.tax otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("21","22","23","24","25","26","27","28", "47","48","49","50","52","53","55")] diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group) diagdds$Group <- relevel(diagdds$Group, "Group8") diagdds = DESeq(diagdds, test="Wald", fitType="parametric") resultsNames(diagdds) res = results(diagdds, cooksCutoff = FALSE) alpha = 0.05 sigtab = res[which(res$padj < alpha), ] sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix")) sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ] kable(sigtab) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) library("ggplot2") theme_set(theme_bw()) scale_fill_discrete <- function(palname = "Set1", ...) { scale_fill_brewer(palette = palname, ...) } x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x)) x = sort(x) sigtab$Order = factor(as.character(sigtab$Order), levels=names(x)) x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x)) x = sort(x) sigtab$Family = factor(as.character(sigtab$Family), levels=names(x)) ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+ theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5)) ``` ## Group6 vs Group8 ```{r, echo=TRUE, warning=FALSE} ps.ng.tax_sel <- ps.ng.tax otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")] diagdds = phyloseq_to_deseq2(ps.ng.tax_sel, ~Group) diagdds$Group <- relevel(diagdds$Group, "Group8") diagdds = DESeq(diagdds, test="Wald", fitType="parametric") resultsNames(diagdds) res = results(diagdds, cooksCutoff = FALSE) alpha = 0.05 sigtab = res[which(res$padj < alpha), ] sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(ps.ng.tax_sel)[rownames(sigtab), ], "matrix")) sigtab <- sigtab[rownames(sigtab) %in% rownames(tax_table(ps.ng.tax_most_copied)), ] kable(sigtab) %>% kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive")) library("ggplot2") theme_set(theme_bw()) scale_fill_discrete <- function(palname = "Set1", ...) { scale_fill_brewer(palette = palname, ...) } x = tapply(sigtab$log2FoldChange, sigtab$Order, function(x) max(x)) x = sort(x) sigtab$Order = factor(as.character(sigtab$Order), levels=names(x)) x = tapply(sigtab$log2FoldChange, sigtab$Family, function(x) max(x)) x = sort(x) sigtab$Family = factor(as.character(sigtab$Family), levels=names(x)) ggplot(sigtab, aes(x=log2FoldChange, y=Family, color=Order)) + geom_point(aes(size=padj)) + scale_size_continuous(name="padj",range=c(8,4))+ theme(axis.text.x = element_text(angle = -25, hjust = 0, vjust=0.5)) ``` -
MicrobiotaProcess.R
# ----------------------------------- # ---- prepare the R environment ---- #Rscript MicrobiotaProcess.R #NOTE: exit R script, then login again R-environment; rm -rf Phyloseq*_cache rmarkdown::render('Phyloseq.Rmd',output_file='Phyloseq.html') # ----------------------------- # ---- 3.1. bridges other tools ##https://github.com/YuLab-SMU/MicrobiotaProcess ##https://www.bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc/MicrobiotaProcess.html ##https://chiliubio.github.io/microeco_tutorial/intro.html#framework ##https://yiluheihei.github.io/microbiomeMarker/reference/plot_cladogram.html #BiocManager::install("MicrobiotaProcess") #install.packages("microeco") #install.packages("ggalluvial") #install.packages("ggh4x") library(MicrobiotaProcess) library(microeco) library(ggalluvial) library(ggh4x) library(gghalves) ## Convert the phyloseq object to a MicrobiotaProcess object #mp <- as.MicrobiotaProcess(ps.ng.tax) #mt <- phyloseq2microeco(ps.ng.tax) #--> ERROR #abundance_table <- mt$abun_table #taxonomy_table <- mt$tax_table #ps.ng.tax_abund <- phyloseq::filter_taxa(ps.ng.tax, function(x) sum(x > total*0.01) > 0, TRUE) #ps.ng.tax_most = phyloseq::filter_taxa(ps.ng.tax_rel, function(x) mean(x) > 0.001, TRUE) ##OPTION1: take all samples, prepare mpse_abund! ##mpse <- ps.ng.tax %>% as.MPSE() #mpse_abund <- ps.ng.tax_abund %>% as.MPSE() ##OPTION2: take partial samples, prepare mpse_abund ps.ng.tax_sel <- ps.ng.tax #IMPORTANT ##otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("1","2","5","6","7", "15","16","17","18","19","20", "29","30","31","32", "40","41","42","43","44","46")] ##NOTE: Only choose Group2, Group4, Group6, Group8 #> ps.ng.tax_sel #otu_table() OTU Table: [ 37465 taxa and 29 samples ] #sample_data() Sample Data: [ 29 samples by 10 sample variables ] #tax_table() Taxonomy Table: [ 37465 taxa by 7 taxonomic ranks ] #phy_tree() Phylogenetic Tree: [ 37465 tips and 37461 internal nodes ] otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")] #For quick calculation #otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax_abund)[,c("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")] mpse_abund <- ps.ng.tax_sel %>% as.MPSE() # ----------------------------------- # ---- 3.2. alpha diversity analysis # Rarefied species richness + RareAbundance mpse_abund %<>% mp_rrarefy() # 'chunks' represent the split number of each sample to calculate alpha # diversity, default is 400. e.g. If a sample has total 40000 # reads, if chunks is 400, it will be split to 100 sub-samples # (100, 200, 300,..., 40000), then alpha diversity index was # calculated based on the sub-samples. # '.abundance' the column name of abundance, if the '.abundance' is not be # rarefied calculate rarecurve, user can specific 'force=TRUE'. # + RareAbundance mpse_abund %<>% mp_cal_rarecurve( .abundance = RareAbundance, chunks = 400 ) # The RareAbundanceRarecurve column will be added the colData slot # automatically (default action="add") mpse_abund %>% print(width=180, n=100) # default will display the confidence interval around smooth. # se=TRUE p1 <- mpse_abund %>% mp_plot_rarecurve( .rare = RareAbundanceRarecurve, .alpha = Observe, ) p2 <- mpse_abund %>% mp_plot_rarecurve( .rare = RareAbundanceRarecurve, .alpha = Observe, .group = pre_post_stroke ) + scale_color_manual(values=c("#00A087FF", "#3C5488FF")) + scale_fill_manual(values=c("#00A087FF", "#3C5488FF"), guide="none") # combine the samples belong to the same groups if plot.group=TRUE p3 <- mpse_abund %>% mp_plot_rarecurve( .rare = RareAbundanceRarecurve, .alpha = "Observe", .group = pre_post_stroke, plot.group = TRUE ) + scale_color_manual(values=c("#00A087FF", "#3C5488FF")) + scale_fill_manual(values=c("#00A087FF", "#3C5488FF"),guide="none") png("rarefaction_of_samples_or_groups.png", width=1080, height=600) p1 + p2 + p3 dev.off() # ------------------------------------------ # 3.3. calculate alpha index and visualization library(ggplot2) library(MicrobiotaProcess) mpse_abund %<>% mp_cal_alpha(.abundance=RareAbundance) mpse_abund f1 <- mpse_abund %>% mp_plot_alpha( .group=pre_post_stroke, .alpha=c(Observe, Chao1, ACE, Shannon, Simpson, Pielou) ) + scale_fill_manual(values=c("#00A087FF", "#3C5488FF"), guide="none") + scale_color_manual(values=c("#00A087FF", "#3C5488FF"), guide="none") f2 <- mpse_abund %>% mp_plot_alpha( .alpha=c(Observe, Chao1, ACE, Shannon, Simpson, Pielou) ) png("alpha_diversity_comparison.png", width=1000, height=1000) f1 / f2 dev.off() # ------------------------------------------- # 3.4. The visualization of taxonomy abundance (Class) mpse_abund %<>% mp_cal_abundance( # for each samples .abundance = RareAbundance ) %>% mp_cal_abundance( # for each groups .abundance=RareAbundance, .group=pre_post_stroke ) mpse_abund # visualize the relative abundance of top 20 phyla for each sample. p1 <- mpse_abund %>% mp_plot_abundance( .abundance=RareAbundance, .group=time, taxa.class = Class, topn = 20, relative = TRUE ) # visualize the abundance (rarefied) of top 20 phyla for each sample. p2 <- mpse_abund %>% mp_plot_abundance( .abundance=RareAbundance, .group=time, taxa.class = Class, topn = 20, relative = FALSE ) png("relative_abundance_and_abundance.png", width= 1200, height=600) #NOT PRODUCED! p1 / p2 dev.off() h1 <- mpse_abund %>% mp_plot_abundance( .abundance = RareAbundance, .group = pre_post_stroke, taxa.class = Class, relative = TRUE, topn = 20, geom = 'heatmap', features.dist = 'euclidean', features.hclust = 'average', sample.dist = 'bray', sample.hclust = 'average' ) h2 <- mpse_abund %>% mp_plot_abundance( .abundance = RareAbundance, .group = pre_post_stroke, taxa.class = Class, relative = FALSE, topn = 20, geom = 'heatmap', features.dist = 'euclidean', features.hclust = 'average', sample.dist = 'bray', sample.hclust = 'average' ) # the character (scale or theme) of figure can be adjusted by set_scale_theme # refer to the mp_plot_dist png("relative_abundance_and_abundance_heatmap.png", width= 1200, height=600) aplot::plot_list(gglist=list(h1, h2), tag_levels="A") dev.off() # visualize the relative abundance of top 20 class for each .group (pre_post_stroke) p3 <- mpse_abund %>% mp_plot_abundance( .abundance=RareAbundance, .group=pre_post_stroke, taxa.class = Class, topn = 20, plot.group = TRUE ) # visualize the abundance of top 20 phyla for each .group (time) p4 <- mpse_abund %>% mp_plot_abundance( .abundance=RareAbundance, .group= pre_post_stroke, taxa.class = Class, topn = 20, relative = FALSE, plot.group = TRUE ) png("relative_abundance_and_abundance_groups.png", width= 1000, height=1000) p3 / p4 dev.off() # --------------------------- # 3.5. Beta diversity analysis # --------------------------------------------- # 3.5.1 The distance between samples or groups # standardization # mp_decostand wraps the decostand of vegan, which provides # many standardization methods for community ecology. # default is hellinger, then the abundance processed will # be stored to the assays slot. mpse_abund %<>% mp_decostand(.abundance=Abundance) mpse_abund # calculate the distance between the samples. # the distance will be generated a nested tibble and added to the # colData slot. mpse_abund %<>% mp_cal_dist(.abundance=hellinger, distmethod="bray") mpse_abund # mp_plot_dist provides there methods to visualize the distance between the samples or groups # when .group is not provided, the dot heatmap plot will be return p1 <- mpse_abund %>% mp_plot_dist(.distmethod = bray) png("distance_between_samples.png", width= 1000, height=1000) p1 dev.off() # when .group is provided, the dot heatmap plot with group information will be return. p2 <- mpse_abund %>% mp_plot_dist(.distmethod = bray, .group = pre_post_stroke) # The scale or theme of dot heatmap plot can be adjusted using set_scale_theme function. p2 %>% set_scale_theme( x = scale_fill_manual( values=c("orange", "deepskyblue"), guide = guide_legend( keywidth = 1, keyheight = 0.5, title.theme = element_text(size=8), label.theme = element_text(size=6) ) ), aes_var = pre_post_stroke # specific the name of variable ) %>% set_scale_theme( x = scale_color_gradient( guide = guide_legend(keywidth = 0.5, keyheight = 0.5) ), aes_var = bray ) %>% set_scale_theme( x = scale_size_continuous( range = c(0.1, 3), guide = guide_legend(keywidth = 0.5, keyheight = 0.5) ), aes_var = bray ) png("distance_between_samples_with_group_info.png", width= 1000, height=1000) p2 dev.off() # when .group is provided and group.test is TRUE, the comparison of different groups will be returned p3 <- mpse_abund %>% mp_plot_dist(.distmethod = bray, .group = pre_post_stroke, group.test=TRUE, textsize=2) png("comparison_of_distance.png", width= 1000, height=1000) p3 dev.off() # ----------------------- # 3.5.2 The PCoA analysis #install.packages("corrr") library(corrr) #install.packages("ggside") library(ggside) mpse_abund %<>% mp_cal_pcoa(.abundance=hellinger, distmethod="bray") # The dimensions of