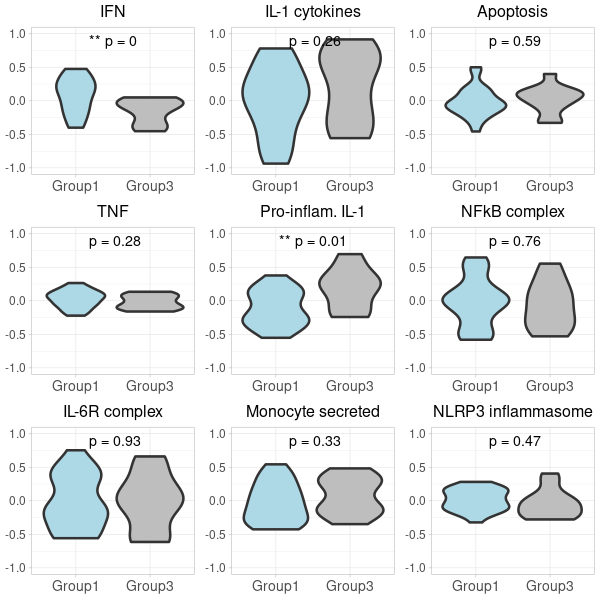
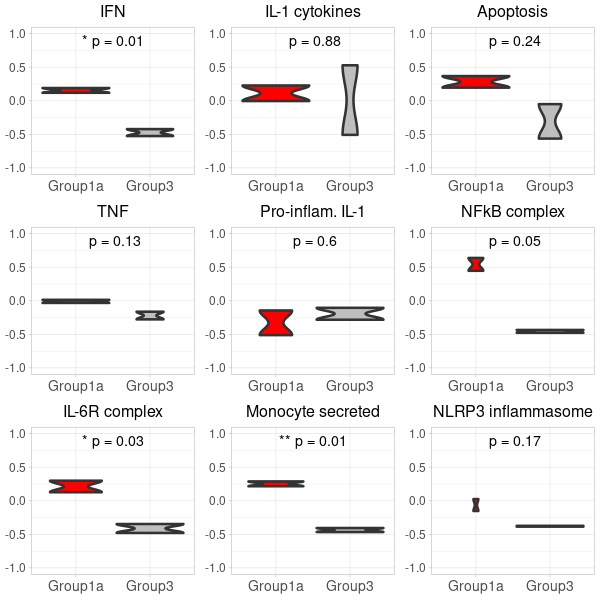
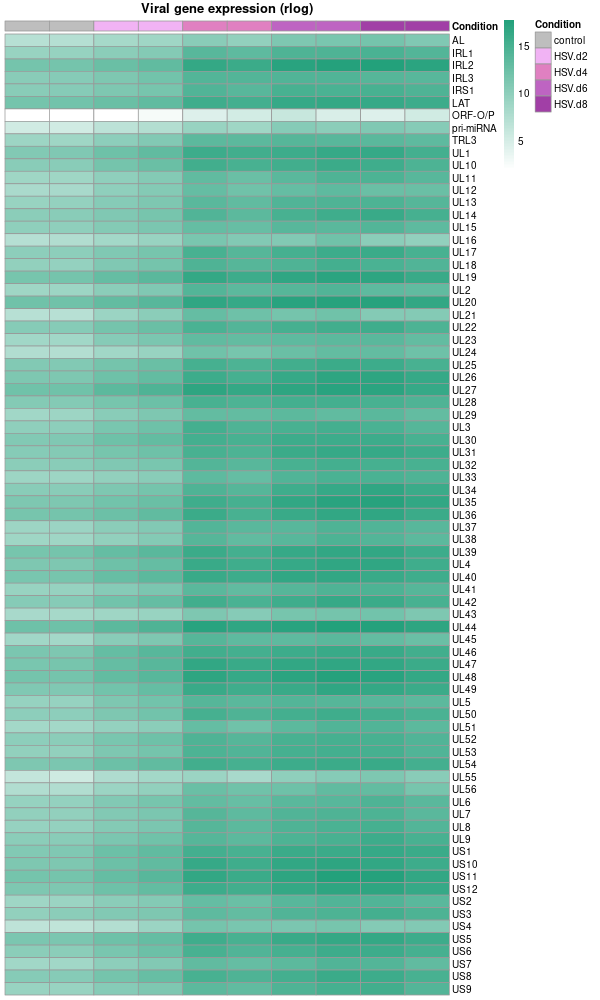
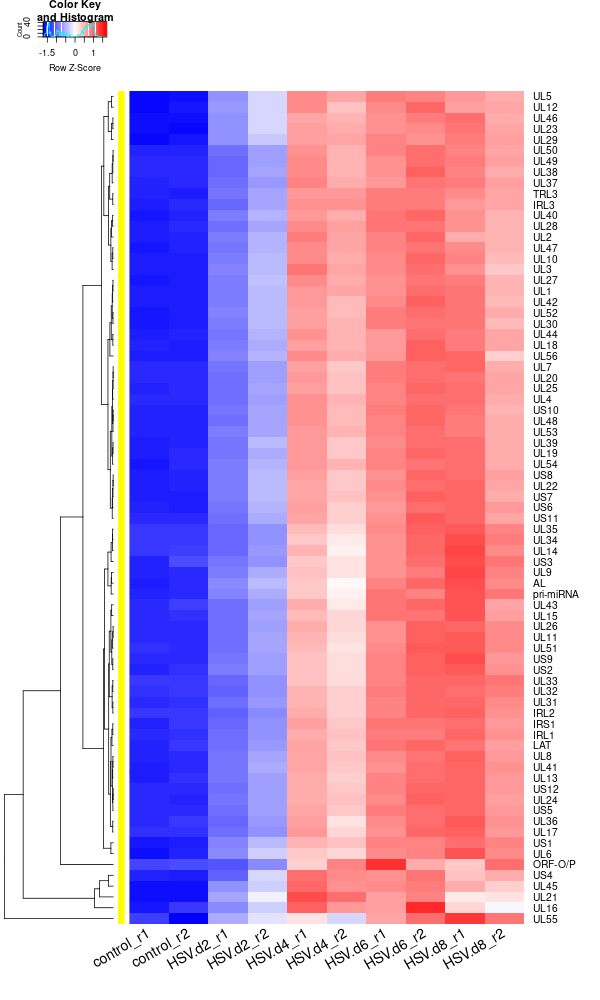
-
import data and pca-plot
# Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) setwd("~/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon") # Define paths to your Salmon output quantification files files <- c("control_r1" = "./control_r1/quant.sf", "control_r2" = "./control_r2/quant.sf", "HSV.d2_r1" = "./HSV.d2_r1/quant.sf", "HSV.d2_r2" = "./HSV.d2_r2/quant.sf", "HSV.d4_r1" = "./HSV.d4_r1/quant.sf", "HSV.d4_r2" = "./HSV.d4_r2/quant.sf", "HSV.d6_r1" = "./HSV.d6_r1/quant.sf", "HSV.d6_r2" = "./HSV.d6_r2/quant.sf", "HSV.d8_r1" = "./HSV.d8_r1/quant.sf", "HSV.d8_r2" = "./HSV.d8_r2/quant.sf") # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) # Define the replicates and condition of the samples replicate <- factor(c("r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2")) condition <- factor(c("control", "control", "HSV.d2", "HSV.d2", "HSV.d4", "HSV.d4", "HSV.d6", "HSV.d6", "HSV.d8", "HSV.d8")) # Define the colData for DESeq2 colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) # In the context of your new code which is using tximport and DESeq2, you don't necessarily need this step. The reason is that DESeq2 performs its own filtering of low-count genes during the normalization and differential expression steps. # Filter data to retain only genes with more than 2 counts > 3 across all samples # dds <- dds[rowSums(counts(dds) > 3) > 2, ] # Output raw count data to a CSV file write.csv(counts(dds), file="transcript_counts.csv") # -- gene-level count data -- # Read in the tx2gene map from salmon_tx2gene.tsv #tx2gene <- read.csv("salmon_tx2gene.tsv", sep="\t", header=FALSE) tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) # Continue with the DESeq2 workflow as before... colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #60605-->26543 write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv") #~/Tools/csv2xls-0.4/csv_to_xls.py gene_counts.csv -d',' -o gene_counts.xls #TODO: why a lot of reads were removed due to the too_short? #STAR --runThreadN 4 --genomeDir /path/to/GenomeDir --readFilesIn /path/to/read1.fastq /path/to/read2.fastq --outFilterMatchNmin 50 --outSAMtype BAM SortedByCoordinate --outFileNamePrefix /path/to/output dim(counts(dds)) head(counts(dds), 10) #DEBUG: DESeq should not used here!? #TODO_NEXT_WEEK: rerun without fistly DESeq(dds) to compare if the results is the same to process_1 #dds <- DESeq(dds) rld <- rlogTransformation(dds) # draw simple pca and heatmap library(gplots) library("RColorBrewer") #mat <- assay(rld) #mm <- model.matrix(~condition, colData(rld)) #mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm) #assay(rld) <- mat # -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
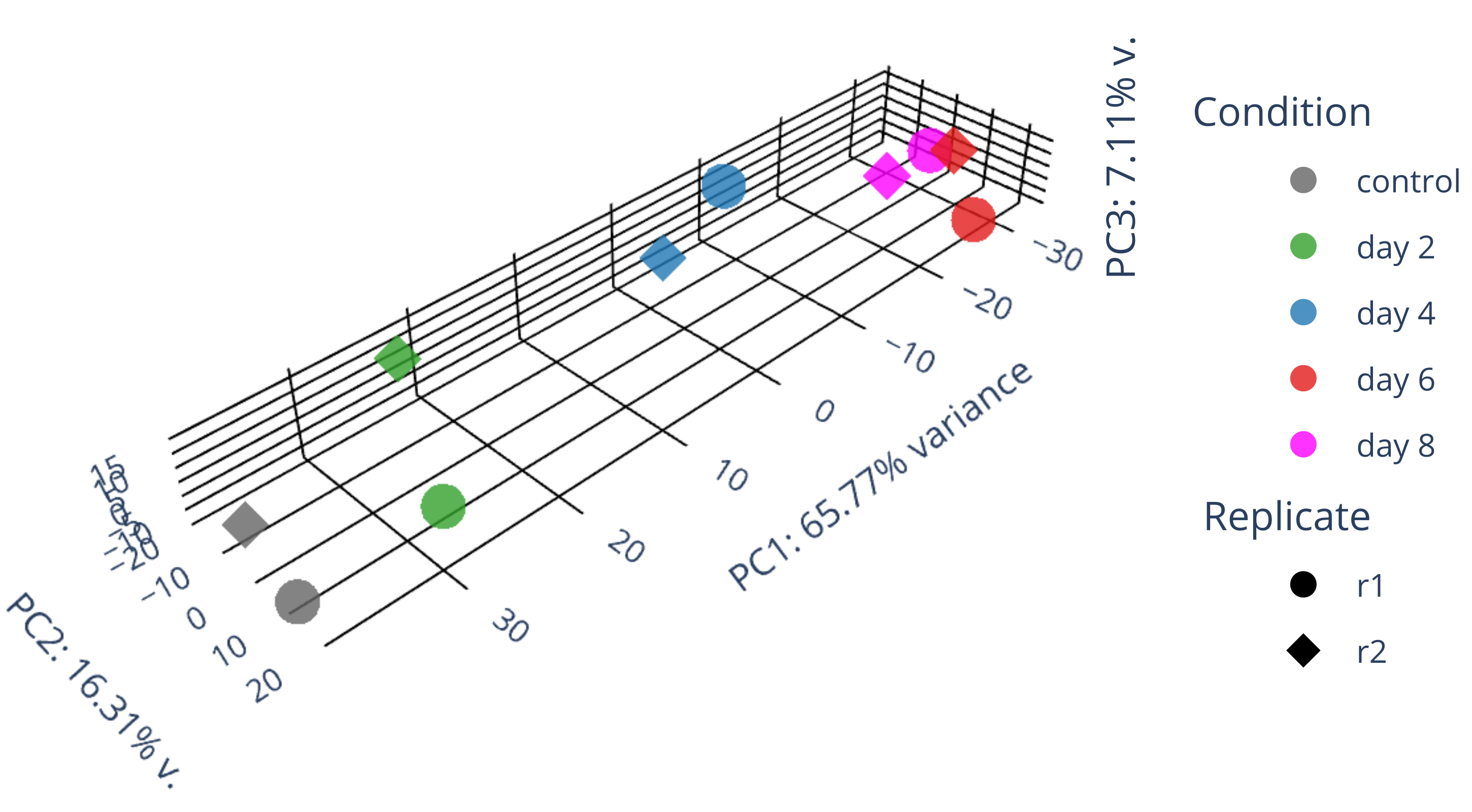
draw 3D PCA plots.
library(gplots) library("RColorBrewer") library(ggplot2) data <- plotPCA(rld, intgroup=c("condition", "replicate"), returnData=TRUE) write.csv(data, file="plotPCA_data.csv") #calculate all PCs including PC3 with the following codes library(genefilter) ntop <- 500 rv <- rowVars(assay(rld)) select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))] mat <- t( assay(rld)[select, ] ) pc <- prcomp(mat) pc$x[,1:3] #df_pc <- data.frame(pc$x[,1:3]) df_pc <- data.frame(pc$x) identical(rownames(data), rownames(df_pc)) #-->TRUE data$PC1 <- NULL data$PC2 <- NULL merged_df <- merge(data, df_pc, by = "row.names") #merged_df <- merged_df[, -1] row.names(merged_df) <- merged_df$Row.names merged_df$Row.names <- NULL # remove the "name" column merged_df$name <- NULL merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","group","condition","replicate")] write.csv(merged_df, file="merged_df_10PCs.csv") summary(pc) #0.5333 0.2125 0.06852 #0.8026 0.09042 0.06578 draw_3D.py # adjust proportion to real values in the following plot import plotly.graph_objects as go import pandas as pd from sklearn.decomposition import PCA import numpy as np # Read in data as a pandas dataframe #df = pd.DataFrame({ # 'PC1': [-13.999925, -12.504291, -12.443057, -13.065235, -17.316215], # 'PC2': [-1.498823, -3.342411, -6.067055, -8.205809, 3.293993], # 'PC3': [-3.335085, 15.207755, -14.725450, 15.078469, -6.917358], # 'condition': ['GFP d3', 'GFP d3', 'GFP d8', 'GFP d8', 'GFP+mCh d9/12'], # 'replicate': ['DI', 'DII', 'DI', 'DII', 'DI'] #}) df = pd.read_csv('merged_df_10PCs.csv', index_col=0, header=0) df['condition'] = df['condition'].replace("control", "control") df['condition'] = df['condition'].replace("HSV.d2", "day 2") df['condition'] = df['condition'].replace("HSV.d4", "day 4") df['condition'] = df['condition'].replace("HSV.d6", "day 6") df['condition'] = df['condition'].replace("HSV.d8", "day 8") # Fit PCA model to reduce data dimensions to 3 pca = PCA(n_components=3) pca.fit(df.iloc[:, :-3]) X_reduced = pca.transform(df.iloc[:, :-3]) # Get variance ratios explained_variance_ratio = pca.explained_variance_ratio_ # Add reduced data back to dataframe df['PC1'] = X_reduced[:, 0] df['PC2'] = X_reduced[:, 1] df['PC3'] = X_reduced[:, 2] # Create PCA plot with 3D scatter fig = go.Figure() ##ff7f00 condition_color_map = { 'control': 'rgb(100, 100, 100)', 'day 2': '#33a02c', 'day 4': '#1f78b4', 'day 6': '#e31a1c', 'day 8': 'magenta' } replicate_symbol_map = {'r1': 'circle', 'r2': 'diamond'} for replicate, replicate_symbol in replicate_symbol_map.items(): for condition, condition_color in condition_color_map.items(): mask = (df['condition'] == condition) & (df['replicate'] == replicate) fig.add_trace(go.Scatter3d(x=df.loc[mask, 'PC1'], y=df.loc[mask, 'PC2'], z=df.loc[mask, 'PC3'], mode='markers', name=f'{condition}' if replicate == 'r1' else None, legendgroup=f'{condition}', showlegend=True if replicate == 'r1' else False, marker=dict(size=6 if replicate_symbol in ['diamond'] else 10, opacity=0.8, color=condition_color, symbol=replicate_symbol))) for replicate, replicate_symbol in replicate_symbol_map.items(): fig.add_trace(go.Scatter3d(x=[None], y=[None], z=[None], mode='markers', name=replicate, legendgroup=f'{replicate}', showlegend=True, marker=dict(size=10, opacity=1, color='black', symbol=replicate_symbol), hoverinfo='none')) # Annotations for the legend blocks #TODO: calculate the PC values. #TODO: adjust the axis length according to the actual size of axis! fig.update_layout( annotations=[ dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False, text='Condition', font=dict(size=15)), dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False, text='Replicate', font=dict(size=15)) ], scene=dict( #aspectmode='cube', #xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 53% v.', scaleratio=0.53), #yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 21% v.', scaleratio=0.21), #zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 7% variance', scaleratio=0.07), aspectmode='manual', aspectratio=dict(x=explained_variance_ratio[0], y=explained_variance_ratio[1], z=explained_variance_ratio[2]), xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 53% v.', range=[min(df['PC1']), max(df['PC1'])]), yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 21% v.', range=[min(df['PC2']), max(df['PC2'])]), zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 7% variance', range=[min(df['PC3']), max(df['PC3'])]), bgcolor='white' ), margin=dict(l=5, r=5, b=5, t=0) # Adjust the margins to prevent clipping of axis titles ) #fig.show() fig.write_image("fig1.svg") fig.update_layout( annotations=[ dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False, text='Condition', font=dict(size=15)), dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False, text='Donor', font=dict(size=15)), dict(x=1.08, y=0.2, xref='paper', yref='paper', showarrow=False, text=f'PC3: {explained_variance_ratio[2]*100:.2f}% v.', font=dict(size=15), textangle=-90) ], scene=dict( aspectmode='manual', aspectratio=dict(x=explained_variance_ratio[0]*2, y=explained_variance_ratio[1]*2, z=explained_variance_ratio[2]*2), #, range=[min(df['PC1']), max(df['PC1'])] #, range=[min(df['PC2']), max(df['PC2'])] #, range=[min(df['PC3']), max(df['PC3'])] xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=f'PC1: {explained_variance_ratio[0]*100:.2f}% variance'), yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=f'PC2: {explained_variance_ratio[1]*100:.2f}% v.'), zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=''), bgcolor='white' ), margin=dict(l=0, r=0, b=0, t=0) # Adjust the margins to prevent clipping of axis titles ) fig.write_image("PCA_3D.svg") #/usr/bin/convert g235.png -crop 3250x1680+1+750 PCA_3D_.png -
(optional) estimate size factors
> head(dds) class: DESeqDataSet dim: 6 10 metadata(1): version assays(6): counts avgTxLength ... H cooks rownames(6): ENSG00000000003 ENSG00000000005 ... ENSG00000000460 ENSG00000000938 rowData names(34): baseMean baseVar ... deviance maxCooks colnames(10): control_r1 control_r2 ... HSV.d8_r1 HSV.d8_r2 colData names(2): condition replicate #convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor sizeFactors(dds) #NULL dds <- estimateSizeFactors(dds) > sizeFactors(dds) raw_counts <- counts(dds) normalized_counts <- counts(dds, normalized=TRUE) #write.table(raw_counts, file="raw_counts.txt", sep="\t", quote=F, col.names=NA) #write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA) # ---- DEBUG sizeFactors(dds) always NULL, see https://support.bioconductor.org/p/97676/ ---- nm <- assays(dds)[["avgTxLength"]] sf <- estimateSizeFactorsForMatrix(counts(dds), normMatrix=nm) assays(dds)$counts # for count data assays(dds)$avgTxLength # for average transcript length, etc. assays(dds)$normalizationFactors In normal circumstances, the size factors should be stored in the DESeqDataSet object itself and not in the assays, so they are typically not retrievable via the assays() function. However, due to the issues you're experiencing, you might be able to manually compute the size factors and assign them back to the DESeqDataSet. To calculate size factors manually, DESeq2 uses the median ratio method. Here's a very simplified version of how you could compute this manually: > assays(dds) List of length 6 names(6): counts avgTxLength normalizationFactors mu H cooks To calculate size factors manually, DESeq2 uses the median ratio method. Here's a very simplified version of how you could compute this manually: geoMeans <- apply(assays(dds)$counts, 1, function(row) if (all(row == 0)) 0 else exp(mean(log(row[row != 0])))) sizeFactors(dds) <- median(assays(dds)$counts / geoMeans, na.rm = TRUE) # ---- DEBUG END ---- #unter konsole # control_r1 ... # 1/0.9978755 ... > sizeFactors(dds) HeLa_TO_r1 HeLa_TO_r2 0.9978755 1.1092227 1/0.9978755=1.002129023 1/1.1092227= #bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r1Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r1.bw --binSize 10 --scaleFactor 1.002129023 --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r2Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r2.bw --binSize 10 --scaleFactor 0.901532217 --effectiveGenomeSize 2864785220 -
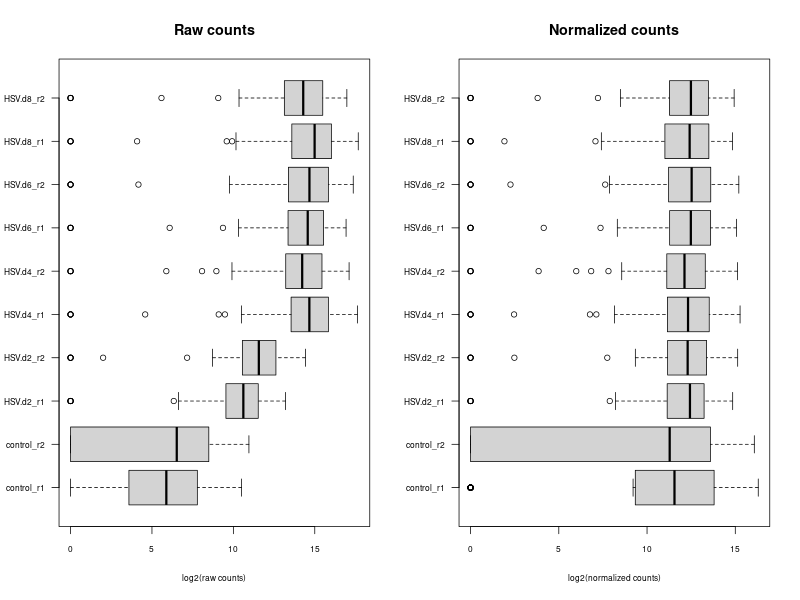
compare the normalization methods
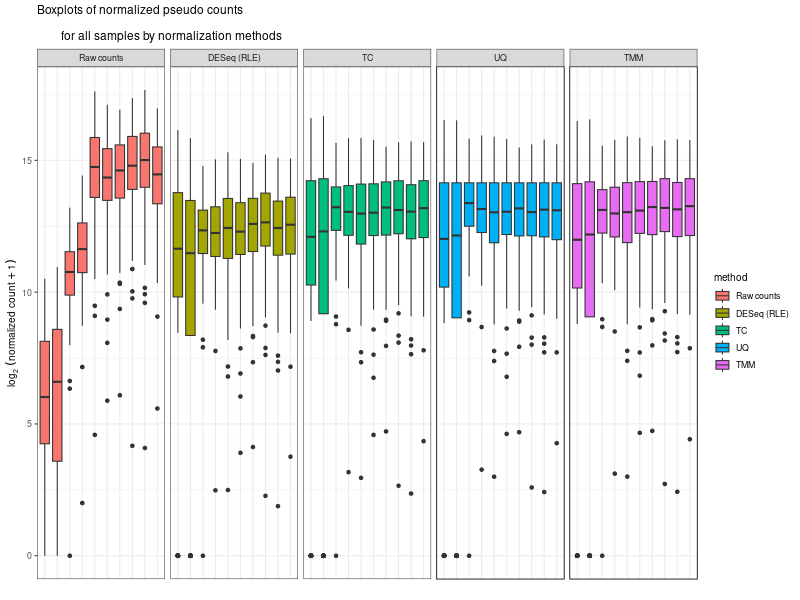
# ---- draw normalization before and after ---- ### Let's implement such a function ### cds is a countDataset estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) head(estimSf(dds)) all(round(estimSf(dds),6) == round(sizeFactors(dds), 6)) ## Checking the normalization png("normalization.png", width=800, height=600) epsilon <- 1 # pseudo-count to avoid problems with log(0) par(mfrow=c(1,2),cex.lab=0.7) boxplot(log2(raw_counts+epsilon), cex.axis=0.7, las=1, xlab="log2(raw counts)", horizontal=TRUE, main="Raw counts") boxplot(log2(normalized_counts+epsilon), cex.axis=0.7, las=1, xlab="log2(normalized counts)", horizontal=TRUE, main="Normalized counts") #boxplot(log10(assays(dds)[["cooks"]]), range=0, las=2) #plotDensity(log2(counts(dds.norm)+epsilon), col=col.pheno.selected, # xlab="log2(counts)", cex.lab=0.7, panel.first=grid()) #plotDensity(log2(counts(dds.norm, normalized=TRUE)+epsilon), col=col.pheno.selected, # xlab="log2(normalized counts)", cex.lab=0.7, panel.first=grid()) dev.off() # since we Gene-level differential expression analysis with DESeq2, the splicing plays no role in the analysis! # 用nanopore 可以 compare transcript length distribution. 有可能Cellline很长,Extracellular vesicles (EVs)很短! library(ggplot2) library(gridExtra) library(reshape2) library(mixOmics) library(RColorBrewer) library(DESeq) library(edgeR) library(VennDiagram) library(devtools) raw_counts_wn <- raw_counts[rowSums(raw_counts) > 0, ] dim(raw_counts_wn) #--Raw counts-- pseudo_counts <- log2(raw_counts_wn + 1) head(pseudo_counts) df_raw <- melt(pseudo_counts, id = rownames(raw_counts_wn)) names(df_raw)[1:2]<- c("id", "sample") df_raw$method <- rep("Raw counts", nrow(df_raw)) head(df_raw) #--DESeq-- cData = data.frame(row.names=colnames(raw_counts_wn), replicates=replicates, ids=ids) dge<-DESeqDataSetFromMatrix(countData=raw_counts_wn, colData=cData, design=~replicates) dge <- estimateSizeFactors(dge) sizeFactors(dge) deseq_normcount <- counts(dge, normalized = TRUE) test_normcount <- sweep(raw_counts_wn, 2, sizeFactors(dge), "/") sum(test_normcount != deseq_normcount) pseudo_deseq <- log2(deseq_normcount + 1) df_deseq <- melt(pseudo_deseq, id = rownames(raw_counts_wn)) names(df_deseq)[1:2]<- c("id", "sample") df_deseq$method <- rep("DESeq (RLE)", nrow(df_raw)) #--edgeR-- dge2 <- DGEList(raw_counts_wn) dge2 dge2$samples #--Total count-- pseudo_TC <- log2(cpm(dge2) + 1) df_TC <- melt(pseudo_TC, id = rownames(raw_counts_wn)) names(df_TC)[1:2] <- c ("id", "sample") df_TC$method <- rep("TC", nrow(df_TC)) ##--RPKM-- #gene_lengths_wn <- gene_lengths[rowSums(raw_counts) > 0] #pseudo_RPKM <- log2(rpkm(dge2, gene.length = gene_lengths_wn) + 1) #df_RPKM <- melt(pseudo_RPKM, id = rownames(raw_counts_wn)) #names(df_RPKM)[1:2] <- c ("id", "sample") #df_RPKM$method <- rep("RPKM", nrow(df_RPKM)) #--Upper quartile-- dge2 <- calcNormFactors(dge2, method = "upperquartile") dge2$samples test_normcount <- sweep(dge2$counts, 2, dge2$samples$lib.size*dge2$samples$norm.factors / 10^6, "/") range(as.vector(test_normcount - cpm(dge2))) pseudo_UQ <- log2(cpm(dge2) + 1) df_UQ <- melt(pseudo_UQ, id = rownames(raw_counts_wn)) names(df_UQ)[1:2] <- c ("id", "sample") df_UQ$method <- rep("UQ", nrow(df_UQ)) #--TMM-- dge2 <- calcNormFactors(dge2, method = "TMM") dge2$samples pseudo_TMM <- log2(cpm(dge2) + 1) df_TMM <- melt(pseudo_TMM, id = rownames(raw_counts_wn)) names(df_TMM)[1:2] <- c ("id", "sample") #MODIFIED! df_TMM$method <- rep("DESeq (RLE)", nrow(df_TMM)) #TMM #--Comparison-- png("normalization.png", width=800, height=600) #df_allnorm <- rbind(df_raw, df_deseq, df_TC, df_UQ, df_TMM) #df_allnorm$method <- factor(df_allnorm$method, levels = c("Raw counts", "DESeq (RLE)", "TC", "TMM", "UQ")) df_allnorm <- rbind(df_raw, df_TMM) df_allnorm$method <- factor(df_allnorm$method, levels = c("Raw counts", "DESeq (RLE)")) p <- ggplot(data=df_allnorm, aes(x=sample, y=value, fill=method)) p <- p + geom_boxplot() p <- p + theme_bw() p <- p + ggtitle("Boxplots of normalized pseudo counts\n for all samples by normalization methods") p <- p + facet_grid(. ~ method) p <- p + ylab(expression(log[2] ~ (normalized ~ count + 1))) + xlab("") p <- p + theme(title = element_text(size=10), axis.text.x = element_blank(), axis.ticks.x = element_blank()) print(p) dev.off() -
select the differentially expressed genes
#https://galaxyproject.eu/posts/2020/08/22/three-steps-to-galaxify-your-tool/ #https://www.biostars.org/p/282295/ #https://www.biostars.org/p/335751/ #> condition # [1] control control HSV.d2 HSV.d2 HSV.d4 HSV.d4 HSV.d6 HSV.d6 HSV.d8 HSV.d8 #Levels: control HSV.d2 HSV.d4 HSV.d6 HSV.d8 #CONSOLE: mkdir /home/jhuang/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon/degenes setwd("/home/jhuang/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon/degenes") #---- relevel to control ---- dds$condition <- relevel(dds$condition, "control") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d2_vs_control","HSV.d4_vs_control", "HSV.d6_vs_control", "HSV.d8_vs_control") dds$condition <- relevel(dds$condition, "HSV.d2") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d4_vs_HSV.d2", "HSV.d6_vs_HSV.d2", "HSV.d8_vs_HSV.d2") dds$condition <- relevel(dds$condition, "HSV.d4") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d6_vs_HSV.d4", "HSV.d8_vs_HSV.d4") dds$condition <- relevel(dds$condition, "HSV.d6") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d8_vs_HSV.d6") for (i in clist) { contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=1) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-1) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) } echo "contrast = paste(\"condition\", \"${i}\", sep=\"_\")" echo "res = results(dds, name=contrast)" #echo "res <- res[!is.na(res$log2FoldChange),]" echo "res <- na.omit(res)" ##https://github.com/kevinblighe/EnhancedVolcano #BiocManager::install("EnhancedVolcano") library("EnhancedVolcano") for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do #for i in HSV.d8_vs_control; do echo "res <- read.csv(file = paste(\"${i}\", \"all.txt\", sep=\"-\"), row.names = 1)" echo "res_df <- as.data.frame(res)" echo "png(\"${i}.png\",width=800, height=600)" #legendPosition = 'right',legendLabSize = 12, arrowheads = FALSE, #echo "EnhancedVolcano(res, lab = rownames(res),x = 'log2FoldChange',y = 'padj', pCutoff=5e-2, FCcutoff=1.2, title='', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha=1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle=expression(~Delta*\"$(echo $i | cut -d'_' -f1) versus \" *~Delta*\"$(echo $i | cut -d'_' -f3)\"))" echo "EnhancedVolcano(res, lab = rownames(res),x = 'log2FoldChange',y = 'padj', pCutoff=5e-2, FCcutoff=1.0, title='', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha=1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle=expression(\"$(echo $i | cut -d'_' -f1) versus $(echo $i | cut -d'_' -f3)\"))" echo "dev.off()" done #DEBUG: why some genes in HSV.d8 in control high regulated --> ERROR! We should keep the number of reads in the raw counts, leading all genes low regulated! Not using the default normalization method!!!! #res <- read.csv(file = paste("HSV.d8_vs_control", "all.txt", sep="-"), row.names = 1) #res_df <- as.data.frame(res) #png("HSV.d8_vs_control.png",width=800, height=600) #EnhancedVolcano(res, lab = rownames(res),x = 'log2FoldChange',y = 'padj', pCutoff=5e-2, FCcutoff=1.0, title='', subtitleLabSize = 18, pointSize = 3.0, labSize = 5.0, colAlpha=1, legendIconSize = 4.0, drawConnectors = TRUE, widthConnectors = 0.5, colConnectors = 'black', subtitle=expression("HSV.d8 versus control")) #dev.off() #under DIR degenes under KONSOLE for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "~/Tools/csv2xls-0.4/csv_to_xls.py ${i}-all.txt ${i}-up.txt ${i}-down.txt -d$',' -o ${i}.xls;"; done -
clustering the genes and draw heatmap
