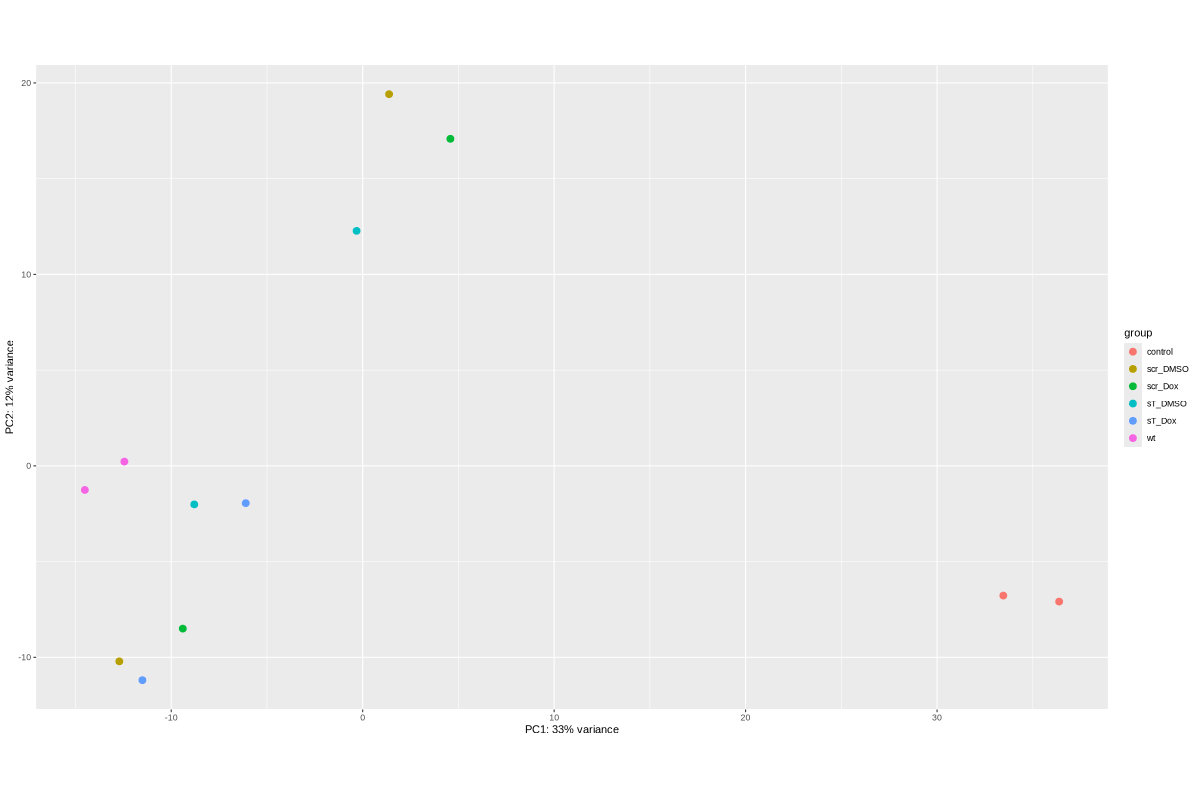
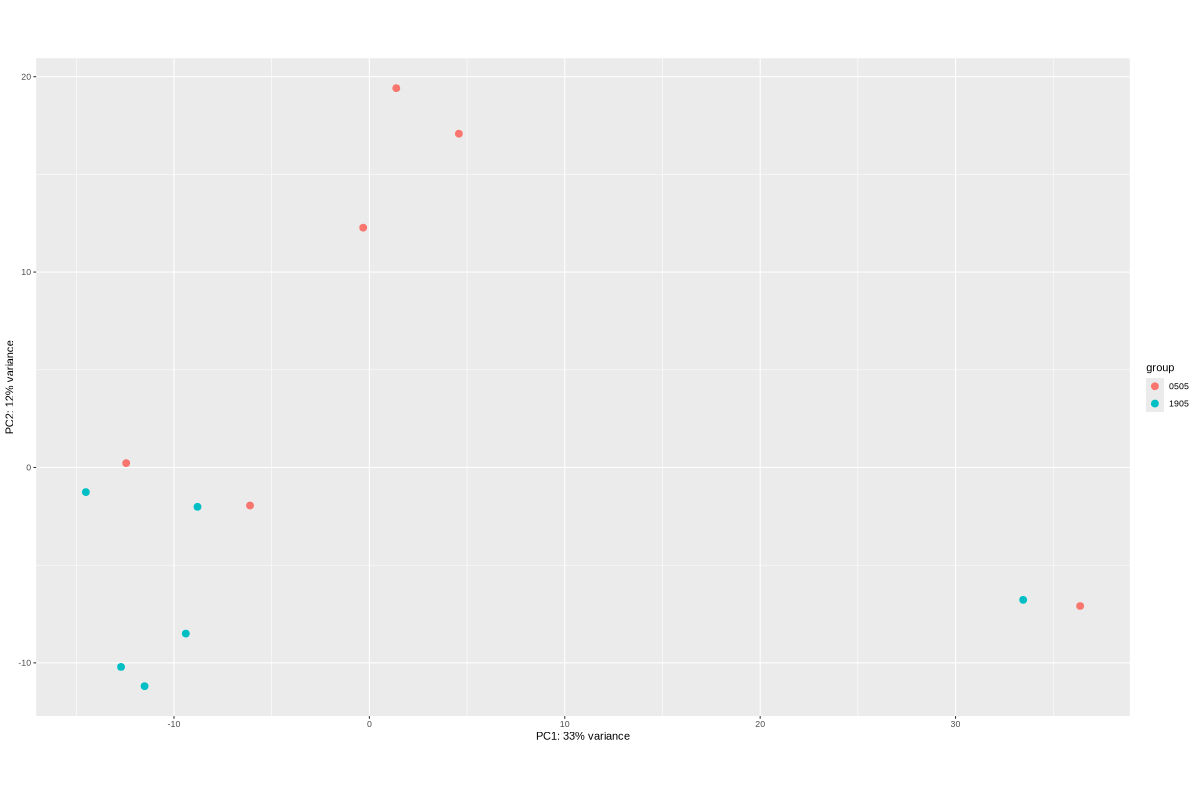
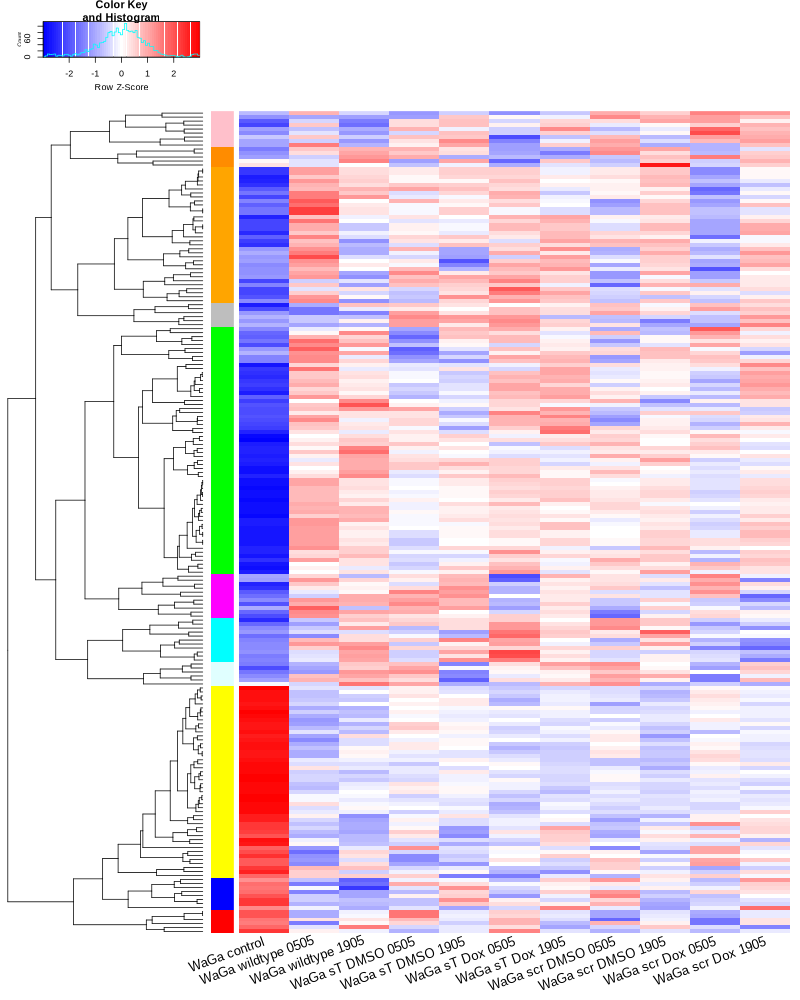
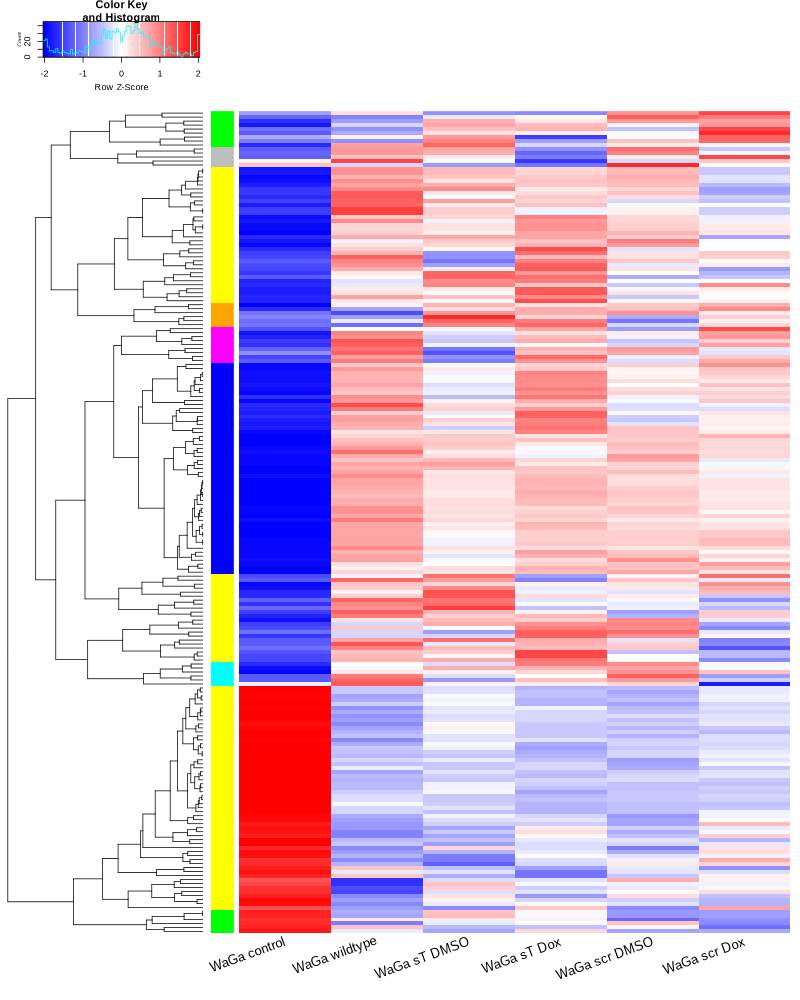
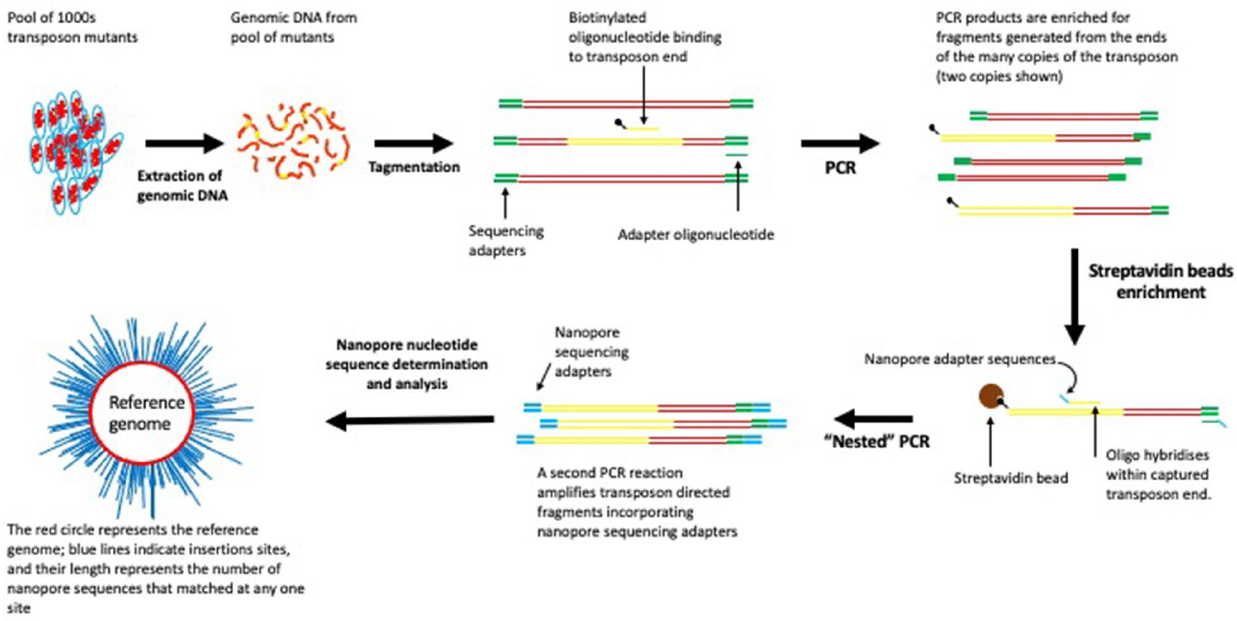
TODO: edit and sort the following text after vacation
# Define the number of reads to sample
for i in {1..9}; do
num_reads=$((i * 1000000))
# Sample the reads
seqtk sample -s100 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R1_001.fastq $num_reads > ./samples/initial_mutants_rep1_S25_R1_${num_reads}.fastq