ordination analysis will be added the colData slot (default). mpse_abund #> methods(class=class(mpse_abund)) # [1] [ [[<- [<- # [4] $ $<- arrange # [7] as_tibble as.data.frame as.phyloseq #[10] coerce coerce<- colData<- #[13] distinct filter group_by #[16] left_join mp_adonis mp_aggregate_clade #[19] mp_aggregate mp_anosim mp_balance_clade #[22] mp_cal_abundance mp_cal_alpha mp_cal_cca #[25] mp_cal_clust mp_cal_dca mp_cal_dist #[28] mp_cal_nmds mp_cal_pca mp_cal_pcoa #[31] mp_cal_pd_metric mp_cal_rarecurve mp_cal_rda #[34] mp_cal_upset mp_cal_venn mp_decostand #[37] mp_diff_analysis mp_diff_clade mp_envfit #[40] mp_extract_abundance mp_extract_assays mp_extract_dist #[43] mp_extract_feature mp_extract_internal_attr mp_extract_rarecurve #[46] mp_extract_refseq mp_extract_sample mp_extract_taxonomy #[49] mp_extract_tree mp_filter_taxa mp_mantel #[52] mp_mrpp mp_plot_abundance mp_plot_alpha #[55] mp_plot_diff_boxplot mp_plot_diff_res mp_plot_dist #[58] mp_plot_ord mp_plot_rarecurve mp_plot_upset #[61] mp_plot_venn mp_rrarefy mp_select_as_tip #[64] mp_stat_taxa mutate otutree #[67] otutree<- print pull #[70] refsequence refsequence<- rename #[73] rownames<- select show # [ reached getOption("max.print") -- omitted 6 entries ] #see '?methods' for accessing help and source code # We also can perform adonis or anosim to check whether it is significant to the dissimilarities of groups. mpse_abund %<>% mp_adonis(.abundance=hellinger, .formula=~Group, distmethod="bray", permutations=9999, action="add") mpse_abund %>% mp_extract_internal_attr(name=adonis) # ("1","2","5","6","7", "15","16","17","18","19","20", "29","30","31","32", "40","41","42","43","44","46") #div.df2[div.df2 == "Group1"] <- "aged.post" #div.df2[div.df2 == "Group3"] <- "young.post" #div.df2[div.df2 == "Group5"] <- "aged.post" #div.df2[div.df2 == "Group7"] <- "young.post" # ("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55") #div.df2[div.df2 == "Group2"] <- "aged.pre" #div.df2[div.df2 == "Group4"] <- "young.pre" #div.df2[div.df2 == "Group6"] <- "aged.pre" #div.df2[div.df2 == "Group8"] <- "young.pre" #Group1: f.aged and post #Group2: f.aged and pre #Group3: f.young and post #Group4: f.young and pre #Group5: m.aged and post #Group6: m.aged and pre #Group7: m.young and post #Group8: m.young and pre #[,c("1","2","5","6","7", "8","9","10","12","13","14")] #[,c("15","16","17","18","19","20", "21","22","23","24","25","26","27","28")] #[,c("29","30","31","32", "33","34","35","36","37","38","39","51")] #[,c("40","41","42","43","44","46", "47","48","49","50","52","53","55")] p1 <- mpse_abund %>% mp_plot_ord( .ord = pcoa, .group = Group, .color = Group, .size = 2.4, .alpha = 1, ellipse = TRUE, show.legend = FALSE # don't display the legend of stat_ellipse ) + scale_fill_manual( #values = c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#cab2d6", "#6a3d9a"), #values = c("#a6cee3", "#b2df8a", "#fb9a99", "#cab2d6"), values = c("#1f78b4", "#33a02c", "#e31a1c", "#6a3d9a"), guide = guide_legend(keywidth=1.6, keyheight=1.6, label.theme=element_text(size=12)) ) + scale_color_manual( #values=c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#cab2d6", "#6a3d9a"), #values = c("#a6cee3", "#b2df8a", "#fb9a99", "#cab2d6"), values = c("#1f78b4", "#33a02c", "#e31a1c", "#6a3d9a"), guide = guide_legend(keywidth=1.6, keyheight=1.6, label.theme=element_text(size=12)) ) #scale_fill_manual(values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500")) + #scale_color_manual(values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500")) #scale_fill_manual(values=c("#00A087FF", "#3C5488FF")) + #scale_color_manual(values=c("#00A087FF", "#3C5488FF")) #png("PCoA.png", width= 1000, height=1000) svg("PCoA.svg", width= 11, height=10) #svg("PCoA_.svg", width=10, height=10) #svg("PCoA.svg") pdf("PCoA.pdf") p1 