for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "cut -d',' -f1-1 ${i}-up.txt > ${i}-up.id"; echo "cut -d',' -f1-1 ${i}-down.txt > ${i}-down.id"; done cat *.id | sort -u > ids #add Gene_Id in the first line, delete the "" GOI <- read.csv("ids")$Gene_Id RNASeq.NoCellLine <- assay(rld) #install.packages("gplots") library("gplots") #clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman #datamat = RNASeq.NoCellLine[GOI, ] datamat = RNASeq.NoCellLine write.csv(as.data.frame(datamat), file ="gene_expressions.txt") constant_rows <- apply(datamat, 1, function(row) var(row) == 0) if(any(constant_rows)) { cat("Removing", sum(constant_rows), "constant rows.\n") datamat <- datamat[!constant_rows, ] } hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.5) mycol = c("YELLOW", "BLUE", "ORANGE", "MAGENTA", "CYAN", "RED", "GREEN", "MAROON", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN", "LIGHTRED", "LIGHTGREEN"); mycol = mycol[as.vector(mycl)] #png("DEGs_heatmap.png", width=900, height=800) #cex.lab=10, labRow="", png("DEGs_heatmap.png", width=900, height=1000) heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row', scale='row',trace='none',col=bluered(75), RowSideColors = mycol, margins=c(10,20), cexRow=1.5, srtCol=45, lhei = c(2, 8)) #rownames(datamat) #heatmap.2(datamat, Rowv=as.dendrogram(hr), col=bluered(75), scale="row", RowSideColors=mycol, trace="none", margin=c(5,5), sepwidth=c(0,0), dendrogram = 'row', Colv = 'false', density.info='none', labRow="", srtCol=30, lhei=c(0.1,2)) dev.off() #### cluster members ##### write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt') write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt') write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt') #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o DEGs_heatmap_cluster_members.xls ~/Tools/csv2xls-0.4/csv_to_xls.py \ significant_gene_expressions.txt \ -d',' -o DEGs_heatmap_expression_data.xls;
Author Archives: gene_x
Normalization of RNA-seq and ChIP-seq data
Normalization methods for RNA-seq data
-
DESeq (RLE – Relative Log Expression):
- Goal: To normalize for differences in library size and distribution of read counts.
- Method: Uses a median ratio method, where for each gene, the ratio of its read count to the geometric mean of read counts across all samples is calculated. The median of these ratios is used as a size factor for normalization.
- Example: Suppose we have two samples A and B with raw read counts for three genes X, Y, Z as follows: A: X=10, Y=20, Z=40 B: X=20, Y=40, Z=80 The size factor for A would be 1, and for B would be 2 (since all counts in B are double of A). After normalization, the counts in B would be divided by 2, making them equal to A.
-
TMM (Trimmed Mean of M-values):
- Goal: To normalize for differences in library size and distribution, while being less sensitive to extreme values.
- Method: Calculates a weighted trimmed mean of the log expression ratios, after excluding the most extreme values.
- Example: Continuing with the previous example, TMM would also recognize that B has double the counts of A. After normalization, the counts in B would be divided by 2.
-
Upper Quantile:
- Goal: To adjust for differences in library size based on the upper quantile of counts.
- Method: Scales the read counts so that the upper quantiles of the counts are the same across samples.
- Example: In the previous example, the 75% quantile for both A and B is the same, so no further normalization would be required.
-
Total Counts:
- Goal: To normalize based on total counts across samples.
- Method: Divides each read count by the total number of reads in the sample.
- Example: If A had 100 total reads and B had 200, the counts in B would be divided by 2 for normalization.
-
RPKM (Reads Per Kilobase of transcript per Million mapped reads):
- Goal: To normalize for gene length and total read count.
- Method: For each gene, divide the read count by the gene length (in kilobases) and then by the total number of reads (in millions).
- Example: If gene X is 2kb long, in sample A (with 100 total reads), RPKM for X = (10 / 2) / 0.1 = 50.
Normalization methods for ChIP-seq Data
For ChIP-seq data, it is crucial to also consider other factors like input control normalization, and peak calling. Some of the complete methods/tools that incorporate normalization as part of the ChIP-seq analysis pipeline include:
-
MACS (Model-based Analysis of ChIP-Seq):
- Description: A widely used tool for identifying transcription factor binding sites and regions of histone modification.
- Features: Provides robust peak calling with a focus on identifying precise locations of binding sites.
- Normalization Method: It includes a local background correction to account for bias due to local chromatin structure or GC content.
-
SICER (Spatial clustering approach for the Identification of ChIP-Enriched Regions):
- Description: A tool designed to identify broad regions of enrichment that are typically associated with histone modifications.
- Features: Particularly useful for datasets where the regions of enrichment are distributed in broader domains rather than sharp peaks.
- Normalization Method: Uses a spatial clustering approach to differentiate between true signals and background noise, accounting for both local and global variations.
-
ChIPQC:
- Description: A Bioconductor package providing quality control and normalization functionalities for ChIP-seq data.
- Features: Offers comprehensive analysis of ChIP-seq quality, including diagnostic plots and summary statistics.
- Normalization Method: Provides tools for normalization, though it primarily focuses on quality control aspects. Users may integrate ChIPQC with other packages for more advanced normalization procedures.
There are several standalone methods and packages specifically designed for normalization of ChIP-seq data, considering the unique characteristics of these experiments. Below are some of the popular ones:
-
csaw:
- Description: A Bioconductor package that provides functions for normalization and differential binding analysis in ChIP-seq data.
- Features: It is designed for analyzing broad genomic regions and is effective even in the presence of strong sample-to-sample variability.
- Normalization Method: It uses a sliding window approach and models the count data using a negative binomial distribution.
-
DiffBind:
- Description: Another Bioconductor package that performs differential binding analysis on ChIP-seq data.
- Features: It provides extensive functionalities for quality control, normalization, and downstream analysis.
- Normalization Method: It supports several normalization methods including total count scaling, RPKM, and DESeq normalization.
-
ChIPnorm:
- Description: A standalone R package for normalization of ChIP-seq data.
- Features: It is specifically designed for normalization of ChIP-seq data against input controls.
- Normalization Method: It uses quantile normalization to correct for the distribution of read counts.
-
deepTools:
- Description: A suite of python tools particularly used for quality control and normalization of deep-sequencing data.
- Features: It includes a wide variety of tools for assessing correlation between samples, visualizing data, and normalization.
- Normalization Method: It supports normalization methods such as reads per kilobase per million (RPKM), log1p, and z-score normalization.
-
DANPOS:
- Description: A package for dynamic analysis of nucleosome and protein-DNA binding with high resolution.
- Features: It is designed for analyzing positional patterns of regulatory elements in ChIP-seq data.
- Normalization Method: It includes methods for normalizing sequencing depth and background noise.
-
SPP:
- Description: An R package for analyzing ChIP-seq data with a focus on identifying quality metrics.
- Features: It includes functionalities for creating quality control plots, assessing cross-correlation, and normalizing read counts.
- Normalization Method: It provides cross-correlation based normalization.
-
SeqNorm:
- Description: A standalone tool for normalizing ChIP-seq data.
- Features: It is designed to normalize ChIP-seq datasets to a common reference, improving comparability.
- Normalization Method: It uses a scaling factor based on non-enriched regions.
These tools offer a variety of normalization methods tailored for the specific challenges posed by ChIP-seq data. Users can choose the one that best fits their experimental design and analysis needs.
RNA-seq skin organoids on GRCh38+chrHsv1 (final)
-
run nextflow rnaseq
#under sage ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq (rnaseq) nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_GRCh38 --genome GRCh38 --with_umi --umitools_extract_method regex --umitools_bc_pattern '^(?P.{12}).*' -profile docker -resume --max_cpus 54 --max_memory 120.GB --max_time 2400.h --save_align_intermeds --save_unaligned --save_reference --aligner star_salmon --pseudo_aligner salmon --umitools_grouping_method unique #Debug the following error: added "--minAssignedFrags 0 \\" to modules/nf-core/salmon/quant/main.nf option "salmon quant" and added "--min_mapped_reads 0" in the nextflow command (rnaseq) nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_chrHsv1 --fasta chrHsv1_s17.fasta --gtf chrHsv1_s17.gtf --with_umi --umitools_extract_method regex --umitools_bc_pattern '^(?P.{12}).*' --umitools_dedup_stats -profile test_full -resume --max_memory 256.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0 #TODO: why a lot of reads were removed due to the too_short? -
import data and pca-plot
# Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) library("org.Hs.eg.db") library(dplyr) library(tidyverse) setwd("~/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon") # Define paths to your Salmon output quantification files files <- c("control_r1" = "./control_r1/quant.sf", "control_r2" = "./control_r2/quant.sf", "HSV.d2_r1" = "./HSV.d2_r1/quant.sf", "HSV.d2_r2" = "./HSV.d2_r2/quant.sf", "HSV.d4_r1" = "./HSV.d4_r1/quant.sf", "HSV.d4_r2" = "./HSV.d4_r2/quant.sf", "HSV.d6_r1" = "./HSV.d6_r1/quant.sf", "HSV.d6_r2" = "./HSV.d6_r2/quant.sf", "HSV.d8_r1" = "./HSV.d8_r1/quant.sf", "HSV.d8_r2" = "./HSV.d8_r2/quant.sf") # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) # Define the replicates and condition of the samples replicate <- factor(c("r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2")) condition <- factor(c("control", "control", "HSV.d2", "HSV.d2", "HSV.d4", "HSV.d4", "HSV.d6", "HSV.d6", "HSV.d8", "HSV.d8")) # Define the colData for DESeq2 colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) # -- transcript-level count data (x2) -- # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) write.csv(counts(dds), file="transcript_counts.csv") # -- gene-level count data (x2) -- # Read in the tx2gene map from salmon_tx2gene.tsv tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) # Continue with the DESeq2 workflow as before... colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #60605-->26543 write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv") # -- merge the raw counts of human and microbe -- #cat ~/DATA/Data_Manja_RNAseq_Organoids/results_GRCh38_unique/star_salmon/gene_counts.csv ~/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon/gene_counts.csv > merged_gene_counts.csv #DELETE the second line "","control_r1","control_r2","HSV.d2_r1","HSV.d2_r2","HSV.d4_r1","HSV.d4_r2","HSV.d6_r1","HSV.d6_r2","HSV.d8_r1","HSV.d8_r2" #~/Tools/csv2xls-0.4/csv_to_xls.py merged_gene_counts.csv -d',' -o raw_gene_counts.xls; # -- for merged analysis due to false normalization factors wenn alone analyzed on virus data -- setwd("~/DATA/Data_Manja_RNAseq_Organoids_Merged/") d.raw <- read.csv("merged_gene_counts.csv", header=TRUE, row.names=1) dds <- DESeqDataSetFromMatrix(countData=d.raw, colData=colData, design=~condition+replicate) dim(counts(dds)) head(counts(dds), 10) rld <- rlogTransformation(dds) #We don't need to run DESeq(dds) before estimateSizeFactors(dds). In fact, the typical workflow in DESeq2 is the opposite: we usually run estimateSizeFactors(dds) (and other preprocessing functions) before running the main DESeq(dds) function. #The estimateSizeFactors function is used to calculate size factors for normalization, which corrects for differences in library size (i.e., the number of read counts) between samples. This normalization step is crucial to ensure that differences in gene expression aren't merely due to differences in sequencing depth between samples. #The DESeq function, on the other hand, performs the main differential expression analysis, comparing gene expression between different conditions or groups. #So, the typical workflow is: # - Create the DESeqDataSet object. # - Use estimateSizeFactors to normalize for library size. # - (Optionally, estimate dispersion with estimateDispersions if not using the full DESeq function later.) # - Use DESeq for the differential expression analysis. # - However, it's worth noting that if you run the main DESeq function directly after creating the DESeqDataSet object, it will automatically perform the normalization (using estimateSizeFactors) and dispersion estimation steps for you. In that case, there's no need to run estimateSizeFactors separately before DESeq. # draw simple pca and heatmap library(gplots) library("RColorBrewer") #mat <- assay(rld) #mm <- model.matrix(~condition, colData(rld)) #mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm) #assay(rld) <- mat # -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
draw 3D PCA plots.
library(gplots) library("RColorBrewer") library(ggplot2) data <- plotPCA(rld, intgroup=c("condition", "replicate"), returnData=TRUE) write.csv(data, file="plotPCA_data.csv") #calculate all PCs including PC3 with the following codes library(genefilter) ntop <- 500 rv <- rowVars(assay(rld)) select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))] mat <- t( assay(rld)[select, ] ) pc <- prcomp(mat) pc$x[,1:3] #df_pc <- data.frame(pc$x[,1:3]) df_pc <- data.frame(pc$x) identical(rownames(data), rownames(df_pc)) #-->TRUE data$PC1 <- NULL data$PC2 <- NULL merged_df <- merge(data, df_pc, by = "row.names") #merged_df <- merged_df[, -1] row.names(merged_df) <- merged_df$Row.names merged_df$Row.names <- NULL # remove the "name" column merged_df$name <- NULL merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","group","condition","replicate")] write.csv(merged_df, file="merged_df_10PCs.csv") summary(pc) #0.5333 0.2125 0.06852 #0.8026 0.09042 0.06578 #0.6577 0.1631 0.07106 #Proportion of Variance 0.6577 0.1631 0.07106 draw_3D.py #/usr/bin/convert PCA_3D.png -crop 2900x1600+250+700 PCA_3D_cropped.png # adjust proportion to real values in the following plot import plotly.graph_objects as go import pandas as pd from sklearn.decomposition import PCA import numpy as np # Read in data as a pandas dataframe #df = pd.DataFrame({ # 'PC1': [-13.999925, -12.504291, -12.443057, -13.065235, -17.316215], # 'PC2': [-1.498823, -3.342411, -6.067055, -8.205809, 3.293993], # 'PC3': [-3.335085, 15.207755, -14.725450, 15.078469, -6.917358], # 'condition': ['GFP d3', 'GFP d3', 'GFP d8', 'GFP d8', 'GFP+mCh d9/12'], # 'replicate': ['DI', 'DII', 'DI', 'DII', 'DI'] #}) df = pd.read_csv('merged_df_10PCs.csv', index_col=0, header=0) df['condition'] = df['condition'].replace("control", "control") df['condition'] = df['condition'].replace("HSV.d2", "day 2") df['condition'] = df['condition'].replace("HSV.d4", "day 4") df['condition'] = df['condition'].replace("HSV.d6", "day 6") df['condition'] = df['condition'].replace("HSV.d8", "day 8") # Fit PCA model to reduce data dimensions to 3 pca = PCA(n_components=3) pca.fit(df.iloc[:, :-3]) X_reduced = pca.transform(df.iloc[:, :-3]) # Get variance ratios explained_variance_ratio = pca.explained_variance_ratio_ # Add reduced data back to dataframe df['PC1'] = X_reduced[:, 0] df['PC2'] = X_reduced[:, 1] df['PC3'] = X_reduced[:, 2] # Create PCA plot with 3D scatter fig = go.Figure() ##ff7f00 condition_color_map = { 'control': 'rgb(100, 100, 100)', 'day 2': '#33a02c', 'day 4': '#1f78b4', 'day 6': '#e31a1c', 'day 8': 'magenta' } replicate_symbol_map = {'r1': 'circle', 'r2': 'diamond'} for replicate, replicate_symbol in replicate_symbol_map.items(): for condition, condition_color in condition_color_map.items(): mask = (df['condition'] == condition) & (df['replicate'] == replicate) fig.add_trace(go.Scatter3d(x=df.loc[mask, 'PC1'], y=df.loc[mask, 'PC2'], z=df.loc[mask, 'PC3'], mode='markers', name=f'{condition}' if replicate == 'r1' else None, legendgroup=f'{condition}', showlegend=True if replicate == 'r1' else False, marker=dict(size=6 if replicate_symbol in ['diamond'] else 10, opacity=0.8, color=condition_color, symbol=replicate_symbol))) for replicate, replicate_symbol in replicate_symbol_map.items(): fig.add_trace(go.Scatter3d(x=[None], y=[None], z=[None], mode='markers', name=replicate, legendgroup=f'{replicate}', showlegend=True, marker=dict(size=10, opacity=1, color='black', symbol=replicate_symbol), hoverinfo='none')) # Annotations for the legend blocks #TODO: calculate the PC values. #TODO: adjust the axis length according to the actual size of axis! fig.update_layout( annotations=[ dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False, text='Condition', font=dict(size=15)), dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False, text='Replicate', font=dict(size=15)) ], scene=dict( #aspectmode='cube', #xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 53% v.', scaleratio=0.53), #yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 21% v.', scaleratio=0.21), #zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 7% variance', scaleratio=0.07), aspectmode='manual', aspectratio=dict(x=explained_variance_ratio[0], y=explained_variance_ratio[1], z=explained_variance_ratio[2]), xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 53% v.', range=[min(df['PC1']), max(df['PC1'])]), yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 21% v.', range=[min(df['PC2']), max(df['PC2'])]), zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 7% variance', range=[min(df['PC3']), max(df['PC3'])]), bgcolor='white' ), margin=dict(l=5, r=5, b=5, t=0) # Adjust the margins to prevent clipping of axis titles ) #fig.show() fig.write_image("fig1.svg") fig.update_layout( annotations=[ dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False, text='Condition', font=dict(size=15)), dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False, text='Donor', font=dict(size=15)), dict(x=1.08, y=0.2, xref='paper', yref='paper', showarrow=False, text=f'PC3: {explained_variance_ratio[2]*100:.2f}% v.', font=dict(size=15), textangle=-90) ], scene=dict( aspectmode='manual', aspectratio=dict(x=explained_variance_ratio[0]*2, y=explained_variance_ratio[1]*2, z=explained_variance_ratio[2]*2), #, range=[min(df['PC1']), max(df['PC1'])] #, range=[min(df['PC2']), max(df['PC2'])] #, range=[min(df['PC3']), max(df['PC3'])] xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=f'PC1: {explained_variance_ratio[0]*100:.2f}% variance'), yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=f'PC2: {explained_variance_ratio[1]*100:.2f}% v.'), zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=''), bgcolor='white' ), margin=dict(l=0, r=0, b=0, t=0) # Adjust the margins to prevent clipping of axis titles ) fig.write_image("PCA_3D.svg") #/usr/bin/convert g235.png -crop 3250x1680+1+750 PCA_3D_.png -
(optional) estimate size factors and dispersion values.