seqtk sample -s100 ./240405_VH00358_89_AAFC5MTM5/kr1/initial_mutants_rep1_S25_R2_001.fastq $num_reads > ./samples/initial_mutants_rep1_S25_R2_${num_reads}.fastq
done
#set the parameters in ./lib/python3.10/site-packages/pytpp/tpp_tools.py
#set primer_start_window
vars.primer_start_window = 0,159
#we didn't give primer-parameter, therefore the prefix is defined as TAAGAGACAG. We have only set a condition, namely prefix (see below).
if "primer" not in kwargs and vars.transposon == "Tn5":
vars.prefix = "TAAGAGACAG"
#Thank you for your feedback. In my last analysis, I focused on verifying whether read1 contains the end segment "TAAGAGACAG" of the 95-nt transposon sequence, allowing for one mismatch. This step is pivotal as accurately locating this segment in read1 is essential for pinpointing the exact insertion position of the transposon. The presence of "TAAGAGACAG" is critical for downstream analysis because without it, we cannot accurately determine transposon insertion sites. Utilizing this strategy, we found that approximately 6% of read1 in the initial_mutants_rep1 sample contained "TAAGAGACAG" (allowing for one mismatch). Almost all of these reads also start with the sequences "ACCTACAACAAAGCTCTCATCAACC", "CACCTACAACAAAGCTCTCATCAAC", or "CCTACAACAAAGCTCTCATCAACCG".