dev.off() # The size of point also can be mapped to other variables such as Observe, or Shannon # Then the alpha diversity and beta diversity will be displayed simultaneously. p2 <- mpse_abund %>% mp_plot_ord( .ord = pcoa, .group = Group, .color = Group, .size = Shannon, .alpha = Observe, ellipse = TRUE, show.legend = FALSE # don't display the legend of stat_ellipse ) + scale_fill_manual( values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"), guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8)) ) + scale_color_manual( values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"), guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8)) ) + scale_size_continuous( range=c(0.5, 3), guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8)) ) # ------------------------------------------ # 3.5.3 Hierarchical cluster (tree) analysis #input should contain hellinger! mpse_abund %<>% mp_cal_clust( .abundance = hellinger, distmethod = "bray", hclustmethod = "average", # (UPGAE) action = "add" # action is used to control which result will be returned ) mpse_abund # if action = 'add', the result of hierarchical cluster will be added to the MPSE object # mp_extract_internal_attr can extract it. It is a treedata object, so it can be visualized # by ggtree. sample.clust <- mpse_abund %>% mp_extract_internal_attr(name='SampleClust') sample.clust library(ggtree) p_cluster <- ggtree(sample.clust) + geom_tippoint(aes(color=pre_post_stroke)) + geom_tiplab(as_ylab = TRUE) + ggplot2::scale_x_continuous(expand=c(0, 0.01)) png("hierarchical_cluster1.png", width= 1000, height=800) p_cluster dev.off() library(ggtreeExtra) library(ggplot2) # # Extract relative abundance of phyla # phyla.tb <- mouse.time.mpse %>% # mp_extract_abundance(taxa.class=Phylum, topn=30) # # The abundance of each samples is nested, it can be flatted using the unnest of tidyr. # phyla.tb %<>% tidyr::unnest(cols=RareAbundanceBySample) %>% dplyr::rename(Phyla="label") # phyla.tb # # p1 <- p + # geom_fruit( # data=phyla.tb, # geom=geom_col, # mapping = aes(x = RelRareAbundanceBySample, # y = Sample, # fill = Phyla # ), # orientation = "y", # #offset = 0.4, # pwidth = 3, # axis.params = list(axis = "x", # title = "The relative abundance of phyla (%)", # title.size = 4, # text.size = 2, # vjust = 1), # grid.params = list() # ) # png("hierarchical_cluster2.png", width = 1000, height = 800) # p1 # dev.off() # Extract relative abundance of classes mpse_abund %>% print(width=150) class.tb <- mpse_abund %>% mp_extract_abundance(taxa.class = Class, topn = 30) # Flatten and rename the columns class.tb %<>% tidyr::unnest(cols = RareAbundanceBySample) %>% dplyr::rename(Class = "label") # View the data frame class.tb # Create the plot p1 <- p + geom_fruit( data = class.tb, geom = geom_col, mapping = aes(x = RelRareAbundanceBySample, y = Sample, fill = Class ), orientation = "y", pwidth = 3, axis.params = list(axis = "x", title = "The relative abundance of classes (%)", title.size = 4, text.size = 2, vjust = 1), grid.params = list() ) # Save the plot to a file png("hierarchical_cluster2.png", width = 1000, height = 800) print(p1) dev.off() # ----------------------- # 3.6 Biomarker discovery library(ggtree) library(ggtreeExtra) library(ggplot2) library(MicrobiotaProcess) library(tidytree) library(ggstar) library(forcats) mpse_abund %>% print(width=150) mpse_abund %<>% mp_cal_abundance( # for each samples .abundance = RareAbundance ) %>% mp_cal_abundance( # for each groups .abundance=RareAbundance, .group=pre_post_stroke ) mpse_abund mpse_abund %<>% mp_diff_analysis( .abundance = RelRareAbundanceBySample, .group = pre_post_stroke, first.test.alpha = 0.01 ) # The result is stored to the taxatree or otutree slot, you can use mp_extract_tree to extract the specific slot. taxa.tree <- mpse_abund %>% mp_extract_tree(type="taxatree") taxa.tree ## And the result tibble of different analysis can also be extracted ## with tidytree (>=0.3.5) taxa.tree %>% select(label, nodeClass, LDAupper, LDAmean, LDAlower, Sign_pre_post_stroke, pvalue, fdr) %>% dplyr::filter(!is.na(fdr)) taxa.tree %>% print(width=150, n=100) #.data, layout, tree.type, .taxa.class, tiplab.size, offset.abun, pwidth.abun, offset.effsize, pwidth.effsize, group.abun, tiplab.linetype p <- mpse_abund %>% mp_plot_diff_res( group.abun = TRUE, pwidth.abun=0.05, offset.abun=0.02, pwidth.effsize=0.3, offset.effsize=0.46, tiplab.size = 4.9 ) + scale_fill_manual(values=c("deepskyblue", "orange")) + scale_fill_manual( aesthetics = "fill_new", # The fill aes was renamed to "fill_new" for the abundance dotplot layer values = c("deepskyblue", "orange") ) + scale_fill_manual( aesthetics = "fill_new_new", # The fill aes for hight light layer of tree was renamed to 'fill_new_new' values = c("#E41A1C", "#377EB8", "#4DAF4A", "#984EA3", "#FF7F00", "#FFFF33", "#A65628", "#F781BF", "#00FFFF", "#999999" ) ) + theme( axis.title = element_text(size = 28), # Font size for axis titles axis.text = element_text(size = 28), # Font size for axis text plot.title = element_text(size = 28), # Font size for plot title legend.title = element_text(size = 16), # Font size for legend title legend.text = element_text(size = 14) # Font size for legend text ) #p$layers[[2]]$geom <- geom_tiplab(fontsize = 22) # Change 12 to the desired font size png("differently_expressed_otu.png", width=2000, height=2000) #svg("p7.svg",width=8, height=8) p dev.off() f <- mpse_abund %>% mp_plot_diff_cladogram( label.size = 2.5, hilight.alpha = .3, bg.tree.size = .5, bg.point.size = 2, bg.point.stroke = .25 ) + scale_fill_diff_cladogram( # set the color of different group. values = c('deepskyblue', 'orange') ) + scale_size_continuous(range = c(1, 4)) #png("cladogram.png", width=1000, height=1000) svg("cladogram.svg", width=10, height=10) f dev.off() ## Extract the OTU table and taxonomy table from the phyloseq object #otu_table <- phyloseq::otu_table(ps.ng.tax_abund) %>% as.data.frame() %>% as.matrix() #tax_table <- phyloseq::tax_table(ps.ng.tax_abund) %>% as.data.frame() %>% as.matrix() #write.csv(otu_table, file="otu_table.csv") #write.csv(tax_table, file="tax_table.csv") #~/Tools/csv2xls-0.4/csv_to_xls.py otu_table.csv tax_table.csv -d',' -o otu_tax.xls
3.1. Environment Setup: It sets up a Conda environment named picrust2, using the conda create command and then activates this environment using conda activate picrust2.
#https://github.com/picrust/picrust2/wiki/PICRUSt2-Tutorial-(v2.2.0-beta)#minimum-requirements-to-run-full-tutorial
conda create -n picrust2 -c bioconda -c conda-forge picrust2=2.2.0_b
conda activate picrust23.2. Data Preparation: The script creates a new directory called picrust2_out, then enters it using mkdir and cd commands. It then identifies input files that are needed for the analysis: metadata.tsv, seqs.fna, table.biom. The biom commands are used to inspect and convert the BIOM format files.
mkdir picrust2_out
cd picrust2_out
# Identifying input data
# Note: Replace the paths and filenames with your actual data if different
# metadata.tsv == ../map_corrected.txt
# seqs.fna == ../clustering/seqs.fna
# table.biom == ../core_diversity_e42369/table_even42369.biom
# Inspect and convert the BIOM format files
biom head -i ../core_diversity_e42369/table_even42369.biom
biom summarize-table -i ../core_diversity_e42369/table_even42369.biom
biom convert -i ../core_diversity_e42369/table_even42369.biom -o table_even42369.tsv --to-tsv3.3. Running PiCRUST2: The place_seqs.py command aligns the input sequences to a reference tree. The hsp.py commands generate hidden state prediction for multiple functional categories.