#Size Factors: These are used to normalize the read counts across different samples. The size factor for a sample accounts for differences in sequencing depth (i.e., the total number of reads) and other technical biases between samples. After normalization with size factors, the counts should be comparable across samples. Size factors are usually calculated in a way that they reflect the median or mean ratio of gene expression levels between samples, assuming that most genes are not differentially expressed. #Dispersion: This refers to the variability or spread of gene expression measurements. In RNA-seq data analysis, each gene has its own dispersion value, which reflects how much the counts for that gene vary between different samples, more than what would be expected just due to the Poisson variation inherent in counting. Dispersion is important for accurately modeling the data and for detecting differentially expressed genes. #So in summary, size factors are specific to samples (used to make counts comparable across samples), and dispersion values are specific to genes (reflecting variability in gene expression). sizeFactors(dds) #NULL # Estimate size factors dds <- estimateSizeFactors(dds) # Estimate dispersions dds <- estimateDispersions(dds) #> sizeFactors(dds) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #2.3282468 2.0251928 1.8036883 1.3767551 0.9341929 1.0911693 0.5454526 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #0.4604461 0.5799834 0.6803681 # If alone with virus data, the following BUG occured: #Still NULL --> BUG --> using manual calculation method for sizeFactor calculation! HeLa_TO_r1 HeLa_TO_r2 0.9978755 1.1092227 data.frame(genes = rownames(dds), dispersions = dispersions(dds)) #Given the raw counts, the control_r1 and control_r2 samples seem to have a much lower sequencing depth (total read count) than the other samples. Therefore, when normalization methods are applied, the normalization factors for these control samples will be relatively high, boosting the normalized counts. 1/0.9978755=1.002129023 1/1.1092227= #bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r1Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r1.bw --binSize 10 --scaleFactor 1.002129023 --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r2Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r2.bw --binSize 10 --scaleFactor 0.901532217 --effectiveGenomeSize 2864785220 raw_counts <- counts(dds) normalized_counts <- counts(dds, normalized=TRUE) #write.table(raw_counts, file="raw_counts.txt", sep="\t", quote=F, col.names=NA) #write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA) #convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) -
compare the normalization methods
#The provided code indeed simulates the normalization method used by DESeq, which is known as the "Relative Log Expression (RLE)" normalization. The core idea behind this method is to scale the counts in each sample (column) by a size factor derived from the geometric means across all samples. #Here's a step-by-step breakdown of the code: # 1. estimSf function: # - The counts matrix is retrieved from the DESeq dataset. # - For each gene (row), the geometric mean across all samples is computed. # - Counts are divided by their respective gene's geometric mean. # - For each sample (column), the median of these ratios is computed. This median serves as the size factor for the sample. # 2. Once size factors are computed, the counts in the original matrix are then divided by these size factors to get the normalized counts. ### cds is a countDataset estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) # round(estimSf(dds),6) is manually calculated sizeFactors of head(estimSf(dds)) all(round(estimSf(dds),6) == round(sizeFactors(dds), 6)) ## Checking the normalization png("normalization_small.png", width=800, height=600) epsilon <- 1 # pseudo-count to avoid problems with log(0) par(mfrow=c(1,2),cex.lab=0.7) boxplot(log2(raw_counts+epsilon), cex.axis=0.7, las=1, xlab="log2(raw counts)", horizontal=TRUE, main="Raw counts") boxplot(log2(normalized_counts+epsilon), cex.axis=0.7, las=1, xlab="log2(normalized counts)", horizontal=TRUE, main="Normalized counts") #boxplot(log10(assays(dds)[["cooks"]]), range=0, las=2) #plotDensity(log2(counts(dds.norm)+epsilon), col=col.pheno.selected, # xlab="log2(counts)", cex.lab=0.7, panel.first=grid()) #plotDensity(log2(counts(dds.norm, normalized=TRUE)+epsilon), col=col.pheno.selected, # xlab="log2(normalized counts)", cex.lab=0.7, panel.first=grid()) dev.off() # since we Gene-level differential expression analysis with DESeq2, the splicing plays no role in the analysis! # 用nanopore 可以 compare transcript length distribution. 有可能Cellline很长,Extracellular vesicles (EVs)很短! #In the folloing code, we compare different normalization methods, however, the function estimateSizeFactors(dge) doesn't work. I want replace the methods with the method implemented above. Please update the code #uding five norm factors from the following five normalization methods. How to replace the last command " sizeFactors(dds) <- dge2" for the five method? library(ggplot2) library(gridExtra) library(reshape2) library(mixOmics) library(RColorBrewer) library(DESeq) library(edgeR) library(VennDiagram) library(devtools) raw_counts_wn <- raw_counts[rowSums(raw_counts) > 0, ] dim(raw_counts_wn) #--Raw counts-- pseudo_counts <- log2(raw_counts_wn + 1) head(pseudo_counts) df_raw <- melt(pseudo_counts, id = rownames(raw_counts_wn)) names(df_raw)[1:2]<- c("id", "sample") df_raw$method <- rep("Raw counts", nrow(df_raw)) head(df_raw) #--DESeq-- cData = data.frame(row.names=colnames(raw_counts_wn), condition=condition) dge<-DESeqDataSetFromMatrix(countData=raw_counts_wn, colData=cData, design=~condition) dge <- estimateSizeFactors(dge) sizeFactors(dge) # Use your function to get the size factors sizeFactorsDGE <- estimSf(dge) isTRUE(all.equal(sizeFactors(dge), sizeFactorsDGE)) # Test the explained method using sweep to simulate the internal process of dge deseq_normcount <- counts(dge, normalized = TRUE) test_normcount <- sweep(raw_counts_wn, 2, sizeFactors(dge), "/") sum(test_normcount != deseq_normcount) pseudo_deseq <- log2(deseq_normcount + 1) df_deseq <- melt(pseudo_deseq, id = rownames(raw_counts_wn)) names(df_deseq)[1:2]<- c("id", "sample") df_deseq$method <- rep("DESeq (RLE)", nrow(df_raw)) #--edgeR-- dge2 <- DGEList(raw_counts_wn) dge2 dge2$samples #--Total count-- pseudo_TC <- log2(cpm(dge2) + 1) df_TC <- melt(pseudo_TC, id = rownames(raw_counts_wn)) names(df_TC)[1:2] <- c ("id", "sample") df_TC$method <- rep("TC", nrow(df_TC)) ##--RPKM-- #gene_lengths_wn <- gene_lengths[rowSums(raw_counts) > 0] #pseudo_RPKM <- log2(rpkm(dge2, gene.length = gene_lengths_wn) + 1) #df_RPKM <- melt(pseudo_RPKM, id = rownames(raw_counts_wn)) #names(df_RPKM)[1:2] <- c ("id", "sample") #df_RPKM$method <- rep("RPKM", nrow(df_RPKM)) #--Upper quartile-- dge2 <- calcNormFactors(dge2, method = "upperquartile") dge2$samples test_normcount <- sweep(dge2$counts, 2, dge2$samples$lib.size*dge2$samples$norm.factors / 10^6, "/") range(as.vector(test_normcount - cpm(dge2))) pseudo_UQ <- log2(cpm(dge2) + 1) df_UQ <- melt(pseudo_UQ, id = rownames(raw_counts_wn)) names(df_UQ)[1:2] <- c ("id", "sample") df_UQ$method <- rep("UQ", nrow(df_UQ)) #--TMM-- dge2 <- calcNormFactors(dge2, method = "TMM") dge2$samples pseudo_TMM <- log2(cpm(dge2) + 1) df_TMM <- melt(pseudo_TMM, id = rownames(raw_counts_wn)) names(df_TMM)[1:2] <- c ("id", "sample") #MODIFIED! df_TMM$method <- rep("TMM", nrow(df_TMM)) #TMM #--Comparison-- png("normalization.png", width=800, height=600) df_allnorm <- rbind(df_raw, df_deseq, df_TC, df_UQ, df_TMM) df_allnorm$method <- factor(df_allnorm$method, levels = c("Raw counts", "DESeq (RLE)", "TC", "UQ", "TMM")) #df_allnorm <- rbind(df_raw, df_TMM) #df_allnorm$method <- factor(df_allnorm$method, levels = c("Raw counts", "DESeq (RLE)")) p <- ggplot(data=df_allnorm, aes(x=sample, y=value, fill=method)) p <- p + geom_boxplot() p <- p + theme_bw() p <- p + ggtitle("Boxplots of normalized pseudo counts\n for all samples by normalization methods") p <- p + facet_grid(. ~ method) p <- p + ylab(expression(log[2] ~ (normalized ~ count + 1))) + xlab("") p <- p + theme(title = element_text(size=10), axis.text.x = element_blank(), axis.ticks.x = element_blank()) print(p) dev.off() #Assign a normalization size factor for downstream analysis from "Raw counts", "DESeq (RLE)", "TC", "UQ", "TMM") #You will need to repeat the DESeq2 analysis steps (like DESeq()) for each set of size factors to see how the results change with different normalization methods. #Note: This approach allows you to apply normalization factors from various methods to DESeq2. But keep in mind that each normalization method was developed with a specific intent and assumptions. Combining normalization methods from one package (like edgeR's TMM) with differential analysis from another package (like DESeq2) might not always be theoretically sound. Always interpret results with caution and preferably in consultation with domain experts. #DESeq (RLE) normalization: sizeFactors(dds) <- sizeFactors(dge) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #0.01996676 0.03367626 0.33493034 0.65395381 4.96825596 4.14671012 4.07416461 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #4.43844964 5.96130192 3.75356239 #Total Count normalization: This is actually just scaling by the total library size. In DESeq2, this would be equivalent to: sizeFactors(dds) <- colSums(raw_counts_wn) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 # 14594 18992 182152 374750 3404540 2521691 2646741 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 # 3207708 3873254 2430625 identical(colnames(dds), rownames(dge2$samples)) #Upper Quartile (UQ) normalization: uq_factors <- dge2$samples$norm.factors * dge2$samples$lib.size / 10^6 names(uq_factors) <- rownames(dge2$samples) sizeFactors(dds) <- uq_factors #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #0.01578156 0.02060450 0.19559938 0.39108853 3.27858657 2.38221969 2.61381808 # HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #3.03441572 3.65683812 2.30405047 #TMM normalization: tmm_factors <- dge2$samples$norm.factors names(tmm_factors) <- rownames(dge2$samples) sizeFactors(dds) <- tmm_factors #> tmm_factors #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #1.0813729 1.0849043 1.0738250 1.0435985 0.9630043 0.9446914 0.9875610 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #0.9459763 0.9441256 0.9479251 #Given the raw counts, the control_r1 and control_r2 samples seem to have a much lower sequencing depth (total read count) than the other samples. Therefore, when normalization methods such as TMM (Trimmed Mean of M-values) or UQ (Upper Quartile normalization) are applied, the normalization factors for these control samples will be relatively high, boosting the normalized counts. #To better understand the situation: # - Check the sequencing depth of each sample. Sum the raw counts across all genes for each sample to get the total read count. If the control samples have a substantially lower total read count, then normalization methods will try to adjust for this discrepancy. # - Consider the normalization method: Different normalization methods might provide slightly different results. TMM normalization, for example, tries to adjust for the compositional differences between samples. It's common to observe larger normalization factors for samples with a lower total read count. # - Visualize the data: MA plots, box plots, or density plots of the raw and normalized counts can help understand the distribution of counts and the effect of normalization. # ---- Adapt the following code to print the normalized_counts using tmm_factors ---- # RLE size factors rle_factors <- sizeFactors(dge) # Print side by side data.frame(RLE = rle_factors, TMM = tmm_factors) sizeFactors(dds) <- rle_factors normalized_counts_rle <- counts(dds, normalized=TRUE) sizeFactors(dds) <- tmm_factors #is it possible in the following command, the calculate de novo normalization factors and ignore the given tmm normalization factors? normalized_counts_tmm <- counts(dds, normalized=TRUE) difference_matrix <- normalized_counts_tmm - normalized_counts_rle all(difference_matrix == 0) -
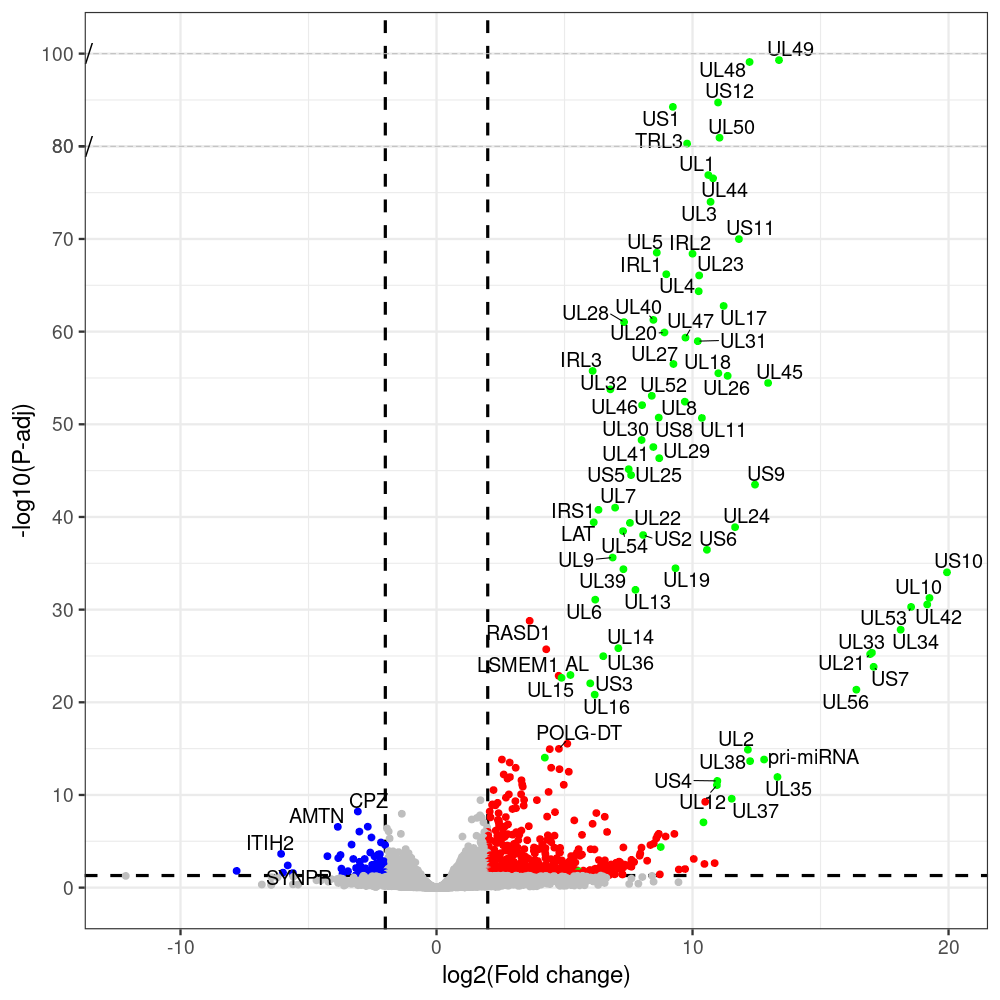
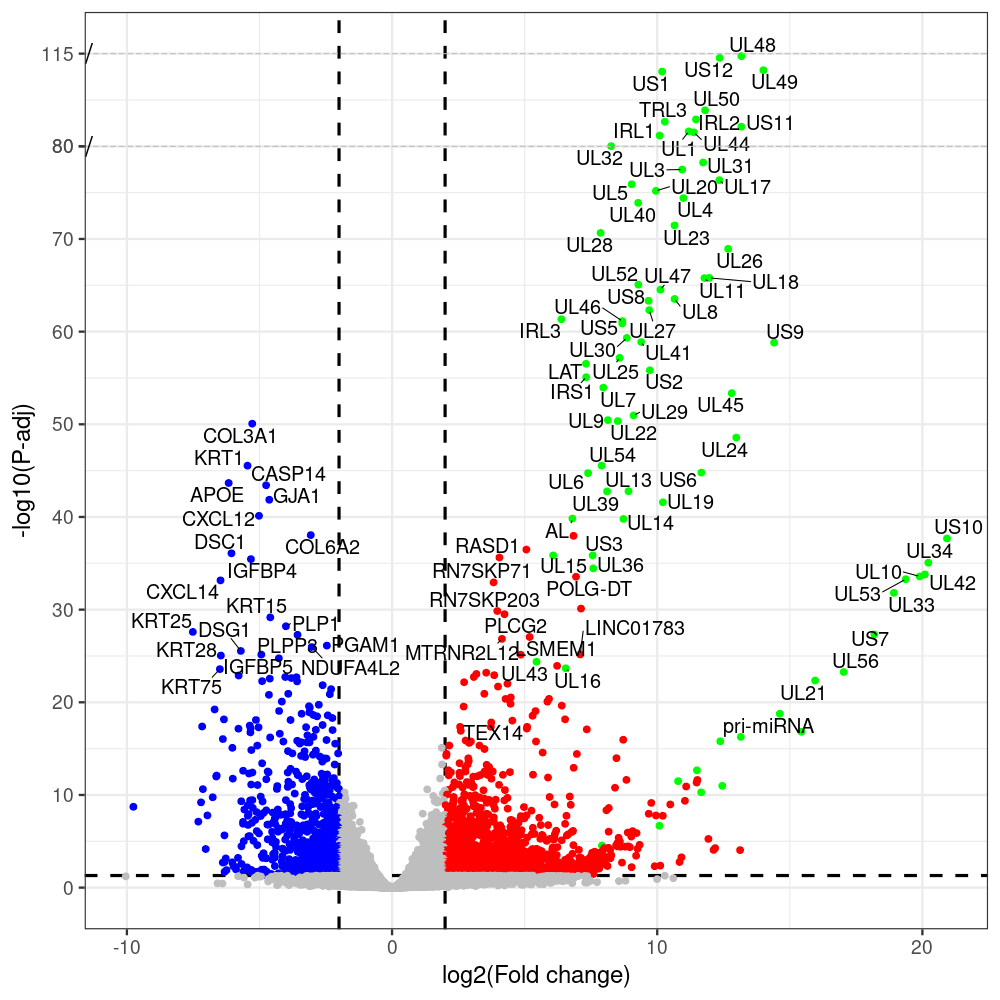
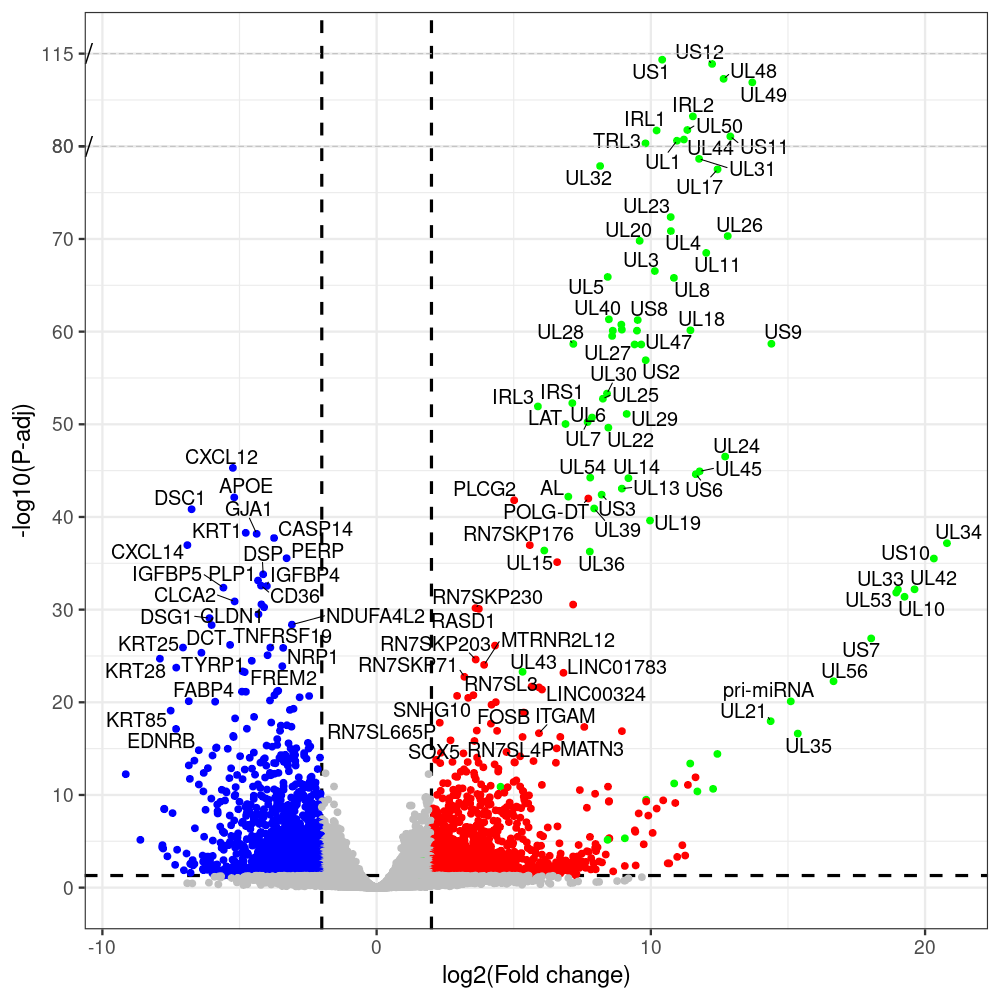
select the differentially expressed genes
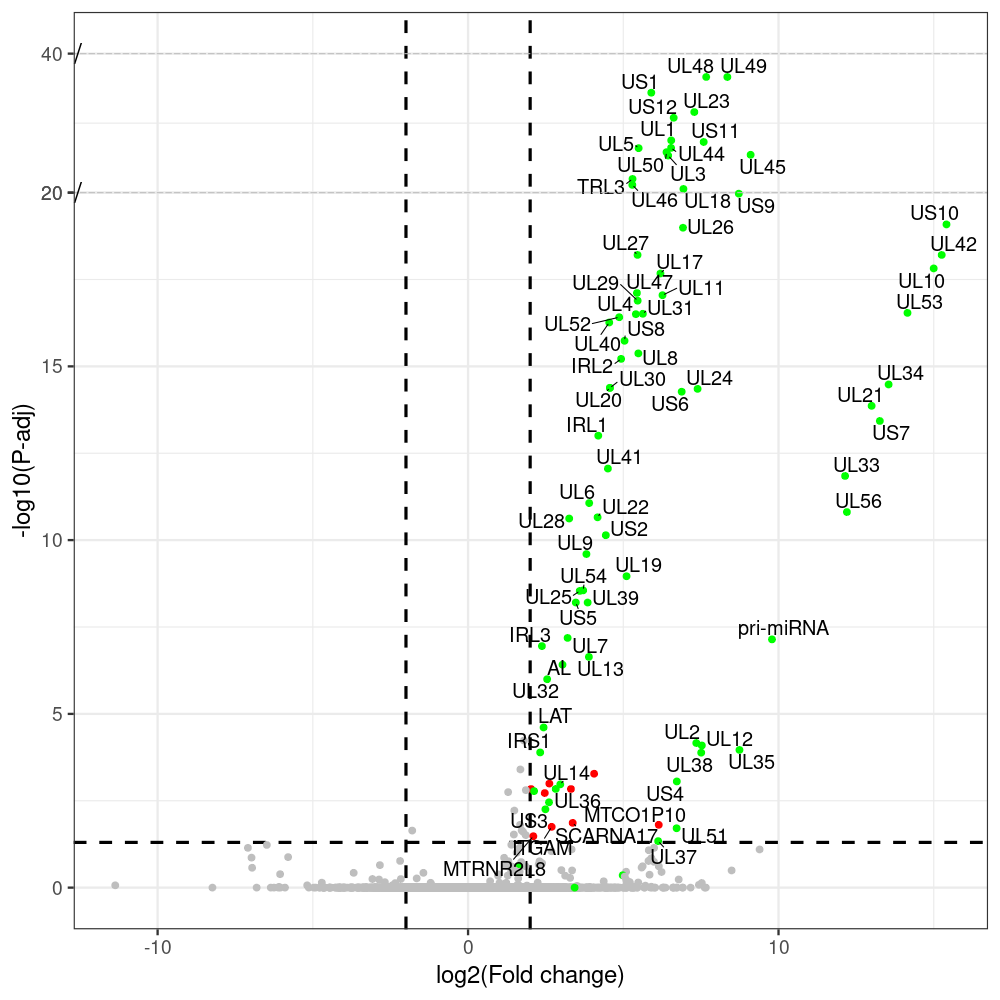
#https://galaxyproject.eu/posts/2020/08/22/three-steps-to-galaxify-your-tool/ #https://www.biostars.org/p/282295/ #https://www.biostars.org/p/335751/ #> condition # [1] control control HSV.d2 HSV.d2 HSV.d4 HSV.d4 HSV.d6 HSV.d6 HSV.d8 HSV.d8 #Levels: control HSV.d2 HSV.d4 HSV.d6 HSV.d8 #CONSOLE: mkdir /home/jhuang/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon/degenes setwd("/home/jhuang/DATA/Data_Manja_RNAseq_Organoids_Merged") #---- relevel to control ---- dds$condition <- relevel(dds$condition, "control") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d2_vs_control","HSV.d4_vs_control", "HSV.d6_vs_control", "HSV.d8_vs_control") dds$condition <- relevel(dds$condition, "HSV.d2") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d4_vs_HSV.d2", "HSV.d6_vs_HSV.d2", "HSV.d8_vs_HSV.d2") dds$condition <- relevel(dds$condition, "HSV.d4") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d6_vs_HSV.d4", "HSV.d8_vs_HSV.d4") dds$condition <- relevel(dds$condition, "HSV.d6") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d8_vs_HSV.d6") library(biomaRt) listEnsembl() listMarts() #--> total 69, 27 GRCh38.p7 and 39 GRCm38.p4 ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="104") datasets <- listDatasets(ensembl) # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(20, 40) compressed_range <- c(20.0, 24.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 20, by=5) y_breaks_compressed <- c(20.0, 24.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 20, by=5) y_labels_compressed <- c(20, 40) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- HSV.d8_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # -- HSV.d6_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # -- HSV.d4_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 100) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 100) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # -- HSV.d2_vs_control.png -- # Define the original and compressed ranges original_range <- c(20, 40) compressed_range <- c(20.0, 24.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 20, by=5) y_breaks_compressed <- c(20.0, 24.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 20, by=5) y_labels_compressed <- c(20, 40) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) #under DIR degenes under KONSOLE for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "~/Tools/csv2xls-0.4/csv_to_xls.py ${i}-all_annotated.txt ${i}-up_annotated.txt ${i}-down_annotated.txt -d$',' -o ${i}.xls;"; done -
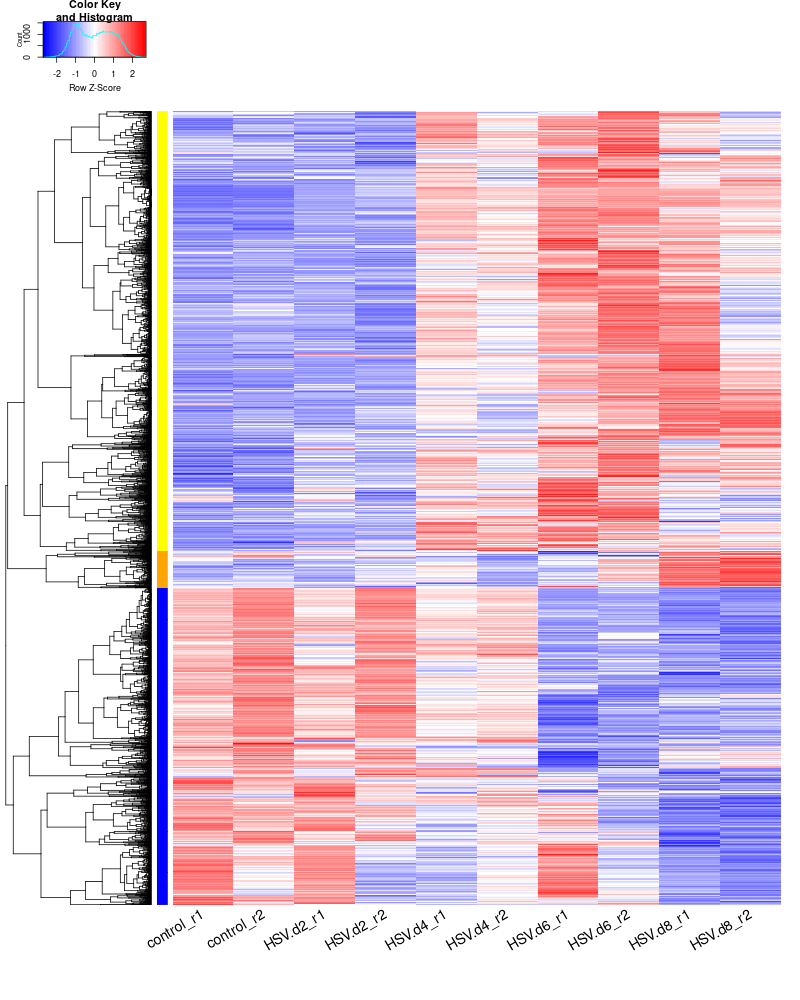
clustering the genes and draw heatmap
for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "cut -d',' -f1-1 ${i}-up_annotated.txt > ${i}-up.id"; echo "cut -d',' -f1-1 ${i}-down_annotated.txt > ${i}-down.id"; done cat *.id | sort -u > ids #add Gene_Id in the first line, delete the "" GOI <- read.csv("ids")$Gene_Id #4647 RNASeq.NoCellLine <- assay(rld) #install.packages("gplots") library("gplots") #clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman datamat = RNASeq.NoCellLine[GOI, ] #datamat = RNASeq.NoCellLine write.csv(as.data.frame(datamat), file ="gene_expressions.txt") constant_rows <- apply(datamat, 1, function(row) var(row) == 0) if(any(constant_rows)) { cat("Removing", sum(constant_rows), "constant rows.\n") datamat <- datamat[!constant_rows, ] } hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.05) mycol = c("YELLOW", "BLUE", "ORANGE", "MAGENTA", "CYAN", "RED", "GREEN", "MAROON", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN", "LIGHTRED", "LIGHTGREEN"); mycol = mycol[as.vector(mycl)] #png("DEGs_heatmap.png", width=900, height=800) #cex.lab=10, labRow="", png("DEGs_heatmap.png", width=800, height=1000) heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',labRow="", scale='row',trace='none',col=bluered(75), cexCol=1.8, RowSideColors = mycol, margins=c(10,2), cexRow=1.5, srtCol=30, lhei = c(1, 8), lwid=c(2, 8)) #rownames(datamat) #heatmap.2(datamat, Rowv=as.dendrogram(hr), col=bluered(75), scale="row", RowSideColors=mycol, trace="none", margin=c(5,5), sepwidth=c(0,0), dendrogram = 'row', Colv = 'false', density.info='none', labRow="", srtCol=30, lhei=c(0.1,2)) dev.off() #### cluster members ##### write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt') write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt') write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt') write.csv(names(subset(mycl, mycl == '4')),file='cluster4.txt') #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o DEGs_heatmap_cluster_members.xls ~/Tools/csv2xls-0.4/csv_to_xls.py \ significant_gene_expressions.txt \ -d',' -o DEGs_heatmap_expression_data.xls; #### cluster members (advanced) ##### subset_1<-names(subset(mycl, mycl == '1')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_1, ]) #2575 subset_2<-names(subset(mycl, mycl == '2')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_2, ]) #1855 subset_3<-names(subset(mycl, mycl == '3')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_3, ]) #217 subset_4<-names(subset(mycl, mycl == '4')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_4, ]) # subset_5<-names(subset(mycl, mycl == '5')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_5, ]) # # Initialize an empty data frame for the annotated data annotated_data <- data.frame() # Determine total number of genes total_genes <- length(rownames(data)) # Loop through each gene to annotate for (i in 1:total_genes) { gene <- rownames(data)[i] result <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = gene, mart = ensembl) # If multiple rows are returned, take the first one if (nrow(result) > 1) { result <- result[1, ] } # Check if the result is empty if (nrow(result) == 0) { result <- data.frame(ensembl_gene_id = gene, external_gene_name = NA, gene_biotype = NA, entrezgene_id = NA, chromosome_name = NA, start_position = NA, end_position = NA, strand = NA, description = NA) } # Transpose expression values expression_values <- t(data.frame(t(data[gene, ]))) colnames(expression_values) <- colnames(data) # Combine gene information and expression data combined_result <- cbind(result, expression_values) # Append to the final dataframe annotated_data <- rbind(annotated_data, combined_result) # Print progress every 100 genes if (i %% 100 == 0) { cat(sprintf("Processed gene %d out of %d\n", i, total_genes)) } } # Save the annotated data to a new CSV file #write.csv(annotated_data, "cluster1_YELLOW.csv", row.names=FALSE) write.csv(annotated_data, "cluster2_DARKBLUE.csv", row.names=FALSE) write.csv(annotated_data, "cluster3_DARKORANGE.csv", row.names=FALSE) write.csv(annotated_data, "cluster4_DARKMAGENTA.csv", row.names=FALSE) write.csv(annotated_data, "cluster5_DARKCYAN.csv", row.names=FALSE) #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.csv -d',' -o DEGs_heatmap_clusters.xls -
code of differential gene analysis for clist <- c("HSV.d2_vs_control","HSV.d4_vs_control", "HSV.d6_vs_control", "HSV.d8_vs_control")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d2_vs_control.png -- # Define the original and compressed ranges original_range <- c(20, 40) compressed_range <- c(20.0, 24.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 20, by=5) y_breaks_compressed <- c(20.0, 24.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 20, by=5) y_labels_compressed <- c(20, 40) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d4_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 100) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 100) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[3] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d6_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[4] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d8_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() -
code of differential gene analysis for clist <- c("HSV.d4_vs_HSV.d2", "HSV.d6_vs_HSV.d2", "HSV.d8_vs_HSV.d2")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(15, 20) compressed_range <- c(15.0, 20.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 15, by=5) y_breaks_compressed <- c(15.0, 20.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 15, by=5) y_labels_compressed <- c(15, 20) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=10) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=10) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[3] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=10) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=10) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() -