#For reference, I have listed the 95-nt sequences from the transposon as follows: ACCTACAACAAAGCTCTCATCAAC CGTGGCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTCAGGGTTGAGATGTGTA TAAGAGACAG.
conda deactivate
# ------ for 1000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_1000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_1000000.fastq -output 1000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
# Break-down of total reads (1000000):
# 939516 reads (94.0%) lack the expected Tn prefix
# Break-down of trimmed reads with valid Tn prefix (60484), some of them are not starting with the sequences "ACCTACAACAAAGCTCTCATCAACC", "CACCTACAACAAAGCTCTCATCAAC", or "CCTACAACAAAGCTCTCATCAACCG". --> only 47994 are in the Excel-table.
mv tpp.cfg 1000000.tpp.cfg
mkdir 1M
mv 1000000* 1M
cd 1M
cp 1000000.tn_stats 1000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 1000000.tn_stats_ 1000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 47994 and 44916, 29258
# ------ for 2000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_2000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_2000000.fastq -output 2000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 2000000.tpp.cfg
mkdir 2M
mv 2000000* 2M
cd 2M
cp 2000000.tn_stats 2000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 2000000.tn_stats_ 2000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 96067 84578 43293
# ------ for 3000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_3000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_3000000.fastq -output 3000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 3000000.tpp.cfg
mkdir 3M
mv 3000000* 3M
cd 3M
cp 3000000.tn_stats 3000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 3000000.tn_stats_ 3000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 143700 119668 51657
# ------ for 4000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_4000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_4000000.fastq -output 4000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 4000000.tpp.cfg
mkdir 4M
mv 4000000* 4M
cd 4M
cp 4000000.tn_stats 4000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 4000000.tn_stats_ 4000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 191626 151444 57684
# ------ for 5000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_5000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_5000000.fastq -output 5000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 5000000.tpp.cfg
mkdir 5M
mv 5000000* 5M
cd 5M
cp 5000000.tn_stats 5000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 5000000.tn_stats_ 5000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 239814 180369 62089
# ------ for 6000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_6000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_6000000.fastq -output 6000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 6000000.tpp.cfg
mkdir 6M
mv 6000000* 6M
cd 6M
cp 6000000.tn_stats 6000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 6000000.tn_stats_ 6000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 287396 206325 65438
# ------ for 7000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_7000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_7000000.fastq -output 7000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 7000000.tpp.cfg
mkdir 7M
mv 7000000* 7M
cd 7M
cp 7000000.tn_stats 7000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 7000000.tn_stats_ 7000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 335593 230313 68150