#insert reads into reference tree using EPA-NG
cp ../clustering/rep_set.fna ./
grep ">" rep_set.fna | wc -l #44238
vim table_even42369.tsv #40596-2
samtools faidx rep_set.fna
cut -f1-1 table_even42369.tsv > table_even42369.id
#manually modify table_even42369.id by replacing "\n" with " >> seqs.fna\nsamtools faidx rep_set.fna "
run table_even42369.id
rm -rf intermediate/
place_seqs.py -s seqs.fna -o out.tre -p 64 --intermediate intermediate/place_seqs
#castor: Efficient Phylogenetics on Large Trees
#https://github.com/picrust/picrust2/wiki/Hidden-state-prediction
hsp.py -i 16S -t out.tre -o 16S_predicted_and_nsti.tsv.gz -p 100 -n
hsp.py -i COG -t out.tre -o COG_predicted.tsv.gz -p 100
hsp.py -i PFAM -t out.tre -o PFAM_predicted.tsv.gz -p 100
hsp.py -i KO -t out.tre -o KO_predicted.tsv.gz -p 100
hsp.py -i EC -t out.tre -o EC_predicted.tsv.gz -p 100
hsp.py -i TIGRFAM -t out.tre -o TIGRFAM_predicted.tsv.gz -p 100
hsp.py -i PHENO -t out.tre -o PHENO_predicted.tsv.gz -p 100
>In this table the predicted copy number of all Enzyme Classification (EC) numbers is shown for each ASV. The NSTI values per ASV are not in this table since we did not specify the -n option. EC numbers are a type of gene family defined based on the chemical reactions they catalyze. For instance, EC:1.1.1.1 corresponds to alcohol dehydrogenase. In this tutorial we are focusing on EC numbers since they can be used to infer MetaCyc pathway levels (see below).
zless -S EC_predicted.tsv.gz
sequence EC:1.1.1.1 EC:1.1.1.10 EC:1.1.1.100 ...
20e568023c10eaac834f1c110aacea18 2 0 3 ...
23fe12a325dfefcdb23447f43b6b896e 0 0 1 ...
288c8176059111c4c7fdfb0cd5afce64 1 0 1 ...
...
##Why import the tsv file to MyData?
#MyData <- read.csv(file="./COG_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 4598 e.g. COG5665
#MyData <- read.csv(file="./PFAM_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 11089 e.g. PF17225
#MyData <- read.csv(file="./KO_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 10543 e.g. K19791
#MyData <- read.csv(file="./EC_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 2913 e.g. EC.6.6.1.2
#MyData <- read.csv(file="./16S_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 1 e.g. X16S_rRNA_Count
#MyData <- read.csv(file="./TIGRFAM_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 4287 e.g. TIGR04571
#MyData <- read.csv(file="./PHENO_predicted.tsv", header=TRUE, sep="\t", row.names=1) #6806 41 e.g. Use_of_nitrate_as_electron_acceptor, Xylose_utilizing3.4. The metagenome_pipeline.py commands perform metagenomic prediction for several functional categories. Predicted gene families weighted by the relative abundance of ASVs in their community. In other words, we are interested in inferring the metagenomes of the communities.
#Generate metagenome predictions using EC numbers https://en.wikipedia.org/wiki/List_of_enzymes#Category:EC_1.1_(act_on_the_CH-OH_group_of_donors)
metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f COG_predicted.tsv.gz -o COG_metagenome_out --strat_out
metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f EC_predicted.tsv.gz -o EC_metagenome_out --strat_out
metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f KO_predicted.tsv.gz -o KO_metagenome_out --strat_out
metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f PFAM_predicted.tsv.gz -o PFAM_metagenome_out --strat_out
metagenome_pipeline.py -i ../core_diversity_e42369/table_even42369.biom -m 16S_predicted_and_nsti.tsv.gz -f TIGRFAM_predicted.tsv.gz -o TIGRFAM_metagenome_out --strat_out3.5. Pathway-level inference: By default this script infers MetaCyc pathway abundances based on EC number abundances, although different gene families and pathways can also be optionally specified. This script performs a number of steps by default, which are based on the approach implemented in HUMAnN2:
- Regroups EC numbers to MetaCyc reactions.
- Infers which MetaCyc pathways are present based on these reactions with MinPath.