code of differential gene analysis for clist <- c("HSV.d6_vs_HSV.d4", "HSV.d8_vs_HSV.d4")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(15, 20) compressed_range <- c(15.0, 20.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 15, by=5) y_breaks_compressed <- c(15.0, 20.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 15, by=5) y_labels_compressed <- c(15, 20) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=5) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=5) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() -
code of differential gene analysis for clist <- c("HSV.d8_vs_HSV.d6")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=5) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=5) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off()
Plotting Alpha Diversities from 16S rRNA Sequencing Data
Plot Chao1 richness estimator, Observed OTUs, Shannon index, and Phylogenetic diversity. Regroup together samples from the same group.
library("readxl") # necessary to import the data from Excel file
library("ggplot2") # graphics
library("picante")
library("microbiome") # data analysis and visualisation
library("phyloseq") # also the basis of data object. Data analysis and visualisation
library("ggpubr") # publication quality figures, based on ggplot2
library("dplyr") # data handling, filter and reformat data frames
library("RColorBrewer") # nice color options
library("heatmaply")
library(vegan)
library(gplots)
ps.ng.tax <- readRDS("ps.ng.tax.rds")
hmp.meta <- meta(ps.ng.tax)
hmp.meta$sam_name <- rownames(hmp.meta)
hmp.div_qiime <- read.csv("adiv_even.txt", sep="\t")
colnames(hmp.div_qiime) <- c("sam_name", "chao1", "observed_otus", "shannon", "PD_whole_tree")
row.names(hmp.div_qiime) <- hmp.div_qiime$sam_name
div.df <- merge(hmp.div_qiime, hmp.meta, by = "sam_name")
div.df2 <- div.df[, c("Group", "chao1", "shannon", "observed_otus", "PD_whole_tree")]
colnames(div.df2) <- c("Group", "Chao-1", "Shannon", "OTU", "Phylogenetic Diversity")
#options(max.print=999999)
stat.test.Shannon <- compare_means(
Shannon ~ Group, data = div.df2,
method = "t.test"
)
div_df_melt <- reshape2::melt(div.df2)
p <- ggboxplot(div_df_melt, x = "Group", y = "value",
facet.by = "variable",
scales = "free",
width = 0.5,
fill = "gray", legend= "right")
p3 <- p +
stat_compare_means(
method="t.test",
comparisons = list(c("P16-P20.Foot", "P16-P20.Nose"), c("AH-XN.LH", "AH-XN.Nose"), c("AH-XN.NLH", "AH-XN.Nose")),
label = "p.signif",
symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")),
) +
theme(axis.text.x = element_text(angle = 30, hjust = 1)) # add this line to rotate x-axis text
png("alpha_diversity.png", width=1000, height=800)
print(p3)
dev.off()
# Filter the data frame for each plot
groups_plot_1 <- c("AH-XN.LH", "AH-XN.NLH", "AH-XN.Nose")
groups_plot_2 <- setdiff(unique(div_df_melt$Group), groups_plot_1)
div_df_melt_plot_1 <- div_df_melt %>% filter(Group %in% groups_plot_1)
div_df_melt_plot_2 <- div_df_melt %>% filter(Group %in% groups_plot_2)
# Create and save Plot 1
div_df_melt_plot_1 <- div_df_melt_plot_1 %>% rename(Value = value)
p <- ggboxplot(div_df_melt_plot_1, x = "Group", y = "Value",
facet.by = "variable",
scales = "free",
width = 0.5,
fill = "gray", legend= "right")
p1 <- p +
stat_compare_means(
method="t.test",
comparisons = list(c("AH-XN.LH", "AH-XN.Nose"), c("AH-XN.NLH", "AH-XN.Nose")),
label = "p.signif",
symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")),
) +
theme(axis.text.x = element_text(angle = 15, hjust = 1)) # add this line to rotate x-axis text
#png("alpha_diversity1.png", width=800, height=600)
print(p1)
#dev.off()
ggsave("alpha_diversity1.png", device="png", height = 8, width = 8)
#/usr/bin/convert alpha_diversity1.png -resize 800x800 alpha_diversity1_resized.png
# Create and save Plot 2
div_df_melt_plot_2 <- div_df_melt_plot_2 %>% rename(Value = value)
p <- ggboxplot(div_df_melt_plot_2, x = "Group", y = "Value",
facet.by = "variable",
scales = "free",
width = 0.5,
fill = "gray", legend= "right")
p2 <- p +
stat_compare_means(
method="t.test",
comparisons = list(c("P16-P20.Foot", "P16-P20.Nose")),
label = "p.signif",
symnum.args <- list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns")),
) +
theme(axis.text.x = element_text(angle = 15, hjust = 1)) # add this line to rotate x-axis text
ggsave("alpha_diversity2.png", device="png", height = 8, width = 8)
#/usr/bin/convert alpha_diversity2.png -resize 800x800 alpha_diversity2_resized.png
#Using the function create_plot (Deprecated!)
#create_plot <- function(data, comparisons = NULL) {
# p <- ggboxplot(data, x = "Group", y = "Value",
# facet.by = "variable",
# scales = "free",
# width = 0.5,
# fill = "gray", legend = "right") +
# theme(axis.text.x = element_text(angle = 30, hjust = 1), strip.text = element_text(size = 15))
# #theme(axis.title.x = element_text(angle=30, hjust=1, size = 15), # Increase size of X-axis title
# # axis.title.y = element_text(size = 15), # Increase size of Y-axis title
# # strip.text = element_text(size = 15), # Increase size of Facet labels
# # axis.text.x = element_text(size = 12), # Increase size of X-axis text
# # axis.text.y = element_text(size = 12)) # Increase size of Y-axis text
#
# if (!is.null(comparisons)) {
# for (comp in comparisons) {
# if (all(comp %in% unique(data$Group))) {
# p <- p +
# stat_compare_means(
# method = "t.test",
# comparisons = list(comp),
# label = "p.signif",
# symnum.args = list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, 1), symbols = c("****", "***", "**", "*", "ns"))
# )
# } else {
# warning("One or more groups in the comparison are not present in the data: ", paste(comp, collapse = ", "))
# }
# }
# }
#
# return(p)
#}
#div_df_melt_plot_2 <- div_df_melt_plot_2 %>% rename(Value = value)
#p2 <- create_plot(div_df_melt_plot_2, list(c("P16-P20.Foot", "P16-P20.Nose")))
#png("alpha_diversity_plot_2.png", width=1000, height=800)
#print(p2)
#dev.off()GSVA-plot for carotis nanoString data
-
preparing exprs for gsva(exprs, selected_geneSets, method=”gsva”)
#Input: Images/*png and (hard-coded) /mnt/Samsung_T5/Data_Susanne_spatialRNA/M666_UKE_Hamburg_box/ dccs/*.dcc, pkcs3/all.zip, and annotation/M666_All_Data_SP.xlsx library("rmarkdown") library("tidyverse") library(rmarkdown) library("GeomxTools") library("GeoMxWorkflows") library("NanoStringNCTools") setwd("/home/jhuang/DATA/Data_Susanne_Carotis_spatialRNA_PUBLISHING/run_2023_2_GSVA") render("run.Rmd", c("html_document")) #identical(exprs(target_m666Data), assay(target_m666Data, "exprs")): contains the gene expression values that have been both normalized and log-transformed, which are often used for downstream analysis and visualization in many genomic studies. #assay(target_m666Data, "log_q"): These are likely log2-transformed values of the quantile-normalized data. Quantile normalization ensures that the distributions of gene expression values across samples are the same, and taking the log2 of these normalized values is a common step in the analysis of high-throughput gene expression data. It's a way to make the data more suitable for downstream statistical analysis and visualization. #assay(target_m666Data, "q_norm"): This typically represents the quantile-normalized gene expression values. Quantile normalization adjusts the expression values in each sample so that they have the same distribution. This normalization method helps remove systematic biases and makes the data more comparable across samples. # For the following calculation, the GSVA input requires a gene expression matrix 'exprs' where rows are genes and columns are samples. This matrix must be in non-log space. exprs <- exprs(target_m666Data) #> dim(exprs) #[1] 18677 45 #exprs <- exprs(filtered_or_neg_target_m666Data) -
preparing selected_geneSets in gsva(exprs, selected_geneSets, method=”gsva”)
#Input: Signatures.xls library(readxl) library(gridExtra) library(ggplot2) library(GSVA) # Paths to the Excel files file_paths <- list("Signatures.xls", "Signatures_additional.xls") # Get sheet names for each file sheet_names_list <- lapply(file_paths, excel_sheets) # Initialize an empty list to hold gene sets geneSets <- list() # Loop over each file path and its corresponding sheet names for (i in 1:length(file_paths)) { file_path <- file_paths[[i]] sheet_names <- sheet_names_list[[i]] # Loop over each sheet, extract the ENSEMBL IDs, and add to the list for (sheet in sheet_names) { # Read the sheet data <- read_excel(file_path, sheet = sheet) # Process the GeneSet names (replacing spaces with underscores, for example) gene_set_name <- gsub(" ", "_", unique(data$GeneSet)[1]) # Add ENSEMBL IDs to the list geneSets[[gene_set_name]] <- unique(as.character(data$geneSymbol)) } } # Print the result to check summary(geneSets) #desired_geneSets <- c("Monocytes", "Plasma_cells", "T_regs", "Cyt._act._T_cells", "Neutrophils", "Inflammatory_neutrophils", "Suppressive_neutrophils", "LDG", "CD40_activated") desired_geneSets <- c("IFN", "TNF", "IL-6R_complex", "IL-1_cytokines", "Pro-inflam._IL-1", "Monocyte_secreted", "Apoptosis", "NFkB_complex", "NLRP3_inflammasome") selected_geneSets <- geneSets[desired_geneSets] # Print the selected gene sets print(selected_geneSets) -
prepare violin plots for 1_vs_3
# 1. Compute GSVA scores: gsva_scores <- gsva(exprs, selected_geneSets, method="gsva") # 2. Convert to data.frame for ggplot: gsva_df <- as.data.frame(t(gsva_scores)) # 3. Add conditions to gsva_df: identical(rownames(pData(target_m666Data)), rownames(gsva_df)) gsva_df$Condition <- pData(target_m666Data)$Grp #identical(rownames(gsva_df_filtered), rownames(pData(target_m666Data)) ) gsva_df$SampleID <- pData(target_m666Data)$SampleID # 4. Filter the gsva_df to retain only the desired conditions: #group 1 vs. group 3 in the nanostring data gsva_df_filtered <- gsva_df[gsva_df$Condition %in% c("1", "3"), ] # 5. Define a function to plot violin plots: # Define custom colors custom_colors <- c("Group1" = "lightblue", "Group1a" = "red", "Group3" = "grey") #To implement the custom colors, and make the adjustments to abbreviate "Inflammatory" and "Suppressive", as well as increase the font size for the groups on the x-axis, we can modify the plot_violin function as follows: gsva_df_filtered$Condition <- gsub("1", "Group1", gsva_df_filtered$Condition) gsva_df_filtered$Condition <- gsub("3", "Group3", gsva_df_filtered$Condition) gsva_df_filtered$Condition <- factor(gsva_df_filtered$Condition, levels = c("Group1", "Group3")) plot_violin <- function(data, gene_name) { # Calculate the t-test p-value for the two conditions condition1_data <- data[data$Condition == "Group1", gene_name] condition2_data <- data[data$Condition == "Group3", gene_name] p_value <- t.test(condition1_data, condition2_data)$p.value # Convert p-value to annotation p_annotation <- ifelse(p_value < 0.01, "**", ifelse(p_value < 0.05, "*", "")) rounded_p_value <- paste0("p = ", round(p_value, 2)) plot_title <- gsub("_", " ", gene_name) p <- ggplot(data, aes(x=Condition, y=!!sym(gene_name), fill=Condition)) + geom_violin(linewidth=1.2) + scale_fill_manual(values = custom_colors) + labs(title=plot_title, y="GSVA Score") + ylim(-1, 1) + theme_light() + theme( axis.title.x = element_text(size=12), axis.title.y = element_text(size=12), axis.text.x = element_text(size=10), axis.text.y = element_text(size=10), plot.title = element_text(size=12, hjust=0.5), legend.position = "none" # Hide legend since the colors are self-explanatory ) # Add p-value annotation to the plot p <- p + annotate("text", x=1.5, y=0.9, label=paste0(p_annotation, " ", rounded_p_value), size=5, hjust=0.5) return(p) } # 6. Generate the list of plots in a predefined order: #desired_order <- c("Monocytes", "Plasma_cells", "T_regs", "Cyt._act._T_cells", "Neutrophils", "Inflammatory_neutrophils", "Suppressive_neutrophils", "LDG", "CD40_activated") desired_order <- c("IFN", "TNF", "IL-6R_complex", "IL-1_cytokines", "Pro-inflam._IL-1", "Monocyte_secreted", "Apoptosis", "NFkB_complex", "NLRP3_inflammasome") genes <- colnames(gsva_df_filtered)[!colnames(gsva_df_filtered) %in% "Condition"] genes <- genes[match(desired_order, genes)] genes <- genes[!is.na(genes)] second_row_plots <- lapply(genes, function(gene) plot_violin(gsva_df_filtered, gene)) -
prepare violin plots for 1a_vs_3
# 2. Convert to data.frame for ggplot: gsva_df <- as.data.frame(t(gsva_scores)) # 3. Add conditions to gsva_df: identical(rownames(pData(target_m666Data)), rownames(gsva_df)) gsva_df$Condition <- pData(target_m666Data)$Group #identical(rownames(gsva_df_filtered), rownames(pData(target_m666Data)) ) gsva_df$SampleID <- pData(target_m666Data)$SampleID # 4. Filter the gsva_df to retain only the desired conditions: #group 1 vs. group 3 in the nanostring data gsva_df_filtered <- gsva_df[gsva_df$Condition %in% c("1a", "3"), ] # 5. Define a function to plot violin plots: # Update the condition levels in gsva_df_filtered to ensure the desired order on x-axis: gsva_df_filtered$Condition <- gsub("1a", "Group1a", gsva_df_filtered$Condition) gsva_df_filtered$Condition <- gsub("3", "Group3", gsva_df_filtered$Condition) gsva_df_filtered$Condition <- factor(gsva_df_filtered$Condition, levels = c("Group1a", "Group3")) plot_violin <- function(data, gene_name) { # Calculate the t-test p-value for the two conditions condition1_data <- data[data$Condition == "Group1a", gene_name] condition2_data <- data[data$Condition == "Group3", gene_name] p_value <- t.test(condition1_data, condition2_data)$p.value # Convert p-value to annotation p_annotation <- ifelse(p_value < 0.01, "**", ifelse(p_value < 0.05, "*", "")) rounded_p_value <- paste0("p = ", round(p_value, 2)) plot_title <- gsub("_", " ", gene_name) p <- ggplot(data, aes(x=Condition, y=!!sym(gene_name), fill=Condition)) + geom_violin(linewidth=1.2) + scale_fill_manual(values = custom_colors) + labs(title=plot_title, y="GSVA Score") + ylim(-1, 1) + theme_light() + theme( axis.title.x = element_text(size=12), axis.title.y = element_text(size=12), axis.text.x = element_text(size=10), axis.text.y = element_text(size=10), plot.title = element_text(size=12, hjust=0.5), legend.position = "none" # Hide legend since the colors are self-explanatory ) # Add p-value annotation to the plot p <- p + annotate("text", x=1.5, y=0.9, label=paste0(p_annotation, " ", rounded_p_value), size=5, hjust=0.5) return(p) } # 6. Generate the list of plots in a predefined order: genes <- colnames(gsva_df_filtered)[!colnames(gsva_df_filtered) %in% "Condition"] genes <- genes[match(desired_order, genes)] genes <- genes[!is.na(genes)] first_row_plots <- lapply(genes, function(gene) plot_violin(gsva_df_filtered, gene)) -
(option 1 for plot 1) merge first_row_plots and second_row_plots to a final plot using cowplot
library(cowplot) library(ggplot2) # Start by extracting the 1-8 plots from each list first_row_plots <- first_row_plots[1:9] second_row_plots <- second_row_plots[1:9] # Function to modify the individual plots based on position in the grid modify_plot <- function(p, row, col) { #if (col > 1) { p <- p + theme(axis.title.y = element_blank()) # remove y-axis title if not the first plot #} #if (row == 2) { # p <- p + theme(plot.title = element_blank()) # remove plot title for second row, commented because it has alreay been done below. #} p <- p + theme( axis.title.x = element_blank(), # remove x-axis title for all plots axis.title.y = element_blank(), # remove x-axis title for all plots axis.text = element_text(size = 14), # Increase axis text size axis.title = element_text(size = 16), # Increase axis title size plot.title = element_text(size = 16) # Increase plot title size ) return(p) } # Abbreviate titles for specific plots first_row_plots[[6]]$labels$title <- "Inflam. neutrophils" first_row_plots[[7]]$labels$title <- "Suppr. neutrophils" second_row_plots[[6]]$labels$title <- "Inflam. neutrophils" second_row_plots[[7]]$labels$title <- "Suppr. neutrophils" # Apply the modifications to the plots for (i in 1:9) { first_row_plots[[i]] <- modify_plot(first_row_plots[[i]], 1, i) second_row_plots[[i]] <- modify_plot(second_row_plots[[i]], 2, i) } # Increase the font size of x-axis labels for each plot for (i in 1:9) { first_row_plots[[i]] <- first_row_plots[[i]] + theme(axis.text.x = element_text(size = 14), axis.text.y = element_text(size = 12)) second_row_plots[[i]] <- second_row_plots[[i]] + theme(axis.text.x = element_text(size = 14), axis.text.y = element_text(size = 12), plot.title= element_blank()) } # Now, combine the modified plots into a single list all_plots <- c(first_row_plots, second_row_plots) # Generate the 2x9 grid final_plot <- plot_grid(plotlist = all_plots, ncol = 9) # Save the plot to a PNG file png("Carotis_NanoString2.png", width=1400, height=380) print(final_plot) dev.off() -
(option 2 for plot 2) generating two 3×3 grid plots
# Start by extracting the 1-9 plots from each list first_row_plots <- first_row_plots[1:9] second_row_plots <- second_row_plots[1:9] # Function to modify the individual plots based on position in the grid modify_plot <- function(p, row, col) { p <- p + theme(axis.title.y = element_blank()) # remove y-axis title if not the first plot p <- p + theme( axis.title.x = element_blank(), # remove x-axis title for all plots axis.title.y = element_blank(), # remove x-axis title for all plots axis.text = element_text(size = 14), # Increase axis text size axis.title = element_text(size = 16), # Increase axis title size plot.title = element_text(size = 16) # Increase plot title size ) return(p) } # Apply the modifications to the plots for (i in 1:9) { first_row_plots[[i]] <- modify_plot(first_row_plots[[i]], 1, i) second_row_plots[[i]] <- modify_plot(second_row_plots[[i]], 2, i) } # Increase the font size of x-axis labels for each plot for (i in 1:9) { first_row_plots[[i]] <- first_row_plots[[i]] + theme(axis.text.x = element_text(size = 14), axis.text.y = element_text(size = 12)) second_row_plots[[i]] <- second_row_plots[[i]] + theme(axis.text.x = element_text(size = 14), axis.text.y = element_text(size = 12)) } # Pad first_row_plots to have 9 plots remaining_plots <- 9 - length(first_row_plots) if (remaining_plots > 0) { first_row_plots <- c(first_row_plots, rep(list(NULL), remaining_plots)) } # Pad second_row_plots to have 9 plots remaining_plots2 <- 9 - length(second_row_plots) if (remaining_plots2 > 0) { second_row_plots <- c(second_row_plots, rep(list(NULL), remaining_plots2)) } # Convert the first_row_plots to a matrix and draw plots_matrix_1 <- matrix(first_row_plots, ncol=3, byrow=TRUE) png("Carotis_NanoString_grid_1.png", width=600, height=600) do.call("grid.arrange", c(plots_matrix_1, list(ncol=3))) dev.off() # Convert the second_row_plots to a matrix and draw plots_matrix_2 <- matrix(second_row_plots, ncol=3, byrow=TRUE) png("Carotis_NanoString_grid_2.png", width=600, height=600) do.call("grid.arrange", c(plots_matrix_2, list(ncol=3))) dev.off()
GSVA-plot for carotis RNA-seq data
-
preparing gene expression matrix: calculate DESeq2 results
#Input: merged_gene_counts.txt setwd("/home/jhuang/DATA/Data_Susanne_Carotis_RNASeq/run_2023_GSVA/") library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") #BiocManager::install("org.Hs.eg.db") library("org.Hs.eg.db") library(DESeq2) library(gplots) d.raw<- read.delim2("merged_gene_counts.txt",sep="\t", header=TRUE, row.names=1) colnames(d.raw)<-c("gene_name", "leer_mock_2h_r2", "Ace2_mock_2h_r2", "leer_inf_24h_r1", "Ace2_inf_2h_r1", "leer_inf_24h_r2", "leer_inf_2h_r1", "leer_mock_2h_r1", "leer_inf_2h_r2", "Ace2_inf_2h_r2", "Ace2_mock_2h_r1", "Ace2_inf_24h_r2", "Ace2_inf_24h_r1") col_order <- c("gene_name", "leer_mock_2h_r1","leer_mock_2h_r2","leer_inf_2h_r1","leer_inf_2h_r2","leer_inf_24h_r1","leer_inf_24h_r2","Ace2_mock_2h_r1","Ace2_mock_2h_r2","Ace2_inf_2h_r1","Ace2_inf_2h_r2","Ace2_inf_24h_r1","Ace2_inf_24h_r2") reordered.raw <- d.raw[,col_order] reordered.raw$gene_name <- NULL #d <- d.raw[rowSums(reordered.raw>3)>2,] condition = as.factor(c("leer_mock_2h","leer_mock_2h","leer_inf_2h","leer_inf_2h","leer_inf_24h","leer_inf_24h","Ace2_mock_2h","Ace2_mock_2h","Ace2_inf_2h","Ace2_inf_2h","Ace2_inf_24h","Ace2_inf_24h")) ids = as.factor(c("leer_mock_2h_r1","leer_mock_2h_r2","leer_inf_2h_r1","leer_inf_2h_r2","leer_inf_24h_r1","leer_inf_24h_r2","Ace2_mock_2h_r1","Ace2_mock_2h_r2","Ace2_inf_2h_r1","Ace2_inf_2h_r2","Ace2_inf_24h_r1","Ace2_inf_24h_r2")) #cData = data.frame(row.names=colnames(reordered.raw), condition=condition, batch=batch, ids=ids) #dds<-DESeqDataSetFromMatrix(countData=reordered.raw, colData=cData, design=~batch+condition) cData = data.frame(row.names=colnames(reordered.raw), condition=condition, ids=ids) dds<-DESeqDataSetFromMatrix(countData=reordered.raw, colData=cData, design=~condition) #----more detailed and specific with the following code!---- dds$condition <- relevel(dds$condition, "Ace2_mock_2h") dds = DESeq(dds, betaPrior=FALSE) # betaPrior default value is FALSE resultsNames(dds) -
preparing selected_geneSets in gsva(exprs, selected_geneSets, method=”gsva”). Note that methods are different than methods for nanoString, here are ENSEMBL listed.