# ------ for 8000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_8000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_8000000.fastq -output 8000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 8000000.tpp.cfg
mkdir 8M
mv 8000000* 8M
cd 8M
cp 8000000.tn_stats 8000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 8000000.tn_stats_ 8000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 383769 252191 70463
# ------ for 9000000 paired reads ------
python3 ~/.local/bin/tpp -bwa /usr/bin/bwa -protocol Tn5 -ref WA-314_m.fna -reads1 ./samples/initial_mutants_rep1_S25_R1_9000000.fastq -reads2 ./samples/initial_mutants_rep1_S25_R2_9000000.fastq -output 9000000 -mismatches 1 -bwa-alg mem -replicon-ids contig_2_10,contig_2_9,contig_2_8,contig_2_7,contig_2_6,contig_2_5,contig_2_3,contig_2_2,contig_5_10,contig_5_11,contig_5_12,contig_5_13,contig_5_15,contig_5_16,contig_5_17,contig_5_18,contig_5_9,contig_5_8,contig_5_7,contig_5_6,contig_5_5,contig_5_4,contig_5_3,contig_5_2,contig_5_1,contig_4_2,contig_4_1,contig_3_59,contig_3_58,contig_3_57,contig_3_56,contig_3_55,contig_3_54,contig_3_53,contig_3_52,contig_3_51,contig_3_50,contig_3_49,contig_3_48,contig_3_47,contig_3_46,contig_3_44,contig_3_43,contig_3_42,contig_3_41,contig_3_40,contig_3_39,contig_3_38,contig_3_37,contig_3_36,contig_3_35,contig_3_34,contig_3_33,contig_3_32,contig_3_31,contig_3_30,contig_3_29,contig_3_28,contig_3_27,contig_3_26,contig_3_25,contig_3_24,contig_3_23,contig_3_22,contig_3_21,contig_3_20,contig_3_17,contig_3_16,contig_3_15,contig_3_14,contig_3_13,contig_3_12,contig_3_11,contig_3_9,contig_3_8,contig_3_7,contig_3_6,contig_3_5,contig_3_3,contig_3_2,contig_3_1,contig_1_48,contig_1_47,contig_1_46,contig_1_45,contig_1_44,contig_1_43,contig_1_42,contig_1_41,contig_1_40,contig_1_39,contig_1_38,contig_1_37,contig_1_34,contig_1_33,contig_1_32,contig_1_31,contig_1_28,contig_1_27,contig_1_26,contig_1_25,contig_1_24,contig_1_22,contig_1_20,contig_1_19,contig_1_18,contig_1_17,contig_1_16,contig_1_15,contig_1_14,contig_1_13,contig_1_12,contig_1_10,contig_1_8,contig_1_7,contig_1_6,contig_1_5,contig_1_4,contig_1_3,contig_1_2,contig_1_1,contig_C8715,contig_C8943,contig_C9371,contig_C8939,contig_C9357,contig_C8991,contig_C9445,contig_C8689
mv tpp.cfg 9000000.tpp.cfg
mkdir 9M
mv 9000000* 9M
cd 9M
cp 9000000.tn_stats 9000000.tn_stats_
#Delete all general statistics before the table data in initial_mutants_rep1.tn_stats_; delete the content after "# FR_corr (Fwd templates vs. Rev templates):"
sed -i 's/read_count (TA sites only, for Himar1)/read_counts/g' *.tn_stats_
sed -i 's/NZ_mean (among templates)/NZ_mean (mean template count over non-zero TA sites)/g' *.tn_stats_
python3 ../parse_tn_stats.py 9000000.tn_stats_ 9000000.tn_stats.xlsx
#calculate the sum of the first and second columns by "=SUM(B2:B130)" and "=SUM(C2:C130)", =SUM(F2:F130) 432255 272550 72303
# ------ for 9181515 paired reads ------
# ------ for the complete reads (9181515 x 2 reads), we have the folloiwng number of insertion sites: 441057 276060 72595
#------ Draw saturation graph in python3 ------
# Updated y-axis data
template_count = [44916, 84578, 119668, 151444, 180369, 206325, 230313, 252191, 272550, 276060]
TAs_hit = [29258, 43293, 51657, 57684, 62089, 65438, 68150, 70463, 72303, 72595]
# Plotting the graph with updated data
plt.figure(figsize=(10, 6))
# Plot template_count
plt.plot(paired_reads, template_count, 'o-', label='Template Count', color='blue')
# Plot TAs_hit
plt.plot(paired_reads, TAs_hit, 'o-', label='TAs Hit', color='red')
# Labels and title
plt.xlabel('Input Paired Reads Number')
plt.ylabel('Count')
plt.title('Saturation Curve for Template Count and TAs Hit')
plt.legend()
# Show plot
plt.grid(True)
plt.show()