-
Calculates and returns the abundance of pathways identified as present.
pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_contrib.tsv.gz -o pathways_out -p 15 #Note that the path of map files is under /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/pathway_mapfiles pathway_pipeline.py -i COG_metagenome_out/pred_metagenome_contrib.tsv.gz -o KEGG_pathways_out -p 15 --no_regroup --map /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv #Mapping predicted KO abundances to legacy KEGG pathways (with stratified output that represents contributions to community-wide abundances): pathway_pipeline.py -i KO_metagenome_out/pred_metagenome_strat.tsv.gz -o KEGG_pathways_out --no_regroup --map /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv #Map EC numbers to MetaCyc pathways and get stratified output corresponding to contribution of predicted gene family abundances within each predicted genome: pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -o pathways_out_per_seq --per_sequence_contrib --per_sequence_abun EC_metagenome_out/seqtab_norm.tsv.gz --per_sequence_function EC_predicted.tsv.gz
3.6. Add functional descriptions: Finally, it can be useful to have a description of each functional id in the output abundance tables. The below commands will add these descriptions as new column in gene family and pathway abundance tables
#--6.1. Add descriptions in gene family tables
add_descriptions.py -i COG_metagenome_out/pred_metagenome_unstrat.tsv.gz -m COG -o COG_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz
add_descriptions.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -m EC -o EC_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz
add_descriptions.py -i KO_metagenome_out/pred_metagenome_unstrat.tsv.gz -m KO -o KO_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz # EC and METACYC is a pair, EC for gene_annotation and METACYC for pathway_annotation
add_descriptions.py -i PFAM_metagenome_out/pred_metagenome_unstrat.tsv.gz -m PFAM -o PFAM_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz
add_descriptions.py -i TIGRFAM_metagenome_out/pred_metagenome_unstrat.tsv.gz -m TIGRFAM -o TIGRFAM_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz
#--6.2. Add descriptions in pathway abundance tables
add_descriptions.py -i pathways_out/path_abun_unstrat.tsv.gz -m METACYC -o pathways_out/path_abun_unstrat_descrip.tsv.gz
gunzip path_abun_unstrat_descrip.tsv.gz
#Error - no rows remain after regrouping input table. The default pathway and regroup mapfiles are meant for EC numbers. Note that KEGG pathways are not supported since KEGG is a closed-source database, but you can input custom pathway mapfiles if you have access. If you are using a custom function database did you mean to set the --no-regroup flag and/or change the default pathways mapfile used?
#If ERROR --> USE the METACYC for downstream analyses!!!
add_descriptions.py -i pathways_out/path_abun_unstrat.tsv.gz -o KEGG_pathways_out/path_abun_unstrat_descrip.tsv.gz --custom_map_table /home/jhuang/anaconda3/envs/picrust2/lib/python3.6/site-packages/picrust2/default_files/description_mapfiles/KEGG_pathways_info.tsv.gz3.7. Visualization
#7.1 STAMP
#https://github.com/picrust/picrust2/wiki/STAMP-example
conda activate stamp; #import path_abun_unstrat_descrip.tsv to STAMP
conda deactivate
conda install -c bioconda stamp
#conda install -c bioconda stamp
#sudo pip install pyqi
#sudo apt-get install libblas-dev liblapack-dev gfortran
#sudo apt-get install freetype* python-pip python-dev python-numpy python-scipy python-matplotlib
#sudo pip install STAMP
#conda install -c bioconda stamp
conda create -n stamp -c bioconda/label/cf201901 stamp
brew install pyqt
#DEBUG the environment
conda install pyqt=4
#conda install icu=56
e.g. path_abun_unstrat_descrip.tsv.gz and metadata.tsv from the tutorial)
cut -d$'\t' -f1 map_corrected.txt > 1
cut -d$'\t' -f5 map_corrected.txt > 5
cut -d$'\t' -f6 map_corrected.txt > 6
paste -d$'\t' 1 5 > 1_5
paste -d$'\t' 1_5 6 > metadata.tsv
#SampleID --> SampleID
SampleID Facility Genotype
100CHE6KO PaloAlto KO
101CHE6WT PaloAlto WT
#7.2. ALDEx2
https://bioconductor.org/packages/release/bioc/html/ALDEx2.html
#7.3. Convert png to svg and pdf
inkscape error_bar.png --export-plain-svg=error_bar.svg (embbed)
sudo apt update
sudo apt install autotrace
sudo apt-get install -y libpng-dev libtiff-dev imagemagick
git clone https://github.com/autotrace/autotrace.git
cd autotrace
#sudo apt install intltool
#sudo apt install gettext libglib2.0-dev
#sudo apt install libtool libtool-bin
#sudo apt install automake
sudo apt-get install libxml-parser-perl
./autogen.sh
./configure
make
autotrace -output-format svg -output-file error_bar.svg error_bar.png