#Input: "Signatures.xls" + "Signatures_additional.xls" library(readxl) library(gridExtra) library(ggplot2) library(GSVA) # Paths to the Excel files file_paths <- list("Signatures.xls", "Signatures_additional.xls") # Get sheet names for each file sheet_names_list <- lapply(file_paths, excel_sheets) # Initialize an empty list to hold gene sets geneSets <- list() # Loop over each file path and its corresponding sheet names for (i in 1:length(file_paths)) { file_path <- file_paths[[i]] sheet_names <- sheet_names_list[[i]] # Loop over each sheet, extract the ENSEMBL IDs, and add to the list for (sheet in sheet_names) { # Read the sheet data <- read_excel(file_path, sheet = sheet) # Process the GeneSet names (replacing spaces with underscores, for example) gene_set_name <- gsub(" ", "_", unique(data$GeneSet)[1]) # Add ENSEMBL IDs to the list geneSets[[gene_set_name]] <- as.character(data$ENSEMBL) } } # Print the result to check print(geneSets) summary(geneSets) #desired_geneSets <- c("Monocytes", "Plasma_cells", "T_regs", "Cyt._act._T_cells", "Neutrophils", "Inflammatory_neutrophils", "Suppressive_neutrophils", "LDG", "CD40_activated") desired_geneSets <- c("IFN", "TNF", "IL-6R_complex", "IL-1_cytokines", "Pro-inflam._IL-1", "Monocyte_secreted", "Apoptosis", "NFkB_complex", "NLRP3_inflammasome") selected_geneSets <- geneSets[desired_geneSets] # Print the selected gene sets print(selected_geneSets) -
prepare violin plots
# 0. for Nanostring, the GSVA input requires a gene expression matrix 'exprs' where rows are genes and columns are samples. This matrix must be in non-log space. #exprs <- exprs(filtered_or_neg_target_m666Data) # 0. for RNAseq, the GSVA input requires a gene expression matrix where rows are genes and columns are samples. This matrix must be in non-log space. exprs <- counts(dds, normalized=TRUE) # 1. Compute GSVA scores: gsva_scores <- gsva(exprs, selected_geneSets, method="gsva") # 2. Convert to data.frame for ggplot: gsva_df <- as.data.frame(t(gsva_scores)) # 3. Add conditions to gsva_df: gsva_df$Condition <- dds$condition # 4. Filter the gsva_df to retain only the desired conditions: #group 1 vs. group 3 in the nanostring data gsva_df_filtered <- gsva_df[gsva_df$Condition %in% c("Ace2_mock_2h", "Ace2_inf_24h"), ] # 5. Define a function to plot violin plots: # Update the condition levels in gsva_df_filtered to ensure the desired order on x-axis: gsva_df_filtered$Condition <- gsub("Ace2_mock_2h", "Group3", gsva_df_filtered$Condition) #group3=mock gsva_df_filtered$Condition <- gsub("Ace2_inf_24h", "Group1a", gsva_df_filtered$Condition) #group1a=infection gsva_df_filtered$Condition <- factor(gsva_df_filtered$Condition, levels = c("Group1a", "Group3")) plot_violin <- function(data, gene_name) { # Calculate the t-test p-value for the two conditions condition1_data <- data[data$Condition == "Group1a", gene_name] condition2_data <- data[data$Condition == "Group3", gene_name] p_value <- t.test(condition1_data, condition2_data)$p.value # Convert p-value to annotation p_annotation <- ifelse(p_value < 0.01, "**", ifelse(p_value < 0.05, "*", "")) rounded_p_value <- paste0("p = ", round(p_value, 2)) plot_title <- gsub("_", " ", gene_name) p <- ggplot(data, aes(x=Condition, y=!!sym(gene_name), fill=Condition)) + geom_violin(linewidth=1.2) + scale_fill_manual(values = custom_colors) + labs(title=plot_title, y="GSVA Score") + ylim(-1, 1) + theme_light() + theme( axis.title.x = element_text(size=12), axis.title.y = element_text(size=12), axis.text.x = element_text(size=10), axis.text.y = element_text(size=10), plot.title = element_text(size=12, hjust=0.5), legend.position = "none" # Hide legend since the colors are self-explanatory ) # Add p-value annotation to the plot p <- p + annotate("text", x=1.5, y=0.9, label=paste0(p_annotation, " ", rounded_p_value), size=5, hjust=0.5) return(p) } # 6. Generate the list of plots in a predefined order: genes <- colnames(gsva_df_filtered)[!colnames(gsva_df_filtered) %in% "Condition"] genes <- genes[match(desired_order, genes)] genes <- genes[!is.na(genes)] first_row_plots <- lapply(genes, function(gene) plot_violin(gsva_df_filtered, gene)) -
generating two 3×3 grid plots
# Start by extracting the 1-9 plots from each list first_row_plots <- first_row_plots[1:9] # Function to modify the individual plots based on position in the grid modify_plot <- function(p, row, col) { p <- p + theme(axis.title.y = element_blank()) # remove y-axis title if not the first plot p <- p + theme( axis.title.x = element_blank(), # remove x-axis title for all plots axis.title.y = element_blank(), # remove x-axis title for all plots axis.text = element_text(size = 14), # Increase axis text size axis.title = element_text(size = 16), # Increase axis title size plot.title = element_text(size = 16) # Increase plot title size ) return(p) } # Apply the modifications to the plots for (i in 1:9) { first_row_plots[[i]] <- modify_plot(first_row_plots[[i]], 1, i) } # Increase the font size of x-axis labels for each plot for (i in 1:9) { first_row_plots[[i]] <- first_row_plots[[i]] + theme(axis.text.x = element_text(size = 14), axis.text.y = element_text(size = 12)) } # Pad first_row_plots to have 9 plots remaining_plots <- 9 - length(first_row_plots) if (remaining_plots > 0) { first_row_plots <- c(first_row_plots, rep(list(NULL), remaining_plots)) } # Convert the first_row_plots to a matrix and draw plots_matrix_1 <- matrix(first_row_plots, ncol=3, byrow=TRUE) png("Carotis_RNA-seq_grid_1.png", width=600, height=600) do.call("grid.arrange", c(plots_matrix_1, list(ncol=3))) dev.off()
RNA-seq analysis for characterizing HSV-1 infection of human skin organoid
单纯疱疹性脑炎是由单纯疱疹病毒(HSVs)引起的中枢神经系统的致命疾病。在使用抗病毒药物阿昔洛韦进行标准治疗后,大多数患者仍然出现各种神经后遗症。在这里,我们通过结合单细胞RNA测序、电生理和免疫染色来描述人脑器官样本中的HSV-1感染。我们观察到组织完整性、神经元功能和细胞转录组的强烈扰动。在阿昔洛韦治疗下,病毒复制被停止,但并未防止HSV-1引发的缺陷,如神经元过程和神经上皮的损伤。对感染后失调路径的无偏分析揭示了肿瘤坏死因子激活可能是一个潜在的致因因素。结合抗炎药物如necrostatin-1或bardoxolone methyl和抗病毒治疗可以防止感染引起的损害,这表明在急性感染中调整炎症反应可能会改进当前的治疗策略。
https://github.com/rajewsky-lab/HSV1_cerebral_organoids/blob/main/sc_and_bulkRNAseq_analysis.Rmd
---
title: "Code availability for the paper Modelling viral encephalitis caused by herpes simplex virus 1 infection in cerebral organoids"
output: html_notebook
---
```{r}
library(Seurat)
library(ggplot2)
library(pheatmap)
library(dplyr)
library(tidyr)
library(Matrix)
#library(Nebulosa)
library(edgeR)
library(clusterProfiler)
library(msigdbr)
library(AUCell)
library(DESeq2)
library(gprofiler2)
library(RColorBrewer)
options(bitmapType='cairo')
```
# Analysis of control (CTRL) organoids
Read digital gene expression matrices (generated with Spacemake) for CTRL samples and create Seurat objects for each sample keeping barcodes with at least 250 detected genes and genes detected at least 5 cells
```{r eval=FALSE, include=FALSE}
Xmoo1_CTRL_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_CTRL_1.txt.gz', sep = '', row.names = 1, header = T) %>%
CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_CTRL_1', assay = 'RNA')
Xmoo1_CTRL_1$line = 'iPSC line 1'
Xmoo1_CTRL_1$replicate = 'Rep. 1'
Xmoo1_CTRL_1$condition = 'CTRL'
Xmoo1_CTRL_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_CTRL_2.txt.gz', sep = '', row.names = 1, header = T) %>%
CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_CTRL_2', assay = 'RNA')
Xmoo1_CTRL_2$line = 'iPSC line 1'
Xmoo1_CTRL_2$replicate = 'Rep. 2'
Xmoo1_CTRL_2$condition = 'CTRL'
Gline_CTRL_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_CTRL_1.txt.gz', sep = '', row.names = 1, header = T) %>%
CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_CTRL_1', assay = 'RNA')
Gline_CTRL_1$line = 'iPSC line 2'
Gline_CTRL_1$replicate = 'Rep. 1'
Gline_CTRL_1$condition = 'CTRL'
Gline_CTRL_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_CTRL_2.txt.gz', sep = '', row.names = 1, header = T) %>%
CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_CTRL_2', assay = 'RNA')
Gline_CTRL_2$line = 'iPSC line 2'
Gline_CTRL_2$replicate = 'Rep. 2'
Gline_CTRL_2$condition = 'CTRL'
```
Create a single integrated Seurat object with all CTRL samples (2 replicates for 2 cell lines) after correcting for sample- and cell line-specific batch effects
```{r eval=FALSE, include=FALSE}
integration.list <- list(Xmoo1_CTRL_1, Xmoo1_CTRL_2, Gline_CTRL_1, Gline_CTRL_2)
for (i in 1:length(integration.list)) {
integration.list[[i]] <- SCTransform(integration.list[[i]], verbose = T,method= 'glmGamPoi', assay = 'RNA', new.assay.name = 'SCT', vst.flavor='v2')
}
integration.features <- SelectIntegrationFeatures(object.list = integration.list, nfeatures = 3000)
integration.list <- PrepSCTIntegration(object.list = integration.list, anchor.features = integration.features, verbose = T)
integration.anchors <- FindIntegrationAnchors(object.list = integration.list, normalization.method = "SCT", anchor.features = integration.features, verbose = T)
CTRL <- IntegrateData(anchorset = integration.anchors, normalization.method = "SCT", verbose = T)
DefaultAssay(CTRL) <- 'integrated'
remove(i, integration.list, integration.features, integration.anchors)
```
Perform dimensionality reduction and clustering with 30 principal components and default parameters
```{r eval=FALSE, include=FALSE}
CTRL <- CTRL %>% RunPCA( verbose = T) %>% RunUMAP( dims = 1:30, verbose = T) %>% FindNeighbors( dims = 1:30, verbose = T) %>% FindClusters( verbose = T)
```
Inspect transcript counts (nCount) and % of mitochondrial transcripts (percent_MT) to identify low quality clusters
```{r eval=FALSE, include=FALSE}
DefaultAssay(CTRL)= 'RNA'
CTRL= CTRL %>%PercentageFeatureSet( pattern = "^MT-", col.name = "percent_MT")
VlnPlot(CTRL, 'nCount_RNA', y.max = 2000, pt.size = 0)+ NoLegend()
VlnPlot(CTRL, 'percent_MT', pt.size = 0)+ NoLegend()
```
Remove low quality clusters 1 (low nCount) and 10 (low nCount, high mito) and select top 75% cells by nCount per cluster
```{r eval=FALSE, include=FALSE}
CTRL$barcode= colnames(CTRL)
top75= CTRL@meta.data[!CTRL$seurat_clusters %in% c(1, 10),] %>% group_by(seurat_clusters) %>% top_frac(n = 0.75, wt = nCount_RNA_exonic)
CTRL = subset(CTRL, cells=top75$barcode)
```
Re-run integration, dimensionality reduction and clustering
```{r eval=FALSE, include=FALSE}
Idents(CTRL) = 'sample'
integration.list <- SplitObject(CTRL)
for (i in 1:length(integration.list)) {
integration.list[[i]] <- SCTransform(integration.list[[i]],method= 'glmGamPoi', assay = 'RNA', new.assay.name = 'SCT', vst.flavor='v2')
}
integration.features <- SelectIntegrationFeatures(object.list = integration.list, nfeatures = 3000)
integration.list <- PrepSCTIntegration(object.list = integration.list, anchor.features = integration.features)
integration.anchors <- FindIntegrationAnchors(object.list = integration.list, normalization.method = "SCT", anchor.features = integration.features)
CTRL <- IntegrateData(anchorset = integration.anchors, normalization.method = "SCT")
DefaultAssay(CTRL) <- 'integrated'
CTRL <- CTRL %>% RunPCA() %>% RunUMAP( dims = 1:30) %>% FindNeighbors( dims = 1:30) %>% FindClusters(resolution = 0.4)
remove(i, integration.list, integration.features, integration.anchors)
```
Read in processed object
```{r}
CTRL= readRDS('seurat_objects/CTRL.rds')
```
## Extended figure 1.c
```{r}
DimPlot(CTRL, split.by = 'line', group.by = 'replicate', shuffle = T)
```
## Extended figure 1.d
```{r}
FeaturePlot(CTRL, features = c( 'VIM', 'SOX2', 'FOXG1','MKI67', 'DCX', 'EOMES', 'NEUROD2', 'NEUROD6', 'BCL11B', 'TCF7L2', 'GFAP', 'SPARCL1', 'RSPO2','RSPO3', 'TTR', 'DCT', 'SFRP2','COL1A2', 'RELN' ), slot = 'data', max.cutoff = 2, ncol = 5) * NoLegend() * NoAxes()
```
Important and prepare downloaded reference data (REF: Kanton et al, Nature 2019)
```{r eval=FALSE, include=FALSE}
# Read in cell-level metadata
kanton_meta = read.table('Kanton_et_al_2019/metadata_human_cells.tsv', sep = '\t', header = T)
rownames(kanton_meta)= paste0(kanton_meta$Sample, '_',kanton_meta$Barcode)
# Read in gene names
kanton_gene = read.table('Kanton_et_al_2019/genes_GRCh38.txt', sep = '\t', header = F)
# Read in gene expression matrix
kanton_mtx = readMM('Kanton_et_al_2019/human_cell_counts_GRCh38.mtx')
colnames(kanton_mtx) = rownames(kanton_meta)
rownames(kanton_mtx) = kanton_gene$V2
# Create Seurat object and perform SCT normalization
kanton = CreateSeuratObject(counts = kanton_mtx, meta.data = kanton_meta, min.cells = 50, min.features = 3) %>% SCTransform()
remove(kanton_meta, kanton_gene, kanton_mtx)
```
Label transfer from reference to CTRL organoids
```{r eval=FALSE, include=FALSE}
DefaultAssay(CTRL) <- 'integrated'
obj.list= list(kanton, CTRL)
features <- SelectIntegrationFeatures(obj.list)
anchors <- FindTransferAnchors(reference = kanton, query = CTRL, normalization.method = "SCT", reference.assay = 'SCT', query.assay = 'integrated',reduction = 'pcaproject', dims = 1:20, features = features, nn.method= 'rann', eps = 0.5, verbose = T)
CTRL[["kanton_PredCellType"]] <- TransferData(anchorset = anchors, refdata = kanton$PredCellType, prediction.assay = TRUE, weight.reduction = 'pcaproject', dims = 1:20, eps = 0.5 )
CTRL[["kanton_FullLineage"]] <- TransferData(anchorset = anchors, refdata = kanton$cl_FullLineage, prediction.assay = TRUE, weight.reduction = 'pcaproject', dims = 1:20, eps = 0.5 )
remove(kanton, obj.list, features, anchors)
```
## Supplementary figure 1.a
```{r}
FeaturePlot(CTRL, c('cycling dorsal progenitors', 'RGCs 3','IPs and early cortical neurons', 'cortical neurons 1', 'midbrain/hindbrain cells', 'Astrocyte', 'Mural', 'retina progenitors', 'Choroid'), order = T) * NoLegend() * NoAxes()
```
Annotate clusters
```{r eval=FALSE, include=FALSE}
Idents(CTRL) <- 'seurat_clusters'
CTRL <- RenameIdents(
object = CTRL,
'0'= 'Radial Glia G1/S Phase',
'1'= 'Immature Cortical Neurons 1',
'2'= 'Immature Cortical Neurons 2', #need better annotation
'3'= 'Thalamic Neurons',
'4'= 'Intermediate Progenitors',
'5'= 'Radial Glia G2M Phase',
'6'= 'Astroglia',
'7'= 'Mature Cortical Neurons',
'8'= 'Progenitors', #need better annotation
'9'= 'Cortical Hem/Choroid Plexus',
'10'= 'Hindbrain Neurons 1',
'11'= 'Retinal Progenitors',
'12'= 'Hindbrain Neurons 2', #need better annotation
'13'= 'Mural Cells',
'14'= 'Retinal Pigmented Cells',
'15'= 'Cajal Retzius Neurons')
CTRL$legend = Idents(CTRL)
```
## Figure 1.d
```{r}
color_palette=c('Immature Cortical Neurons 1'="#00133E", 'Immature Cortical Neurons 2'="#16365C", 'Mature Cortical Neurons'="#2D597A", 'Hindbrain Neurons 1'="#447C98", 'Thalamic Neurons'="#5A9FB6", 'Hindbrain Neurons 2'="#71C2D4", 'Cajal Retzius Neurons'="#87E5F2", 'Radial Glia G2M Phase'='#b7245c', 'Progenitors'='#8f0034', 'Radial Glia G1/S Phase'='#de598c', 'Intermediate Progenitors'='#7D5AA5', 'Retinal Progenitors'='#f7c548', 'Retinal Pigmented Cells'='#cf9d20', 'Mural Cells'='#060300', 'Cortical Hem/Choroid Plexus'='#FA8405', 'Astroglia'='#808080')
DimPlot(CTRL, cols = color_palette)
```
# Analysis of control (CTRL), HSV1 1 day post infection (1dpi), 3 days post infection (3dpi) and acyclovir (ACV) treated organoids
Read digital gene expression matrices (generated with Spacemake) for CTRL samples and create Seurat objects for each sample keeping barcodes with at least 250 detected genes and genes detected at least 5 cells
```{r eval=FALSE, include=FALSE}
Xmoo1_inf1dpi_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_inf1dpi_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_inf1dpi_1', assay = 'RNA')
Xmoo1_inf1dpi_1$line = 'iPSC line 1'
Xmoo1_inf1dpi_1$replicate = 'Rep. 1'
Xmoo1_inf1dpi_1$condition = 'HSV-1 (1dpi)'
Xmoo1_inf1dpi_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_inf1dpi_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_inf1dpi_2', assay = 'RNA')
Xmoo1_inf1dpi_2$line = 'iPSC line 1'
Xmoo1_inf1dpi_2$replicate = 'Rep. 2'
Xmoo1_inf1dpi_2$condition = 'HSV-1 (1dpi)'
Xmoo1_inf3dpi_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_inf3dpi_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_inf3dpi_1', assay = 'RNA')
Xmoo1_inf3dpi_1$line = 'iPSC line 1'
Xmoo1_inf3dpi_1$replicate = 'Rep. 1'
Xmoo1_inf3dpi_1$condition = 'HSV-1 (3dpi)'
Xmoo1_inf3dpi_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_inf3dpi_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_inf3dpi_2', assay = 'RNA')
Xmoo1_inf3dpi_2$line = 'iPSC line 1'
Xmoo1_inf3dpi_2$replicate = 'Rep. 2'
Xmoo1_inf3dpi_2$condition = 'HSV-1 (3dpi)'
Xmoo1_inf3dpiACV_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_inf3dpiACV_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_inf3dpiACV_1', assay = 'RNA')
Xmoo1_inf3dpiACV_1$line = 'iPSC line 1'
Xmoo1_inf3dpiACV_1$replicate = 'Rep. 1'
Xmoo1_inf3dpiACV_1$condition = 'HSV-1 (3dpi) +ACV'
Xmoo1_inf3dpiACV_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_inf3dpiACV_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_inf3dpiACV_2', assay = 'RNA')
Xmoo1_inf3dpiACV_2$line = 'iPSC line 1'
Xmoo1_inf3dpiACV_2$replicate = 'Rep. 2'
Xmoo1_inf3dpiACV_2$condition = 'HSV-1 (3dpi)'
Gline_inf1dpi_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_inf1dpi_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_inf1dpi_1', assay = 'RNA')
Gline_inf1dpi_1$line = 'iPSC line 2'
Gline_inf1dpi_1$replicate = 'Rep. 1'
Gline_inf1dpi_1$condition = 'HSV-1 (1dpi)'
Gline_inf1dpi_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_inf1dpi_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_inf1dpi_2', assay = 'RNA')
Gline_inf1dpi_2$line = 'iPSC line 1'
Gline_inf1dpi_2$replicate = 'Rep. 2'
Gline_inf1dpi_2$condition = 'HSV-1 (1dpi)'
Gline_inf3dpi_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_inf3dpi_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_inf3dpi_1', assay = 'RNA')
Gline_inf3dpi_1$line = 'iPSC line 2'
Gline_inf3dpi_1$replicate = 'Rep. 1'
Gline_inf3dpi_1$condition = 'HSV-1 (3dpi)'
Gline_inf3dpi_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_inf3dpi_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_inf3dpi_2', assay = 'RNA')
Gline_inf3dpi_2$line = 'iPSC line 1'
Gline_inf3dpi_2$replicate = 'Rep. 2'
Gline_inf3dpi_2$condition = 'HSV-1 (3dpi)'
Gline_inf3dpiACV_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_inf3dpiACV_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_inf3dpiACV_1', assay = 'RNA')
Gline_inf3dpiACV_1$line = 'iPSC line 2'
Gline_inf3dpiACV_1$replicate = 'Rep. 1'
Gline_inf3dpiACV_1$condition = 'HSV-1 (3dpi) +ACV'
Gline_inf3dpiACV_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Gline_inf3dpiACV_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Gline_inf3dpiACV_2', assay = 'RNA')
Gline_inf3dpiACV_2$line = 'iPSC line 1'
Gline_inf3dpiACV_2$replicate = 'Rep. 2'
Gline_inf3dpiACV_2$condition = 'HSV-1 (3dpi) +ACV'
```
Create a single integrated Seurat object with all 16 samples (2 replicates for 2 cell lines per 4 conditions) after correcting for condition- and cell line-specific effects
```{r eval=FALSE, include=FALSE}
all_timepoints= merge(Xmoo1_CTRL_1_exonic, list(Xmoo1_CTRL_2_exonic, Xmoo1_inf1dpi_1, Xmoo1_inf1dpi_2, Xmoo1_inf3dpi_1, Xmoo1_inf3dpi_2, Xmoo1_inf3dpiACV_1, Xmoo1_inf3dpiACV_2,
Gline_CTRL_1_exonic, Gline_CTRL_2_exonic, Gline_inf1dpi_1, Gline_inf1dpi_2, Gline_inf3dpi_1, Gline_inf3dpi_2, Gline_inf3dpiACV_1, Gline_inf3dpiACV_2))
all_timepoints$line_condition= paste0(all_timepoints$line, '_', all_timepoints$condition)
Idents(all_timepoints) = 'line_condition'
integration.list <- SplitObject(all_timepoints)
for (i in 1:length(integration.list)) {
integration.list[[i]] <- SCTransform(integration.list[[i]], verbose = T,method= 'glmGamPoi', assay = 'RNA', new.assay.name = 'SCT', vst.flavor='v2')
}
integration.features <- SelectIntegrationFeatures(object.list = integration.list, nfeatures = 3000)
integration.list <- PrepSCTIntegration(object.list = integration.list, anchor.features = integration.features, verbose = T)
integration.anchors <- FindIntegrationAnchors(object.list = integration.list, normalization.method = "SCT", anchor.features = integration.features, verbose = T)
all_timepoints <- IntegrateData(anchorset = integration.anchors, normalization.method = "SCT", verbose = T)
DefaultAssay(all_timepoints) <- 'integrated'
remove(i, integration.list, integration.features, integration.anchors)
```
Perform dimensionality reduction and clustering with 30 principal components and default parameters
```{r eval=FALSE, include=FALSE}
all_timepoints <- all_timepoints %>% RunPCA( verbose = T) %>% RunUMAP( dims = 1:30, verbose = T) %>% FindNeighbors( dims = 1:30, verbose = T) %>% FindClusters( verbose = T)
```
Inspect transcript counts (nCount) and % of mitochondrial transcripts (percent_MT) to identify low quality clusters
```{r eval=FALSE, include=FALSE}
VlnPlot(all_timepoints, 'nCount_RNA_exonic', y.max = 2000, pt.size = 0)+ NoLegend()
VlnPlot(all_timepoints, 'percent_MT', y.max = 2000, pt.size = 0)+ NoLegend()
```
Remove low quality clusters 6 (low nCount) and 10 (high mito) and select top 75% cells by nCount per cluster
```{r eval=FALSE, include=FALSE}
all_timepoints$barcode= colnames(all_timepoints)
top75= all_timepoints@meta.data[!all_timepoints$seurat_clusters %in% c(6, 10),] %>% group_by(seurat_clusters) %>% top_frac(n = 0.75, wt = nCount_RNA_exonic)
all_timepoints = subset(all_timepoints, cells=top75$barcode)
```
Re-run integration, dimensionality reduction and clustering
```{r eval=FALSE, include=FALSE}
Idents(all_timepoints) = 'line_condition'
integration.list <- SplitObject(all_timepoints)
for (i in 1:length(integration.list)) {
integration.list[[i]] <- SCTransform(integration.list[[i]],method= 'glmGamPoi', assay = 'RNA', new.assay.name = 'SCT', vst.flavor='v2')
}
integration.features <- SelectIntegrationFeatures(object.list = integration.list, nfeatures = 3000)
integration.list <- PrepSCTIntegration(object.list = integration.list, anchor.features = integration.features)
integration.anchors <- FindIntegrationAnchors(object.list = integration.list, normalization.method = "SCT", anchor.features = integration.features)
all_timepoints <- IntegrateData(anchorset = integration.anchors, normalization.method = "SCT")
DefaultAssay(all_timepoints) <- 'integrated'
all_timepoints <- all_timepoints %>% RunPCA() %>% RunUMAP( dims = 1:30) %>% FindNeighbors( dims = 1:30) %>% FindClusters(resolution = 0.8)
remove(i, integration.list, integration.features, integration.anchors)
```
Quantify the % of CTRL cells (condition) in each cluster
```{r eval=FALSE, include=FALSE}
round(prop.table(table(all_timepoints$seurat_clusters, all_timepoints$condition=='CTRL'), margin=1)*100, digits=1)
```
Add annotations of CTRL cells
```{r eval=FALSE, include=FALSE}
all_timepoints$legend_CTRL = CTRL$legend
```
Quantify the % of CTRL cell type (legend) in each cluster
```{r eval=FALSE, include=FALSE}
round(prop.table(table(all_timepoints$legend_CTRL, all_timepoints$seurat_clusters), margin=1)*100, digits=1)
```
Identify marker genes for each cluster
```{r eval=FALSE, include=FALSE}
all_timepoints_markers= FindAllMarkers(all_timepoints, only.pos = T)
```
Cluster annotation strategy:
1. Clusters composed by at least 33% of CTRL cells (as CTRL cell represent 33% of the dataset) take the annotation of the majority of the CTRL cells
0: Mixed Neurons (35+31%)
3: Cortical Neurons (56 Immature, 34 Mature %)
4: RG (89%)
5: Astroglia (68.5%)
8: RG (76.5%)
12: IPC (86%)
13: RG proliferating (97%)
15: Thalamic (54% Thalamic)
18: Mural (95%)
2. Clusters that contain more than than 50% of the CTRL cell types from a specific cluster and at least 10% of CTRL cells
7: Retinal Progenitors (90%) [10% CTRL]
11: Thalamic Neurons (54%) [26% CTRL]
14: Choroid (94%) [28% CTRL]
16: Hindbrain (65%) [22% CTRL]
17: Retinal Pigmented (81%) [22% CTRL]
3. Missing clusters annotated on markers
1: Infected
2: Infected
6: Infected
9: Infected
10: Hindbrain (HOX genes: HOXA5, B3-5-6)
```{r include=FALSE}
Idents(all_timepoints) <- 'seurat_clusters'
all_timepoints <- RenameIdents(
object = all_timepoints,
'0'= 'Mixed Neurons',
'1'= 'Highly infected 1',
'2'= 'Highly infected 2',
'3'= 'Cortical Neurons',
'4'= 'Radial Glia G1 Phase',
'5'= 'Astroglia',
'6'= 'Highly infected 3',
'7'= 'Retinal Progenitors',
'8'= 'Radial Glia S Phase',
'9'= 'Highly infected 4',
'10'= 'Hindbrain Neurons 1',
'11'= 'Thalamic Neurons 1',
'12'= 'Intermediate Progenitors',
'13'= 'Radial Glia G2M Phase',
'14'= 'Cortical Hem/Choroid Plexus',
'15'= 'Thalamic Neurons 2',
'16'= 'Hindbrain Neurons 2',
'17'= 'Retinal Pigmented Cells',
'18'= 'Mural Cells')
all_timepoints$legend = Idents(all_timepoints)
```
CTRL cells in infected clusters have lower UMIs than CTRL cells in other clusters
```{r eval=FALSE, include=FALSE}
ggplot(all_timepoints@meta.data[all_timepoints$condition=='CTRL',], aes(x= legend, y= nCount_RNA)) + geom_boxplot() + ylim(c(0, 3000)) + RotatedAxis()
```
CTRL cells in infected clusters have lower UMIs than infected cells in Highly infected clusters
```{r eval=FALSE, include=FALSE}
ggplot(all_timepoints@meta.data, aes(x= seurat_clusters, y= nCount_RNA, fill=condition)) + geom_boxplot() + ylim(c(0, 3000))
```
Removing CTRL cells from 'Highly infected' clusters
```{r eval=FALSE, include=FALSE}
all_timepoints =subset(all_timepoints, cells = colnames(all_timepoints)[!(all_timepoints$condition== 'CTRL' & all_timepoints$seurat_clusters %in% c(1, 2, 6, 9) )])
```
Import processed object
```{r}
saveRDS(all_timepoints, 'seurat_objects/all_timepoints.rds')
```
## Figure 2.a
```{r}
color_palette=c( 'Mixed Neurons'="#00133E", 'Thalamic Neurons 2'="#16365C",'Cortical Neurons'="#2D597A",'Hindbrain Neurons 1'="#447C98",'Thalamic Neurons 1'="#5A9FB6",'Hindbrain Neurons 2'="#71C2D4",'Radial Glia G2M Phase'='#b7245c','Radial Glia S Phase'='#8f0034', 'Radial Glia G1 Phase'='#de598c','Intermediate Progenitors'='#7D5AA5','Retinal Progenitors'='#f7c548','Retinal Pigmented Cells'='#cf9d20','Mural Cells'='#060300','Cortical Hem/Choroid Plexus'='#FA8405','Astroglia'='808080','Highly infected 1'='#23A17B','Highly infected 2'='#17B67E','Highly infected 3'='#0CCA80','Highly infected 4'='#005225')
DimPlot(all_timepoints, cols = color_palette)
```
## Extended figure 3.c
```{r}
DimPlot(all_timepoints, split.by = 'line_condition', group.by = 'replicate', shuffle = T, ncol = 4)
```
## Figure 3.b
```{r}
all_timepoints$legend= factor(all_timepoints$legend, levels = names(color_palette))
ggplot(all_timepoints@meta.data, aes(x=condition, fill=legend)) +
geom_bar(position='fill') +
scale_fill_manual(values=color_palette) +
theme_bw() +
xlab('') + ylab('Cluster %') +
scale_y_continuous(labels = scales::percent_format())
```
```{r}
extended_3b= all_timepoints@meta.data %>% group_by(condition, legend) %>% summarise(count= length(condition)) %>% pivot_wider(names_from = legend, values_from = count, values_fill = 0) %>% as.data.frame()
rownames(extended_3b)= extended_3b$condition
extended_3b$condition= NULL
write.csv(round(extended_3b/rowSums(extended_3b)*100, digits = 1), '../gene_lists/source_extended_Figure_3b.csv')
```
## Extended figure 4.a
```{r}
all_timepoints$sample= paste0(all_timepoints$condition, '_', all_timepoints$replicate)
ggplot(all_timepoints@meta.data, aes(x=sample, fill=legend)) +
geom_bar(position='fill') + scale_fill_manual(values=color_palette) +
theme_bw() +
xlab('') + ylab('Cluster %') +
scale_y_continuous(labels = scales::percent_format()) + facet_wrap('line')
```
Compute viral load (% of viral transcripts) per cell
```{r eval=FALSE, include=FALSE}
# Identify viral genes
viral_genes= grep("^US[0-9]{1,2}A{0,1}[0-9]{0,1}|^UL[0-9]{1,2}[A]{0,1}[0-9]{0,1}|^R[LS][12]|^LAT|^GFP$",
rownames(all_timepoints), value="TRUE", perl=TRUE)
# Extract viral gene expression
viral_expression= all_timepoints@assays$RNA[viral_genes, ]
# Count viral transcripts per cell
all_timepoints$nCount_viral= colSums(viral_expression)
# Compute viral load
all_timepoints$viral_load = all_timepoints$nCount_viral/all_timepoints$nCount_RNA*100
```
## Figure 3.c
```{r}
viral_load_matrix= all_timepoints@meta.data %>% group_by(legend, condition) %>% summarise(viral_load= round(median(viral_load), digits = 1) ) %>% tidyr::pivot_wider(names_from = condition, values_from = viral_load) %>% as.data.frame()
rownames(viral_load_matrix) = as.character(viral_load_matrix$legend)
viral_load_matrix$legend= NULL
green_palette= c(colorRampPalette(c("white", "#23A17B"))(10), rep("#23A17B", 60))
pheatmap(viral_load_matrix, cluster_rows = F, cluster_cols = F, display_numbers = T, fontsize_number = 4, number_format = "%.1f",color = green_palette, angle_col = 90, labels_col = c('CTRL', '+HSV-1 1dpi', '+HSV-1 3dpi', '+HSV-1 3dpi +ACV'), na_col = 'white', border_color = 'grey', cellwidth = 12, cellheight = 12, show_rownames = F, main = 'Median viral \n transcript %', fontsize = 10, fontsize_col = 10)
```
## Extended figure 4.c
```{r}
viral_metadata= all_timepoints@meta.data[, c('viral_load', 'condition','legend')]
viral_metadata= viral_metadata[order(viral_metadata$condition, viral_metadata$legend ),]
viral_expression= log10(viral_expression[order(rowSums(viral_expression), decreasing = T) , rownames(viral_metadata)]+1)
condition_cols= c('CTRL'='grey', 'HSV-1 (1dpi)'='#F1B3F4', 'HSV-1 (3dpi)'='#A13FA6', 'HSV-1 (3dpi) +ACV'='#BE64C2')
pheatmap(viral_expression, cluster_rows = F, cluster_cols = F, display_numbers = F, color = colorRampPalette(c("white", "#23A17B"))(100), angle_col = 90, show_rownames = T, main = 'Viral gene expression (log10)', fontsize = 10, fontsize_col = 10, annotation_col = viral_metadata, annotation_colors = list(legend= color_palette, condition= condition_cols ), show_colnames = F)
```
Create a Seurat object only with 'Highly infected' cells
```{r}
infected_clusters= subset(all_timepoints, cells= colnames(all_timepoints)[grep('Highly infected' ,all_timepoints$legend)])
DefaultAssay(infected_clusters) = 'SCT'
```
## Extended figure 4.d
```{r}
ggplot(infected_clusters@meta.data, aes(x=legend, fill= condition))+geom_bar(position = 'fill') +
scale_fill_manual(values = c( '#F1B3F4', '#A13FA6', '#BE64C2')) +
xlab('') + ylab('Condition %') +
scale_y_continuous(labels = scales::percent_format())
```
## Extended figure 4.e
```{r}
DotPlot(infected_clusters , features = c( 'VIM', 'SOX2', 'FOXG1','MKI67', 'DCX', 'EOMES', 'NEUROD2', 'NEUROD6', 'BCL11B', 'TCF7L2' ,'GFAP', 'SPARCL1', 'RSPO2', 'RSPO3', 'TTR', 'DCT', 'SFRP2','COL1A2','RELN'), group.by = 'legend', scale=T )
```
## Extended figure 4.f
```{r}
VlnPlot(infected_clusters, features = c('viral_load'), group.by = 'legend', cols =color_palette) + NoLegend() + ggtitle('') + ylab('Viral load (% UMI)') + xlab('')
remove(infected_clusters)
```
Store condition as a 1-hot encoded assay
```{r}
sample_assay = pivot_wider(data.frame(condition=all_timepoints$condition, values= 1, cells= colnames(all_timepoints)), names_from = 'condition', values_from = values, values_fill = 0 )
sample_assay = data.frame(sample_assay[, 2:5], row.names = sample_assay$cells)
sample_assay= sample_assay/colSums(sample_assay)
all_timepoints[["condition_assay"]] = CreateAssayObject(counts = t(sample_assay) )
```
## Figure 3.d
```{r}
DefaultAssay(all_timepoints) = 'condition_assay'
plot_density(object = all_timepoints, features = rownames(all_timepoints) )
all_timepoints@assays$condition_assay=NULL
```
## Supplementary figure 2.a
```{r}
VlnPlot(all_timepoints, features ='STMN2', group.by = 'condition', cols = condition_cols, assay = 'RNA', log=T, slot = 'data') + NoLegend() +ggtitle('') + ylab('Log normalized counts')
```
Identify differentially expressed genes in each cluster for selected clusters
```{r}
# Prepare dataframe to store results
DE_genes = data.frame(logFC= NA, logCPM= NA,F= NA, PValue = NA, cluster=NA, gene =NA, contrast= NA, adjPvalue= NA )
# Set sample name order
all_timepoints$line_condition_replicate = paste0(all_timepoints$line, '_', all_timepoints$condition, '_', all_timepoints$replicate)
all_timepoints$line_condition_replicate = factor(all_timepoints$line_condition_replicate, levels= c('iPSC line 2_CTRL_Rep. 1', 'iPSC line 2_CTRL_Rep. 2','iPSC line 1_CTRL_Rep. 1', 'iPSC line 1_CTRL_Rep. 2',
'iPSC line 2_HSV-1 (1dpi)_Rep. 1', 'iPSC line 2_HSV-1 (1dpi)_Rep. 2', 'iPSC line 1_HSV-1 (1dpi)_Rep. 1', 'iPSC line 1_HSV-1 (1dpi)_Rep. 2',
'iPSC line 2_HSV-1 (3dpi)_Rep. 1', 'iPSC line 2_HSV-1 (3dpi)_Rep. 2', 'iPSC line 1_HSV-1 (3dpi)_Rep. 1', 'iPSC line 1_HSV-1 (3dpi)_Rep. 2',
'iPSC line 2_HSV-1 (3dpi) +ACV_Rep. 1', 'iPSC line 2_HSV-1 (3dpi) +ACV_Rep. 2', 'iPSC line 1_HSV-1 (3dpi) +ACV_Rep. 1', 'iPSC line 1_HSV-1 (3dpi) +ACV_Rep. 2'))
all_timepoints$barcode= colnames(all_timepoints)
for (i in levels(all_timepoints$legend)[c(1,3, 5, 9)] ){
# Subsample to 100 cells per organoid per cluster to correct for different abundances across conditions
random_100_subset = all_timepoints@meta.data[all_timepoints$legend == i, ] %>% group_by(line_condition_replicate) %>% sample_n(100, replace= T)
cells= random_100_subset$barcode
cluster_group= random_100_subset$line_condition_replicate
# Sum gene expression across cells in each organoid to generate a pseudobulk gene expression matrix
pseudobulk= t(rowsum(x = t(as.matrix(all_timepoints@assays$RNA@counts[, cells])), group = cluster_group))
# Define sample metadata
condition = factor(c('CTRL','CTRL','CTRL','CTRL', 'inf1dpi','inf1dpi','inf1dpi','inf1dpi', 'inf3dpi', 'inf3dpi','inf3dpi', 'inf3dpi', 'inf3dpi_ACV','inf3dpi_ACV', 'inf3dpi_ACV','inf3dpi_ACV'),
levels= c('CTRL','inf1dpi', 'inf3dpi', 'inf3dpi_ACV'))
line= factor(c('Gline','Gline','Xmoo1','Xmoo1', 'Gline','Gline','Xmoo1','Xmoo1', 'Gline', 'Gline','Xmoo1', 'Xmoo1', 'Gline','Gline', 'Xmoo1','Xmoo1'),
levels= c('Gline','Xmoo1'))
# Define experimental design and remove cell-line specific effects
design = model.matrix(~line + condition)
# Define comparisons to assess
my.contrasts <- makeContrasts(
inf1dpivsCTRL = conditioninf1dpi,
inf3dpivsCTRL = conditioninf3dpi,
inf3dpi_ACVvsCTRL = conditioninf3dpi_ACV,
inf3dpivsinf1dpi= conditioninf3dpi - conditioninf1dpi,
inf3dpi_ACVvsinf3dpi= conditioninf3dpi_ACV - conditioninf3dpi,
inf3dpi_ACVvsinf1dpi= conditioninf3dpi_ACV - conditioninf1dpi,
levels = colnames(design))
# Create edgeR object
DE_object = DGEList(counts=pseudobulk,group=condition )
# Filter lowly detected, potentially noisy genes
DE_object = DE_object[filterByExpr(DE_object, min.count= 10, min.total.count= 50), , keep.lib.sizes=FALSE] # remove genes with >100 tpm in at least 2 samples
# Adjust by library size
DE_object$samples$lib.size <- colSums(DE_object$counts)
DE_object <- calcNormFactors(DE_object)
# Fit statistical model
DE_object = estimateDisp(DE_object, design)
DE_object = glmQLFit(DE_object, design)
# Prepare cluster-level dataframe to store results
cluster_DE_genes = data.frame(logFC= NA, logCPM= NA, F =NA, PValue = NA, cluster=NA, gene =NA, contrast= NA, adjPvalue= NA)
for (contrast in colnames(my.contrasts)){
# Compute differentially expressed genes
cluster_comparison= glmQLFTest(DE_object, contrast=my.contrasts[,contrast])$table
# Store metadata
cluster_comparison$cluster = i
cluster_comparison$gene = rownames(cluster_comparison)
cluster_comparison$contrast =contrast
# Compute bonferroni-adjusted p value
cluster_comparison$adjPvalue = p.adjust(cluster_comparison$PValue)
# Add to result data frame
cluster_DE_genes = rbind.data.frame(cluster_DE_genes, cluster_comparison)
cluster_DE_genes= cluster_DE_genes[!is.na(cluster_DE_genes$gene), ]
}
# Add to global result dataframe
DE_genes = rbind.data.frame(DE_genes, cluster_DE_genes)
}
# Exclude viral genes
DE_genes$viral = DE_genes$gene %in% viral_genes
DE_genes= DE_genes[!is.na(DE_genes$gene), ]
# Save results to file
write.csv(DE_genes, 'gene_lists/differentially_expressed_genes.csv')
```
Define function to run gene set enrichment analysis on differential expressed genes in each comparison and for each cluster ad save the results as separate RDS files
```{r}
run_GSEA_edgeR = function(DE_genes_csv, species = 'Homo sapiens', category, subcategory = NULL, cluster, contrasts, cluster_name,scoreType = "std") {
# Check if output directory exist and create it if it doesn't
if (file.exists(paste0('GSEA/', cluster_name))== F){dir.create(paste0('GSEA/', cluster_name))}
# Read in differential expressed genes
differential_genes = read.csv(DE_genes_csv)
# Define gene set database
msigdbr_t2g = msigdbr(species = species, category = category, subcategory = subcategory) %>% dplyr::distinct(gs_name, gene_symbol) %>% as.data.frame()
# Prepare list for storing results of each comparison
contrast_gsea = list()
for (contrast in contrasts) {
# Define differential genes and order them according to decreasing fold change
genes = differential_genes[differential_genes$contrast == contrast & differential_genes$cluster == cluster,]
genes = genes[order(genes$logFC, decreasing = T),]
genes_for_GSEA = genes$logFC
names(genes_for_GSEA) = genes$gene
# Compute gene set enrichments
if(any(genes$gene %in% msigdbr_t2g$gene_symbol)){contrast_gsea[as.character(contrast)] = GSEA(geneList = genes_for_GSEA, TERM2GENE = msigdbr_t2g, scoreType= scoreType)}
}
# Save file to disk
saveRDS(contrast_gsea, paste0('GSEA/', cluster_name,'/', category, '_', subcategory, '.rds'))
}
```
Run GSEA for the HALLMARK database
```{r}
dir.create('GSEA')
for (cluster in levels(all_timepoints$legend)[c(1,3, 5, 9)]){
run_GSEA_edgeR(DE_genes_csv ='gene_lists/differentially_expressed_genes.csv', category = 'H',
contrasts= c('inf1dpivsCTRL', '+HSV-1 3dpi vs CTRL', 'inf3dpi_ACVvsCTRL'),
cluster = cluster, cluster_name = cluster)}
```
Read in GSEA results
```{r}
GSEA_results = data.frame(ID= NA, NES= NA, p.adjust =NA, contrast = NA, cluster=NA)
for (cluster in levels(all_timepoints$legend)[c(1,3, 5, 9)]){
cluster_rds = readRDS(paste0('GSEA/', cluster,'/H_.rds'))
cluster_enrichments= data.frame(ID= NA, NES= NA, p.adjust =NA, contrast = NA)
if(!is.null(cluster_rds$inf1dpivsCTRL)){ if(nrow(cluster_rds$inf1dpivsCTRL@result)>0){cluster_enrichments = rbind.data.frame(cluster_enrichments, data.frame(cluster_rds$inf1dpivsCTR@result[, c('ID', 'NES', 'p.adjust')], contrast= 'inf1dpivsCTRL'))}}
if(!is.null(cluster_rds$inf3dpivsCTRL)){if(nrow(cluster_rds$inf3dpivsCTRL@result)>0){cluster_enrichments = rbind.data.frame(cluster_enrichments, data.frame(cluster_rds$inf3dpivsCTR@result[, c('ID', 'NES', 'p.adjust')], contrast= 'inf3dpivsCTRL' ))}}
if(!is.null(cluster_rds$inf3dpi_ACVvsCTRL)){if(nrow(cluster_rds$inf3dpi_ACVvsCTRL@result)>0){cluster_enrichments = rbind.data.frame(cluster_enrichments, data.frame(cluster_rds$inf3dpi_ACVvsCTRL@result[, c('ID', 'NES', 'p.adjust')], contrast= 'inf3dpi_ACVvsCTRL' ))}}
cluster_enrichments$cluster= cluster
GSEA_results = rbind.data.frame(GSEA_results, cluster_enrichments)
}
# Clean and polish labels
GSEA_results= GSEA_results[!is.na(GSEA_results$ID), ]
GSEA_results$ID= gsub('HALLMARK_', '', GSEA_results$ID)
GSEA_results$ID= gsub('_', ' ', GSEA_results$ID)
GSEA_results$ID= gsub('_', ' ', GSEA_results$ID)
GSEA_results$ID= stringr::str_to_sentence(GSEA_results$ID)
GSEA_results$ID= gsub('nfkb', 'NFkB', GSEA_results$ID)
GSEA_results$ID= gsub('Tnfa', 'TNFa', GSEA_results$ID)
GSEA_results$ID= gsub('Uv', 'UV', GSEA_results$ID)
GSEA_results$ID= gsub('Mtorc', 'mTORC', GSEA_results$ID)
GSEA_results$ID= gsub('Dna', 'DNA', GSEA_results$ID)
```
## Figure 4.a
```{r}
# Define gene set order based on the most frequently up or down regulated clusters
gene_set_stats=GSEA_results %>% group_by(ID) %>% summarise(positive = sum(NES >0), negative= sum(NES<0)) %>% as.data.frame()
gene_set_stats= gene_set_stats[order( gene_set_stats$positive, -gene_set_stats$negative, decreasing = T ),]
GSEA_results$ID= factor(GSEA_results$ID, levels = rev(gene_set_stats$ID))
# Define cluster, gene set and comparison order
GSEA_results$contrast = factor(GSEA_results$contrast, levels= c('inf1dpivsCTRL', 'inf3dpivsCTRL', 'inf3dpi_ACVvsCTRL'))
GSEA_results$cluster= factor(GSEA_results$cluster, levels = c('Radial Glia G1 Phase', 'Cortical Neurons', 'Mixed Neurons', 'Thalamic Neurons 1'))
# Compute -log10 p values
GSEA_results$`-log10 adj. p-value`= -log10(GSEA_results$p.adjust)
ggplot(GSEA_results,
aes(x = contrast, y = ID, col = NES, size = `-log10 adj. p-value`)) +
geom_point() + scale_color_continuous(low = "#00BFC4", high = "#F8766D")+
scale_color_gradientn(colors = c("dodgerblue", "firebrick"))+
facet_wrap('cluster', ncol = 4) +
theme_bw() + RotatedAxis()+ xlab('') + ylab('')
```
## Extended figure 4.a
```{r}
# Retrieve all genes included in the 'TNFA SIGNALLING VIA NFKB' gene set
msigdbr_t2g = msigdbr(species = 'Homo sapiens', category = 'H', subcategory = NA) %>% dplyr::distinct(gs_name, gene_symbol) %>% as.data.frame()
tnfa_genes= msigdbr_t2g[msigdbr_t2g$gs_name == 'HALLMARK_TNFA_SIGNALING_VIA_NFKB',2]
# Keep only genes detected in the scRNAseq data
DefaultAssay(all_timepoints)= 'SCT'
tnfa_genes= tnfa_genes[tnfa_genes %in% rownames(all_timepoints)]
# Retrieve their average gene expression
all_timepoints$legend_condition= paste0(all_timepoints$legend,' ' ,all_timepoints$condition)
gene_expression= AverageExpression(subset(all_timepoints, cells = all_timepoints$barcode[all_timepoints$legend %in% levels(all_timepoints$legend)[c(1,3, 5, 9, 16:20)]]),
assays = 'SCT', slot = 'counts', features = tnfa_genes, group.by = 'legend_condition')
gene_expression= as.data.frame(gene_expression$SCT)
gene_expression$gene= rownames(gene_expression)
# Compute the percentage of expressing cells
pct_expression_data= data.frame(fake_cluster= 1:length(tnfa_genes), row.names = tnfa_genes )
for (cluster in colnames(gene_expression) ){
cells= all_timepoints$barcode[all_timepoints$legend_condition==cluster]
cluster_expression= as.data.frame(all_timepoints@assays$RNA@counts[tnfa_genes, cells]>0)
pct_expression= data.frame(apply(cluster_expression, MARGIN = 1, FUN = function(x){ round(sum(x)/length(x)*100, digits = 0) } ))
colnames(pct_expression)= cluster
pct_expression_data= cbind(pct_expression_data, pct_expression)
}
remove(cluster, pct_expression)
pct_expression_data=pct_expression_data[, 2:29]
pct_expression_data$gene= rownames(pct_expression_data)
pct_expression_data = tidyr::pivot_longer(pct_expression_data, cols = 1:28, values_to = 'pct_expression', names_to = 'cluster')
gene_expression = tidyr::pivot_longer(gene_expression, cols = 1:28, values_to = 'avg_expression', names_to = 'cluster')
plot_data= gene_expression
plot_data$pct_expression= pct_expression_data$pct_expression
cluster_metadata= all_timepoints@meta.data %>% group_by(legend_condition) %>% summarise(cluster= unique(legend_condition), legend= unique(legend), condition= unique(condition) )
plot_data= left_join(plot_data, cluster_metadata[, 2:4])
plot_data= plot_data[plot_data$avg_expression >0.1 & plot_data$gene %in% tnfa_genes,]
plot_data$gene= factor(plot_data$gene, levels = rev(tnfa_genes))
colnames(plot_data)[3:4]= c('Average expression', '% cells expressing')
ggplot(plot_data, aes(y= gene, x= condition, size=`% cells expressing`, col=`Average expression` )) + geom_point() +facet_wrap('legend', nrow = 1) + theme_bw() + RotatedAxis() + scale_color_gradientn(colors = c("orange", "firebrick"))
```
Compute TNFA pathway activity in single cell
```{r eval=FALSE, include=FALSE}
# Rank gene by their expression in each single cell
cells_rankings = AUCell_buildRankings(all_timepoints@assays$SCT@counts, nCores = 40)
# Compute the enrichment of TNFA genes
cells_AUC = AUCell_calcAUC(list(TNFA=tnfa_genes), cells_rankings, aucMaxRank=nrow(cells_rankings)*0.05, nCores = 40)
# Add TNFa score to metadata
all_timepoints$tnfa_score = cells_AUC@assays@data$AUC['TNFA',]
```
## Figure 4.b
```{r}
FeaturePlot(all_timepoints, 'tnfa_score', split.by = 'condition', cols = c('dodgerblue', 'firebrick'), order = T, max.cutoff = 0.15, ncol = 4)
```
## Extended figure 6.b
```{r}
FeaturePlot(all_timepoints, 'tnfa_score', split.by = 'line_condition', cols = c('dodgerblue', 'firebrick'), order = T, max.cutoff = 0.15, ncol = 4)* NoAxes()
```
Importing NFkB direct targets
```{r}
tnfa_direct_targets = read.csv('../gene_lists/nfkb_targets.csv')
tnfa_direct_targets$Human.Gene.Name = gsub("BLIMP1 /PRDM1", "PRDM1", tnfa_direct_targets$Human.Gene.Name)
tnfa_direct_targets$Human.Gene.Name = gsub("SERPINE1, PAI-1", "SERPINE1", tnfa_direct_targets$Human.Gene.Name)
tnfa_genes= unique(tnfa_direct_targets$Human.Gene.Name[tnfa_direct_targets$Human.Gene.Name %in% rownames(all_timepoints)])
```
## Supplementary figure 3.a
```{r}
gene_expression= AverageExpression(subset(all_timepoints, cells = all_timepoints$barcode[all_timepoints$legend %in% levels(all_timepoints$legend)[c(1,3, 5, 9, 16:20)]]), assays = 'SCT', slot = 'counts', features = tnfa_genes, group.by = 'legend_condition')
gene_expression= as.data.frame(gene_expression$SCT)
gene_expression$gene= rownames(gene_expression)
tnfa_genes= tnfa_genes[tnfa_genes %in% rownames(gene_expression)]
pct_expression_data= data.frame(fake_cluster= 1:length(tnfa_genes), row.names = tnfa_genes )
for (cluster in colnames(gene_expression) ){
cells= all_timepoints$barcode[all_timepoints$legend_condition==cluster]
cluster_expression= as.data.frame(all_timepoints@assays$RNA@counts[tnfa_genes, cells]>0)
pct_expression= data.frame(apply(cluster_expression, MARGIN = 1, FUN = function(x){ round(sum(x)/length(x)*100, digits = 0) } ))
colnames(pct_expression)= cluster
pct_expression_data= cbind(pct_expression_data, pct_expression)
}
remove(cluster, pct_expression)
pct_expression_data=pct_expression_data[, 2:29]
pct_expression_data$gene= rownames(pct_expression_data)
pct_expression_data = tidyr::pivot_longer(pct_expression_data, cols = 1:28, values_to = 'pct_expression', names_to = 'cluster')
gene_expression = tidyr::pivot_longer(gene_expression, cols = 1:28, values_to = 'avg_expression', names_to = 'cluster')
#gene_expression$avg_expression = log10(gene_expression$avg_expression+1)
plot_data= gene_expression
plot_data$pct_expression= pct_expression_data$pct_expression
cluster_metadata= all_timepoints@meta.data %>% group_by(legend_condition) %>% summarise(cluster= unique(legend_condition), legend= unique(legend), condition= unique(condition) )
plot_data= left_join(plot_data, cluster_metadata[, 2:4])
plot_data= plot_data[plot_data$avg_expression >0.1 & plot_data$gene %in% tnfa_genes,]
plot_data$gene= factor(plot_data$gene, levels = rev(tnfa_genes))
colnames(plot_data)[3:4]= c('Average expression', '% cells expressing')
plot_data= left_join(plot_data, tnfa_direct_targets[, c('Human.Gene.Name', 'Group')] , by=c('gene'='Human.Gene.Name'))
ggplot(plot_data, aes(y= gene, x= condition, size=`% cells expressing`, col=`Average expression` )) + geom_point() + facet_wrap('legend', nrow = 1) + theme_bw() + RotatedAxis() + scale_color_gradientn(colors = c("orange", "firebrick"))
```
# Analysis of acyclovir treated control (CTRL ACV) organoids
Read digital gene expression matrices (generated with Spacemake) for CTRL samples and create Seurat objects for each sample keeping barcodes with at least 250 detected genes and genes detected at least 5 cells
```{r eval=FALSE, include=FALSE}
Xmoo1_CTRLACV_1 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_CTRLACV_1.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_CTRLACV_1', assay = 'RNA')
Xmoo1_CTRL_1_exonic$line = 'iPSC line 1'
Xmoo1_CTRL_1_exonic$replicate = 'Rep. 1'
Xmoo1_CTRL_1_exonic$condition = 'CTRL ACV'
Xmoo1_CTRLACV_2 = read.table(file = 'digital_gene_expression/scRNAseq_spacemake/Xmoo1_CTRLACV_2.txt.gz', sep = '', row.names = 1, header = T) %>% CreateSeuratObject(min.cells = 5, min.features = 250, project = 'Xmoo1_CTRLACV_2', assay = 'RNA')
Xmoo1_CTRLACV_2$line = 'iPSC line 1'
Xmoo1_CTRLACV_2$replicate = 'Rep. 2'
Xmoo1_CTRLACV_2$condition = 'CTRL ACV'
```
Create a single integrated Seurat object with all CTRL samples (2 replicates for 2 cell lines) after correcting for sample- and cell line-specific batch effects
```{r eval=FALSE, include=FALSE}
integration.list <- list(Xmoo1_CTRLACV_1,Xmoo1_CTRLACV_2, Xmoo1_CTRL_1_exonic, Xmoo1_CTRL_2_exonic, Gline_CTRL_1_exonic, Gline_CTRL_2_exonic)
for (i in 1:length(integration.list)) {
integration.list[[i]] <- SCTransform(integration.list[[i]], verbose = T,method= 'glmGamPoi', assay = 'RNA', new.assay.name = 'SCT', vst.flavor='v2')
}
integration.features <- SelectIntegrationFeatures(object.list = integration.list, nfeatures = 3000)
integration.list <- PrepSCTIntegration(object.list = integration.list, anchor.features = integration.features, verbose = T)
integration.anchors <- FindIntegrationAnchors(object.list = integration.list, normalization.method = "SCT", anchor.features = integration.features, verbose = T)
CTRL_ACV <- IntegrateData(anchorset = integration.anchors, normalization.method = "SCT", verbose = T)
DefaultAssay(CTRL_ACV) <- 'integrated'
remove(i, integration.list, integration.features, integration.anchors)
```
Perform dimensionality reduction and clustering with 30 principal components and default parameters
```{r eval=FALSE, include=FALSE}
CTRL_ACV <- CTRL_ACV %>% RunPCA( verbose = T) %>% RunUMAP( dims = 1:30, verbose = T) %>% FindNeighbors( dims = 1:30, verbose = T) %>% FindClusters( verbose = T)
```
Inspect transcript counts (nCount) and % of mitochondrial transcripts (percent_MT) to identify low quality clusters
```{r eval=FALSE, include=FALSE}
VlnPlot(CTRL_ACV, 'nCount_RNA', y.max = 2000, pt.size = 0)+ NoLegend()
VlnPlot(CTRL_ACV, 'percent_MT', y.max = 2000, pt.size = 0)+ NoLegend()
```
Remove low quality clusters 2 (low nCount) and 11 (low nCount, high mito) and select top 75% cells by nCount per cluster
```{r eval=FALSE, include=FALSE}
CTRL_ACV$barcode= colnames(CTRL_ACV)
top75= CTRL_ACV@meta.data[!CTRL_ACV$seurat_clusters %in% c(2, 11),] %>% group_by(seurat_clusters) %>% top_frac(n = 0.75, wt = nCount_RNA_exonic)
CTRL_ACV = subset(CTRL_ACV, cells=top75$barcode)
```
Re-run integration, dimensionality reduction and clustering
```{r eval=FALSE, include=FALSE}
Idents(CTRL_ACV) = 'sample'
integration.list <- SplitObject(CTRL_ACV)
for (i in 1:length(integration.list)) {
integration.list[[i]] <- SCTransform(integration.list[[i]],method= 'glmGamPoi', assay = 'RNA', new.assay.name = 'SCT', vst.flavor='v2')
}
integration.features <- SelectIntegrationFeatures(object.list = integration.list, nfeatures = 3000)
integration.list <- PrepSCTIntegration(object.list = integration.list, anchor.features = integration.features)
integration.anchors <- FindIntegrationAnchors(object.list = integration.list, normalization.method = "SCT", anchor.features = integration.features)
CTRL_ACV <- IntegrateData(anchorset = integration.anchors, normalization.method = "SCT")
DefaultAssay(CTRL_ACV) <- 'integrated'
CTRL_ACV <- CTRL_ACV %>% RunPCA() %>% RunUMAP( dims = 1:30) %>% FindNeighbors( dims = 1:30) %>% FindClusters()
remove(i, integration.list, integration.features, integration.anchors)
```
Add annotations of CTRL cells
```{r eval=FALSE, include=FALSE}
CTRL_ACV$legend_CTRL = CTRL$legend
```
Annotate clusters accordingly
```{r eval=FALSE, include=FALSE}
Idents(CTRL_ACV) <- 'seurat_clusters'
CTRL_ACV <- RenameIdents(
object = CTRL_ACV,
'0'= 'Immature Cortical Neurons 2',
'1'= 'Radial Glia G1/S Phase',
'2'= 'Immature Cortical Neurons 1',
'3'= 'Radial Glia G2M Phase',
'4'= 'Astroglia',
'5'= 'Mature Cortical Neurons',
'6'= 'Intermediate Progenitors',
'7'= 'Hindbrain Neurons 1',
'8'= 'Radial Glia G1/S Phase',
'9'= 'Cortical Hem/Choroid Plexus',
'10'= 'Thalamic Neurons',
'11'= 'Intermediate Progenitors',
'12'= 'Mature Cortical Neurons',
'13'= 'Retinal Progenitors',
'14'= 'Thalamic Neurons',
'15'= 'Astroglia',
'16'= 'Retinal Pigmented Cells',
'17'= 'Progenitors',
'18'= 'Progenitors',
'19'= 'Mural Cells',
'20'= 'Cortical Hem/Choroid Plexus',
'21'= 'Hindbrain Neurons 2',
'22'= 'Hindbrain Neurons 2',
'23'= 'Cajal Retzius Neurons')
CTRL_ACV$legend = Idents(CTRL_ACV)
```
Import processed object
```{r}
CTRL_ACV= readRDS('seurat_objects/CTRL_ACV.rds')
```
## Extended figure 3.b
```{r}
DimPlot(CTRL_ACV, order = T, split.by = 'condition', cols = color_palette)
```
Compute TNFA pathway activity in single cells
```{r eval=FALSE, include=FALSE}
# Rank gene by their expression in each single cell
cells_rankings = AUCell_buildRankings(CTRL_ACV@assays$SCT@counts, nCores = 40)
# Compute the enrichment of TNFA genes
cells_AUC = AUCell_calcAUC(list(TNFA=tnfa_genes), cells_rankings, aucMaxRank=nrow(cells_rankings)*0.05, nCores = 40)
# Add TNFa score to metadata
CTRL_ACV$tnfa_score = cells_AUC@assays@data$AUC['TNFA',]
remove(cells_rankings, cells_AUC)
```
## Extended figure 6.a
```{r}
rbind(all_timepoints@meta.data[, c('tnfa_score', 'condition')], CTRL_ACV@meta.data[CTRL_ACV$condition== 'CTRL ACV', c('tnfa_score', 'condition')] ) %>%
ggplot(aes(x=condition, y=tnfa_score, fill= condition)) + geom_boxplot() + ylim(c(0, 0.15)) +
theme_bw()
```
# Analysis of bulk RNAseq data from CTRL and HSV1 3 day post infection (3dpi) organoids
Import gene expression counts and sample metadata after alignment with PigX pipeline
```{r}
countData= read.csv('digital_gene_expression/bulkRNAseq_pigx/feature_counts.tsv', sep = '\t')
colData= read.csv('digital_gene_expression/bulkRNAseq_pigx/colData.tsv', sep = '\t')[colnames(countData), ]
```
Run differential gene expression using DESeq2
```{r}
# Prepare DESeq 2 object and exclude cell line specific effects
bulk_data <- DESeqDataSetFromMatrix(countData = countData,
colData = colData,
design = ~ line + group)
# Filter lowly expressed genes
bulk_data <- bulk_data[rowSums(counts(bulk_data)) >= 10,]
# Fit model
bulk_data <- DESeq(bulk_data)
# Extract logFC and expression for all genes
differential_results <- results(bulk_data)
differential_results= data.frame(differential_results@listData, gene = rownames(bulk_data), row.names = rownames(bulk_data))
remove(countData, colData, bulk_data)
```
Identify synaptic genes
```{r}
synaptic.dt = gconvert(query = "GO:0007268")
synaptic.dt= synaptic.dt[synaptic.dt$target %in% rownames(countData),]
```
## Extended figure 2.b
```{r}
syn.labels <- c("ARC", "NPTX2", "EGR2", "SV2B", "HTR2A",
"GRIM3", "SV2A", "SCN2B", "ADCY1", "CALB2",
"CPLX2", "SYT3", "MAP1A", "MAPT", "GABBR2",
"SEPT5", "NLGN3", "RIMS1", "RIMS4", "SST",
"SYP", "VAMP2", "GRIN1", "CHARM3", "SNAP25")
synaptic.selected = differential_results[synaptic.dt$target[synaptic.dt$name %in%syn.labels],]
synaptic.selected$symbol = synaptic.dt$name[synaptic.dt$name %in%syn.labels]
ggplot(differential_results, aes(x= log10(baseMean+1), y= log2FoldChange)) +
geom_point(size=0.01, col= 'lightgrey') +
geom_point(data= differential_results[grepl(pattern = 'HSV', differential_results$gene),],
mapping= aes(x= log10(baseMean+1), y= log2FoldChange),size= 0.5,col= 'darkgreen')+
#geom_density2d(data = differential_results[synaptic.dt$target, ],
# mapping = aes(x= log10(baseMean+1), y= log2FoldChange), col='black') +
geom_hline(yintercept = 0, linetype=2) +
geom_point(data = differential_results[synaptic.dt$target, ],
mapping = aes(x= log10(baseMean+1), y= log2FoldChange), size= 0.5, col= 'red') +
theme(panel.grid.major = element_blank(), panel.background = element_blank(), axis.line = element_line(colour = "black"), panel.border = element_blank())+
ggtitle('HSV1 3dpi vs CTRL (BulkRNAseq)')
remove(synaptic.dt, synaptic.selected)
```
Identify TNF ligands
```{r}
# Import TNF ligands from HGNC database
tnf_ligands= read.table('gene_lists/tnf_ligands_HGNC.csv', header = T, sep=',')[, 'Approved.symbol']
#Convert gene symbols to ensemble ID
library("org.Hs.eg.db")
tnf_ligands_ensembleID <- mapIds(org.Hs.eg.db, keys = tnf_ligands, keytype = "SYMBOL", column="ENSEMBL")
# Add gene symbols for TNF ligands
differential_results$gene_symbol= NA
differential_results[tnf_ligands_ensembleID , 'gene_symbol' ]= names(tnf_ligands_ensembleID)
```
## Extended figure 7.a
```{r}
# Plot FC against mean expression for all genes and highlight viral genes and TNF ligands
ggplot(differential_results, aes(x= log10(baseMean+1), y= log2FoldChange)) +
geom_point(size= 0.01, col= 'lightgrey') +
geom_point(data= differential_results[grepl(pattern = 'HSV', differential_results$gene),],
mapping= aes(x= log10(baseMean+1), y= log2FoldChange),size= 1,col= 'darkgreen')+
geom_point(data= differential_results[differential_results$gene %in% tnf_ligands_ensembleID,],
mapping= aes(x= log10(baseMean+1), y= log2FoldChange),size= 1,col= 'firebrick')+
geom_hline(yintercept = 0, linetype=2) +
ggrepel::geom_text_repel(data= differential_results[differential_results$log2FoldChange>2.5, ],
mapping = aes(x= log10(baseMean+1), y= log2FoldChange, label= gene_symbol), min.segment.length = 0, max.overlaps = 30)+
theme(panel.grid.major = element_blank(), panel.background = element_blank(), axis.line = element_line(colour = "black"), panel.border = element_blank())+
ggtitle('HSV1 3dpi vs CTRL (BulkRNAseq)')
remove(tnf_ligands,tnf_ligands_ensembleID)
```
## Extended figure 7.b
```{r}
# Extract sample level pseudobulk profiles from single cell data
sc_pseudobulk= t(rowsum(x = t(as.matrix(all_timepoints@assays$RNA@counts)), group = paste0(all_timepoints$condition, '_', all_timepoints$line, '_', all_timepoints$replicate) ))
# Define sample level metadata
sc_colData= data.frame(group= c('CTRL','CTRL','CTRL','CTRL', 'inf1dpi','inf1dpi','inf1dpi','inf1dpi', 'inf3dpi', 'inf3dpi','inf3dpi', 'inf3dpi', 'inf3dpi_ACV','inf3dpi_ACV', 'inf3dpi_ACV','inf3dpi_ACV'), line= c('line 1','line 1','line 2','line 2', 'line 1','line 1','line 2','line 2','line 1','line 1','line 2','line 2','line 1','line 1','line 2','line 2'), row.names = colnames(sc_pseudobulk) )
# Define upregulated TNF ligands
upregulated_tnf= names(tnf_ligands_ensembleID)[tnf_ligands_ensembleID %in% differential_results$gene[differential_results$log2FoldChange>2.5]]
# Plot heatmap of upregulated tnf ligands detected in single cell data
pheatmap(sc_pseudobulk[rownames(sc_pseudobulk) %in% upregulated_tnf, rownames(sc_colData) ], annotation_col = sc_colData, cluster_cols = F, show_colnames = F, scale='row',
color= brewer.pal(9,'Blues'),
annotation_colors = list(group= c(CTRL= 'grey' , inf1dpi= 'lightgreen', inf3dpi= 'darkgreen' , inf3dpi_ACV='orange' )),
height = 7, width = 7, main = 'Upreglated TNF ligands detected in scRNAseq (pseudobulk)')
remove(sc_pseudobulk, sc_colData, upregulated_tnf, differential_results)
```pheatmap vs heatmap.2
pheatmap and heatmap.2 are two popular R functions used to generate heatmaps. Both functions are based on the base heatmap function in R, but they have additional features and customizations that make them more flexible and user-friendly. Here are some differences between the two functions:
-
Package: pheatmap is a function in the pheatmap package, while heatmap.2 is part of the gplots package.
-
Clustering: Both pheatmap and heatmap.2 can perform hierarchical clustering on rows and columns. However, pheatmap allows users to directly input custom clustering results, while heatmap.2 requires the data to be pre-clustered and sorted accordingly.
-
Annotations: pheatmap has a more straightforward way of adding annotations to rows and/or columns through the annotation_row and annotation_col parameters. In heatmap.2, you can add annotations through the Rowv and Colv parameters, but it requires more manual manipulation.
-
Color scales: Both functions allow customization of color scales, but pheatmap provides the colorRampPalette function for generating color scales, while heatmap.2 uses the colorpanel function.
-
Output: pheatmap outputs a grid object that can be further customized using the grid package functions. heatmap.2 generates base R graphics, and its output is less flexible for customization.
-
Overall, pheatmap is known for its simplicity and ease of use, whereas heatmap.2 offers more customization options but might be more complex to use.
-
Plot the organoid data using heatmap.2
rld <- rlogTransformation(dds) GOI <- c("AL", "IRL1","IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "pri-miRNA", "TRL3", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20","UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46","UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") RNASeq.NoCellLine <- assay(rld) library("gplots") datamat = RNASeq.NoCellLine[GOI, ] write.csv(as.data.frame(datamat), file ="viral_gene_expressions.txt") constant_rows <- apply(datamat, 1, function(row) var(row) == 0) if(any(constant_rows)) { cat("Removing", sum(constant_rows), "constant rows.\n") datamat <- datamat[!constant_rows, ] } hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.00) mycol = c("YELLOW", "BLUE", "ORANGE", "MAGENTA", "CYAN", "RED", "GREEN", "MAROON", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN", "LIGHTRED", "LIGHTGREEN"); mycol = mycol[as.vector(mycl)] png("viral_deg_heatmap2.png", width=600, height=1000) #labRow="", heatmap.2( as.matrix(datamat), Rowv=as.dendrogram(hr), Colv = NA, dendrogram = 'row', scale='row', trace='none', col=bluered(75), cexCol=1.8, RowSideColors = mycol, margins=c(8,8), cexRow=1.3, # Adjust font size for row labels srtCol=30, lhei = c(1, 10), # Adjust relative height of rows, larger values give more space to the heatmap lwid=c(2, 8) ) dev.off() -
Plot the organoid data using pheatmap
# Extract the colData as metadata metadata <- as.data.frame(colData(dds)) # Select only the 'condition' column for the heatmap annotations and rename it heatmap_metadata <- metadata[ , "condition", drop=FALSE] colnames(heatmap_metadata) <- "Condition" # Rename the column # Set the order of levels for 'Condition' column heatmap_metadata$Condition <- factor(heatmap_metadata$Condition, levels = c('control', 'HSV.d2', 'HSV.d4', 'HSV.d6', 'HSV.d8')) please reorder the color from light to dark # Colors for the heatmap based on the condition # Colors for the heatmap based on the condition condition_cols <- c( 'control' = 'grey', 'HSV.d2' = '#F1B3F4', # Lightest 'HSV.d4' = '#E080C1', 'HSV.d6' = '#BE64C2', 'HSV.d8' = '#A13FA6' # Darkest ) # Drawing the heatmap using pheatmap library(pheatmap) png("viral_deg_pheatmap.png", width=600, height=1000) pheatmap( datamat, # Using rlog-transformed data directly cluster_rows = F, cluster_cols = F, display_numbers = F, color = colorRampPalette(c("white", "#23A17B"))(100), angle_col = 90, show_rownames = T, main = 'Viral gene expression (rlog)', fontsize = 10, fontsize_col = 10, annotation_col = heatmap_metadata, # Using only the 'Condition' column for annotations annotation_colors = list(Condition = condition_cols), # Adjusted to match the new column name show_colnames = F ) dev.off() -
Note that the plot above used rlog rather than log10. While both are types of logarithmic transformations, they serve slightly different purposes and are calculated differently.
-
rlogTransformation (regularized log transformation): This is a transformation used especially with DESeq2 package for RNA-seq data analysis. The “regularized” log transformation stabilizes the variance across genes, especially for genes with low counts, by borrowing information from all genes. It’s a variance stabilizing transformation that aims to make the variance roughly constant across different genes and conditions, without requiring separate normalization to correct for differences in sequencing depth or RNA composition.
-
log10(gene expression): This is a simple logarithm base 10 transformation of the gene expression values. This transformation is used to adjust the dynamic range of the data and make it more interpretable, especially when dealing with large differences in gene expression levels. But it does not have the variance stabilizing properties of the rlogTransformation.
- Original Label rlogTransformation Label (“Regularized log-transformed expression” or “rlog Expression”)
- Viral gene expression (log10) Viral gene expression (rlog)
- Host gene expression (log10) Host gene expression (rlog)
- Differential expression (log10) Differential expression (rlog)
- Transcription level (log10) Transcription level (rlog)
- Expression fold-change (log10) Expression fold-change (rlog)
-
炎症体:先天免疫与细胞自主免疫交汇的关键节点
Inflammasomes at the crossroads of innate and cell-autonomous immunity
炎症体位于先天免疫和细胞自主免疫的交汇点,其在我们体内的免疫防御中发挥着重要作用。
先天免疫是我们身体对抗病原体(如细菌、病毒和其他有害微生物)的第一道防线。它包括一系列的防御机制,如皮肤、粘液层和吞噬细胞等,这些机制可以迅速识别和消灭入侵者。
细胞自主免疫则是发生在细胞内部的免疫反应,其依赖于细胞内部的各种分子和蛋白质来识别和消除有害物质。
炎症体是一种复杂的蛋白质复合体,其在细胞内部形成,并且能够识别一系列的有害信号和入侵者。当炎症体被激活时,它能够引发炎症反应,促使细胞释放信号分子(如细胞因子),这些分子进而吸引免疫细胞前来消灭入侵者。
炎症体在先天免疫和细胞自主免疫中扮演着桥梁的角色。它不仅参与快速的先天免疫反应,帮助身体迅速响应入侵者,还能在细胞内部发挥作用,识别并消除那些可能逃避先天免疫防御的有害物质。
总的来说,炎症体通过在先天免疫和细胞自主免疫之间发挥关键作用,帮助我们的身体维持稳态,抵抗感染。
Genomic Organization of Merkel cell polyomavirus (MCPyV)
Based on the provided image of the MCPyV (Merkel cell polyomavirus) genome, it appears that ALTO is positioned between the LT (Large T antigen) and sT (small T antigen) regions. However, it’s not entirely clear from the diagram alone if ALTO is a distinct domain within sT, or if it’s a separate gene entirely.
In the literature, the ALTO protein (A novel T antigen Open Reading Frame) of MCPyV has been described as a unique feature of the virus. While its specific function and relationship to the other T antigens (LT and sT) are still a topic of research, it is indeed distinct from the canonical LT and sT proteins.
In short, based on the diagram and typical annotations for MCPyV, ALTO does not appear to be a part of sT; rather, it’s a distinct open reading frame (ORF) or protein. However, more detailed genomic annotations and experimental studies would be needed to definitively determine its relationship with other proteins encoded by the virus.
https://www.frontiersin.org/articles/10.3389/fmicb.2021.739695/full
JN707599.gtf
JN707599 Genbank gene 465 1190 . + . gene_id "VP2"; gene_type "protein_coding"
JN707599 Genbank transcript 465 1190 . + . gene_id "VP2"; transcript_id "tx-AEX86628.1"; gene_type "protein_coding"
JN707599 Genbank exon 465 1190 . + 0 gene_id "VP2"; transcript_id "tx-AEX86628.1"; gene_type "protein_coding"
JN707599 Genbank CDS 465 1190 . + 0 gene_id "VP2"; transcript_id "tx-AEX86628.1"; gene_type "protein_coding"
JN707599 Genbank gene 600 1190 . + . gene_id "VP3"; gene_type "protein_coding"
JN707599 Genbank transcript 600 1190 . + . gene_id "VP3"; transcript_id "tx-AEX86629.1"; gene_type "protein_coding"
JN707599 Genbank exon 600 1190 . + 0 gene_id "VP3"; transcript_id "tx-AEX86629.1"; gene_type "protein_coding"
JN707599 Genbank CDS 600 1190 . + 0 gene_id "VP3"; transcript_id "tx-AEX86629.1"; gene_type "protein_coding"
JN707599 Genbank gene 1156 2427 . + . gene_id "VP1"; gene_type "protein_coding"
JN707599 Genbank transcript 1156 2427 . + . gene_id "VP1"; transcript_id "tx-AEX86630.1"; gene_type "protein_coding"
JN707599 Genbank exon 1156 2427 . + 0 gene_id "VP1"; transcript_id "tx-AEX86630.1"; gene_type "protein_coding"
JN707599 Genbank CDS 1156 2427 . + 0 gene_id "VP1"; transcript_id "tx-AEX86630.1"; gene_type "protein_coding"
JN707599 Genbank gene 2503 5387 . - . gene_id "LT"; gene_type "protein_coding"
JN707599 Genbank transcript 5154 5387 . - . gene_id "LT"; transcript_id "tx1-AEX86632.1"; gene_type "protein_coding"
JN707599 Genbank exon 5154 5387 . - 0 gene_id "LT"; transcript_id "tx1-AEX86632.1"; gene_type "protein_coding"
JN707599 Genbank CDS 5154 5387 . - 0 gene_id "LT"; transcript_id "tx1-AEX86632.1"; gene_type "protein_coding"
JN707599 Genbank transcript 2503 4722 . - . gene_id "LT"; transcript_id "tx2-AEX86632.1"; gene_type "protein_coding"
JN707599 Genbank exon 2503 4722 . - 0 gene_id "LT"; transcript_id "tx2-AEX86632.1"; gene_type "protein_coding"
JN707599 Genbank CDS 2503 4722 . - 0 gene_id "LT"; transcript_id "tx2-AEX86632.1"; gene_type "protein_coding"
JN707599 Genbank gene 4827 5387 . - . gene_id "sT"; gene_type "protein_coding"
JN707599 Genbank transcript 4827 5387 . - . gene_id "sT"; transcript_id "tx-AEX86631.1"; gene_type "protein_coding"
JN707599 Genbank exon 4827 5387 . - 0 gene_id "sT"; transcript_id "tx-AEX86631.1"; gene_type "protein_coding"
JN707599 Genbank CDS 4827 5387 . - 0 gene_id "sT"; transcript_id "tx-AEX86631.1"; gene_type "protein_coding"JN707599.fasta
JN707599 CTTGTCTATATGCAGAAGGAGTTTGCAGAAAGAGCAGAGGAGCAAATGAGCTACCTCACTAAGGAGTGGT TTTTATACTGCAGTTTCCCGCCCTTGGGATCTGCCCTTAGATACTGCCTTTTTTGCTAATTAAGCCTCTT AAGCCTCAGAGGCCTCTCTCTTTTTTTTCCAGAGGCCTCGGAGGCTAGGAGCCCCAAGCCTCTGCCAACT TGAAAAAAAAAAGTCACCTAGGCAGCCAAGTTGTGGTTACATGATTGAACTTTTATTGCTGCAGGGTTTC TGGCATTGACTCATTTCCTGGAGAGGCGGAGTTTGACTGATAAACAAAACTTTTTTTCTTTCTGTTTGGG AGGGAGACGGAAGACTCTTAACTTTTTTTCAACAAGGGAGGCCCGGAGGCTTTTTTTTCTCTTACAAAGG GAGGAGGACATTAAAAGAGTAAGTATCCTTATTTATTTTTCAGGATGGGGGGCATCATCACACTGCTGGC CAATATTGGTGAAATTGCTACTGAACTAAGTGCCACCACAGGAGTAACTTTGGAAGCTATTCTTACAGGA GAAGCTTTAGCAGCTTTGGAAGCAGAGATCTCCAGTTTAATGACAATTGAGGGTATTTCTGGCATTGAGG CTTTAGCTCAACTTGGGTTCACAGCTGAACAGTTTTCAAATTTCTCATTAGTGGCTTCTTTGGTTAACCA AGGTTTAACTTATGGCTTCATTCTCCAAACTGTTAGTGGTATAGGCTCTCTAATAACTGTGGGGGTGAGG TTGTCACGCGAGCAAGTGTCACTTGTAAAGAGGGATGTTTCGTGGGTAGGTAGTAATGAGGTTTTGAGGC ATGCACTTATGGCCTTTAGCCTAGATCCTCTGCAGTGGGAAAATAGCTTGCTGCATTCTGTGGGGCAAGA TATTTTTAATTCTTTATCTCCTACCTCTAGGCTGCAGATACAATCAAACCTAGTGAATCTGATACTAAAT AGCCGGTGGGTCTTTCAGACAACTGCTTCTCAGAATCAGGGCCTTTTATCAGGAGAGGCTATATTAATTC CTGAACATATAGGAGGAACTCTGCAGCAGCAAACTCCAGATTGGCTTCTTCCTCTGGTACTAGGCCTTAG TGGATATATTTCTCCTGAATTACAAGTAATTGAAGATGGCACCAAAAAGAAAAGCATCATCCACCTGTAA AACACCCAAAAGGCAATGTATACCTAAGCCGGGATGCTGCCCTAATGTTGCCTCAGTTCCAAAACTGCTT GTTAAAGGAGGAGTGGAAGTATTATCTGTGGTTACTGGAGAAGATAGCATTACCCAAATTGAGTTGTATT TGAATCCAAGAATGGGAGTTAATTCCCCTGATCTTCCTACTACTTCAAACTGGTATACTTATACTTATGA CCTGCAGCCAAAGGGATCATCTCCAGATCAGCCCATCAAGGAAAATTTGCCAGCTTACAGTGTGGCAAGA GTGTCTCTGCCAATGCTAAATGAGGATATTACCTGTGACACATTGCAGATGTGGGAGGCAATATCTGTTA AAACAGAAGTAGTTGGAATAAGTTCTTTAATTAATGTTCATTATTGGGACATGAAAAGAGTTCATGATTA TGGTGCTGGTATTCCTGTGTCAGGGGTAAATTACCATATGTTTGCCATTGGGGGAGAACCTCTGGATTTG CAAGGCCTAGTTTTAGATTACCAGACTGAGTATCCAAAAACTACAAATGGTGGGCCTATTACAATTGAAA CTGTATTGGGAAGAAAAATGACACCTAAAAATCAGGGCCTAGATCCACAAGCTAAAGCAAAATTAGATAA AGATGGAAATTATCCTATAGAAGTATGGTGTCCTGATCCTTCTAAAAATGAAAACAGTAGATACTATGGG TCTATTCAGACAGGCTCTCAGACTCCTACAGTTCTTCAATTTAGTAATACTCTAACTACTGTCCTTTTAG ATGAGAATGGAGTGGGCCCTCTATGCAAAGGAGATGGCCTATTTATTAGCTGTGCAGACATAGTGGGGTT TCTGTTTAAAACCAGTGGAAAAATGGCTCTTCATGGGTTGCCTAGATATTTTAATGTTACTTTGAGAAAA AGATGGGTGAAAAACCCCTACCCAGTAGTTAATTTAATAAACTCACTCTTCAGCAACTTAATGCCAAAAG TGTCAGGCCAACCTATGGAAGGAAAAGATAATCAGGTAGAAGAGGTTAGAATATATGAGGGGTCAGAACA ATTACCTGGTGATCCTGATATTGTCAGATTTTTAGATAAATTTGGGCAGGAGAAAACTGTTTACCCAAAG CCCTCTGTTGCCCCAGCAGCAGTAACATTCCAAAGTAATCAGCAGGATAAGGGCAAGGCGCCACTGAAAG GACCTCAAAAGGCCTCTCAAAAAGAAAGCCAAACACAAGAATTATGAGAATTATTTCATGCATTCCTATT CAGTTAAGTAGGCCCCAGAAAAACAAACACAGGAAATATGAAGCAGATGCCTTTATTGAGAAAAAGTACC AGAATCTTGGGTTTCTTCAGTTTCCTCAGGGCCCTCTTCCTCAATAAGAATATTGAGCAGAGGGTCCTGA CCAGCTTCTACATTTTCTATCATTTGACAAAATTTACCATATGATATTTCACTCTGTAAAATTTGCTTCC AGTTTTTAATTTCTTCTTGTAAGCAAGGCTTAAAGGTTGTATCAGGCAAGCACCAAATAAGACAAAGCAA TAAAGTGGTTCCACTTTGAAGAATTCTTCTTTTTCTTATTTCCATGTTCTGATCCAGGGAATCTCTTAGA TTTGCCTTTGGGGAAAAGTGTAAAGTATAACTAAATCTTGCTATTAATGTTTTGGGAATAAAATAATCAT TAGCAGTAACAATACAAGGAGGAAAAATCTGATGCTTTTTATTCACATGCTTCTTCTCTAAGCTTACAGC TACAGCACCATCTAGATGATCTCTTAAGTTATCAAGGTTATTTATTCCTTGCCCTGGTTGCAGATCTTTA TTTAGGCTATTTTGCCCTTTCACATCCTCAAAAACAACCATAAATTTATCCAAAGCACATCCTAGTTCAA AAGGCAGTTTATCAGATGGACAGTTTATATTCAAGGCCTTCCCTTCTAGCAAATCTATTAAGGCTGCAGC AAAGCTTGTTTTTCCACTGTTAATAGGCCCTTTAAACCAAATGTTTCTATACTTAGGTATATTCTCTGTT AATAATTGAATAATTTTCTGCAGCTTCTTTTCAAACTCTTCAAATAAGCAGCAGTACCAGGCCACACCAC CCATATAATACAGTAGATCTATTGTATCTAAATCTCTTAATCTCTCTAGGTGCTTCTTAAACTTCTTACA TAGCATTTCTGTCCTGGTCATTTCCAGCATCTCTAACCTCCTTTTGGCTAGAACAGTGTCTGCGGCTTGT TGGCAAATGGTTTTCTGAGATTTAGATTCATAAAATAGCTTAGCATTAGAATGATGAGCCTCATGAGCCT TGTGAGGTTTGAGGCGAGATCTGTTTTCACACTTTTGGCAAGGAAATGGTTTTGCAAAGTCTAGATAATG GGCTAAGATAATAAAGTGGTCGTCTAGCTCATATTCACAAGCAAATTCAGCAACTAAATTCCAATTACAG CTGGCCTCTTTTTCTTTTTCTTGAAATTCATAATTGAGCAGTGGCTTATTCTCTTGCAGTAATTTGTAAG GGGGCTTGCATAAATTATTATACATTTCAGGCATCTTATTCACTCCTTTACAAATTAAAAAGCTTATAGT GCAGAAGGTAGAGCAAAAATTCTTAATAGCAGATACTCTATGCTTTGATAAAGTTATAAACAATAAAATA CATCCTAATTCACAGGCATGCCTGCTTTTAAAATCAACTTTAAATTTCTCAATCTTATCATATAACTCTA TAGCTTTATCAGAAGTAGTATAAATGGCAAAACAACTTACTGTTTTATTACTATATACAGCATGGCTAAG ATAATCAGAAAGATCAATAGGAAAATCAGTAGGAACAGGAGTTTCTCTGTTCTTTTTTGGCTTTGGTGGA GTGCTTGTAAAACTTGCTGAACTAGCAGAGCTTGCAGAGCTTCGGGACCCCCCAAATTTTCGCTTTCTTG AGAATGGAGGAGGGGTCTTCGGGGTGGTGAAGGAGGAGGATCTGTATTCCTCATCTGTAAACTGAGATGA CGAGGCCTCCTCGGCAGAGGAAGACGGGGGCTGCCGGGGCGAGCTTCTTGAGGAGGGGGGCTCCTCAGGC TCCTCAGAGGACGAGGGAGGCTCAGGGGAGGAAAGTGATTCATCGCAGAAGAGATCCTCCCAGGTGCCAT CCGTTCTGGAAGAATTTCTAGGTACACTGGTTCCATTGGGTGTGCTGGATTCTCTTCCTGAATTGGTGGT CTCCTCTCTGCTACTGGATCCAGAGGATGAGGTGGGTTCCTCATGGTGTTCGGGAGGTATATCGGGTCCT CTGGACTGGGAGTCTGAAGCCTGGGACGCTGAGAAGGACCCATACCCAGAGGAAGAGCTCTGGCTGTGGG GTGGTGAGCTTCCACTGGGGGCTCCCCTGGATGCATTGGAGGAAGGCTTTCTGGATCTTGAGTTGGTCCC GTGTGGATTGGGCCCATATTCGTATGCCTTCCCGAAGCTGAATCCTCCTGATCTCCACCATTCTTTGAAT TTAGTGGTCCCATATATAGGGGCCTCGTCAACCTAGATGGGAAAGTACAGAAAATCTGTCATAAATAACC TTTCTTTGATATTTTGCCTTATAGACTTTTCCATATCTAATACTTACAGAGGAAGGAAGTAGGAGTCTAG AAAAGGTGCAGATGCAGTAAGCAGTAGTCAGTTTCTTCTAAAGTTTTTTGCCACCAGTCAAAACTTTCCC AAGTAGGAGGAAATCCAAACCAAAGAATAAAGCACTGATAGCAAAAACACTCTCCCCACGTCAGACAGTT TTTTTGCTTTAAAGTTTTTAGACTACAATGCTGGCGAGACAACTTACAGCTAATACAAGCGCACTTAGAA TCTCTAAGTTGCTTAAGCATGCACCCAGGACCTCTGCAAAATCTAGCATTATATCCACTTTGCATATAAT CCTTTAAAGTTCCATATTCTTCCCAAGGAAATTTTGTACTGACCTCATCAAACATAGAGAAGTCACTTCT GAGCTTGTGGATATTTTGCTGGAATTTGCTCCAAAGGGTGTTCAATTCCATCATTATAACAGGATTTCCC CCTTTATCAGGGTGATGCTTTAAGCAGCTTCTTTTGAAAGCAGCTTTCATCAGAGGGATGTTGCCATAAC AATTAGGAGCAATCTCTAAAAGCTTGCAGAGAGCCTCTCTTTCTTTCCTATTTAGGACTAAATCCAT
| title | treatment | time |
|---|---|---|
| untreated_DonorI | untreated | Day 0 |
| untreated_DonorII | untreated | Day 0 |
| p601_d3_DonorII | mCherry control | Day 3 |
| p604_d3_DonorII | sT | Day 3 |
| p601_d8_DonorII | mCherry control | Day 8 |
| p604_d8_DonorII | sT | Day 8 |
| p601_d3_DonorI | mCherry control | Day 3 |
| p604_d3_DonorI | sT | Day 3 |
| p601_d8_DonorI | mCherry control | Day 8 |
| p604_d8_DonorI | sT | Day 8 |
| p600_d3_DonorII | GFP control | Day 3 |
| p605_d3_DonorII | LTtr | Day 3 |
| p600_d8_DonorII | GFP control | Day 8 |
| p605_d8_DonorII | LTtr | Day 8 |
| p600_d3_DonorI | GFP control | Day 3 |
| p605_d3_DonorI | Lttr | Day 3 |
| p600_d8_DonorI | GFP control | Day 8 |
| p605_d8_DonorI | Lttr | Day 8 |
| p602_d8_DonorII | LT | Day 8 |
| p602_d8_DonorI | LT | Day 8 |
| p600and601_d12_DonorI | GFP+mCherry control | Day 12 |
| p604and605_d12_DonorI | sT+LTtr | Day 12 |
| p600and601_d9_DonorII | GFP+mCherry control | Day 9 |
| p604and605_d9_DonorII | sT+LTtr | Day 9 |
| p602_d3_DonorI | LT | Day 3 |
| p602_d3_DonorII | LT | Day 3 |
| p602and604_d3_DonorI | sT+LT | Day 3 |
| p602and604_d3_DonorII | sT+LT | Day 3 |
-
untreated
-
p602 LT *
-
p687 LT K331A *
-
p605 LTtr * (vs. p600 GFP control)
-
p604 sT
-
p602+604 LT+sT (vs. p601 mCherry control)
-
p604+p605 sT+LTtr (vs. GFP+mCherry control p601+p600)
Filtering RNA-seq analysis results and Workbook Creation with R
# Load required libraries
if(!requireNamespace("readxl", quietly = TRUE)) install.packages("readxl")
if(!requireNamespace("dplyr", quietly = TRUE)) install.packages("dplyr")
if(!requireNamespace("writexl", quietly = TRUE)) install.packages("writexl")
if(!requireNamespace("openxlsx", quietly = TRUE)) install.packages("openxlsx")
library(readxl)
library(dplyr)
library(writexl)
library(openxlsx)
setwd("/media/jhuang/Seagate Expansion Drive/Data_Denise_RNASeq/results_24samples/featureCounts/degenes_2021")
# Read the data from the specific sheet
data <- read_excel("degenes_replicates.xls", sheet = "p604_d3_vs_p601_d3-all")
print(data, n = 10, width = Inf)
# Filter the data
#filtered_data <- data %>%
# filter(padj < 0.05, log2FoldChange >= 2)
upregulated_genes <- data %>%
filter(padj < 0.05, log2FoldChange >= 2)
dim(upregulated_genes)
downregulated_genes <- data %>%
filter(padj < 0.05, log2FoldChange <= -2)
dim(downregulated_genes)
# Create a new Excel workbook
wb <- createWorkbook()
# Add sheets with data
addWorksheet(wb, "Up-regulated")
writeData(wb, "Up-regulated", upregulated_genes)
addWorksheet(wb, "Down-regulated")
writeData(wb, "Down-regulated", downregulated_genes)
# Save the workbook
saveWorkbook(wb, "sT_d3_up_and_down.xlsx", overwrite = TRUE)
data <- read_excel("p604_vs_p601_d8.xls", sheet = "p604_d8_vs_p601_d8-all")
print(data, n = 10, width = Inf)
upregulated_genes <- data %>%
filter(padj < 0.05, log2FoldChange >= 2)
dim(upregulated_genes)
downregulated_genes <- data %>%
filter(padj < 0.05, log2FoldChange <= -2)
dim(downregulated_genes)
# Create a new Excel workbook
wb2 <- createWorkbook()
# Add sheets with data
addWorksheet(wb2, "Up-regulated")
writeData(wb2, "Up-regulated", upregulated_genes)
addWorksheet(wb2, "Down-regulated")
writeData(wb2, "Down-regulated", downregulated_genes)
# Save the workbook
saveWorkbook(wb2, "sT_d8_up_and_down.xlsx", overwrite = TRUE)