亚MIC驱动的表型异质与耐药演化:以 A. baumannii RND 外排泵为核心的机制与干预
模型选择与对比:如何在有限功效下回答“two≈one@17h、two>one@24h(尤其ΔadeIJ)”
三种模型的对比
1) 分层按时间做(Option A:每个时间点各跑一个模型)
公式:在 17 h 子集:~ exposure * genotype;在 24 h 子集:~ exposure * genotype
(因为同一子集里 time_h 不变,再加 + time_h 是冗余的)
能回答的问题
- 直接给出同一时间点内的所有核心对比(如 two vs one、基因型差异及其交互)。
- 很适合你现在关心的:“17 h 两次≈一次,而 24 h 两次>一次?”
优点
- 参数少、功效高(不去估计三方交互)。
- 结果直观:每个时间点一套 DEGs,便于和 Fig.3c 的现象逐一对应。
- 抗不平衡设计(两个时间点样本数/离群点不一样时更稳)。
缺点
- 不能在一个模型里“正式检验”时间是否改变了暴露×基因型的效应(缺少三方交互的整体显著性检验)。
- 跨时间比较需要你在结果层面去比对(例如比较 17 h 和 24 h 的 LFC/DEG 数),不是模型内参数。
适用场景:样本量有限、你最关心分时间的效应;先得到清晰、功效高的 per-time 结果。
2) 全局简化模型(Global reduced):~ exposure * genotype + time_h
能回答的问题
- “暴露效应是否被基因型修改?”(有 exposure×genotype 交互)
- 同时控制了一个时间的平均主效应。
优点
- 比全模型参数更少、功效更高。
- 仍可做“差-中-差”式的交互检验(但这是跨两个时间的平均)。
缺点
- 假设暴露×基因型的交互在 17 h 和 24 h 相同/相似;
- 无法告诉你“17 h 小、24 h 大”(时间特异的交互被“平均”掉)。
适用场景:如果 PCA/探索性分析显示时间影响较小而且平行,且你只想得到“平均意义”上的交互结论。
3) 全模型(Full):~ exposure * genotype * time_h
能回答的问题
- 所有主效应与全部交互,尤其是三方交互(检验“暴露×基因型 的差异是否随时间改变”)。
- 可用 LRT(似然比检验) 与简化模型比较,正式证明时间特异。
优点
- 统计上最完整;能一口气检验你提出的“17 h 小、24 h 大”这种时间依赖。
缺点
- 参数最多、功效最低(在基因层面更难达显著;多重比较负担更大)。
- 解释略复杂(系数多,需要合成对比)。
适用场景:样本量足/信号强,或者用于确证“时间改变交互”的结论(比如在 Option A 看到强烈趋势之后)。
给你这套数据的建议(结合你目标与当前信号)
主分析:优先用 Option A(分层按时间)
- 在 17 h 和 24 h 分别拟合
~ exposure * genotype,导出 two vs one(WT 与 ΔadeIJ 各一套)。 - 这和 Fig.3c 的读图是一致的,功效也最好;如果你的预期正确,17 h 的 two vs one 基本不显著,24 h 显著增多,尤其是 ΔadeIJ。
确证时间依赖(可选但推荐):跑一版 全模型
- 用
~ exposure * genotype * time_h做 LRT 对比简化模型~ exposure * genotype + time_h,
若大量基因在 LRT 中显著 ⇒ 说明三方交互真实存在(时间改变了交互)。 - 对重点基因/通路,再报告三方交互的方向与效应量。
若功效更有限或只想给一个“总体交互”的结论:
- 可以只给 Global reduced 的结果(平均的 exposure×genotype 交互),
- 再辅以 Option A 的火山图作为“时间分层的可视化支持”。
简短决策树
- 你要看清 17 h vs 24 h 的差别 → Option A
- 你要在统计上证明“差别随时间改变” → Full + LRT(可在 Option A 之后做)
- 你要功效最大但只要平均结论 → Global reduced
实操小贴士
- 两个时间点样本数尽量均衡;有离群点先处理(PCA/库大小/样本间相关)。
- 需要时加入批次/库制备等协变量(
+ batch),在三种模型里都可以加。 - 报告时同时给:DEG 数、FDR、代表基因 LFC,并用 GSEA/富集做通路层面的对照。
- 图表:每个时间点出 two vs one 的火山图/热图;全模型的 LRT p 值分布或显著通路一览。
一句话结论
为了回答你现在最关心的“17 h 的 two≈one,24 h 的 two>one,尤其在 ΔadeIJ”,Option A 是首选(清晰、功效高);随后用全模型 + LRT补一个“官方盖章”的时间依赖即可。
0. 术语速览(便于快速阅读)
- MDR:多重耐药(Multi-Drug Resistance)
- RND 外排泵:Resistance–Nodulation–Division 家族(A. baumannii 主要为 AdeABC/AdeIJK)
- sub-MIC / 亚MIC:低于最低抑菌浓度(MIC)的暴露/处理
- R/S 亚群:在选择压力下仍能成殖的耐受/适应亚群(R)与不成殖/受损的易感亚群(S)
- PAP:群体分析谱(Population Analysis Profiling)
- Time-kill / MDK:时间杀菌曲线 / 最小杀灭持续时间(MDK99/MDK99.99)
- AUM:人工尿培养基(Artificial Urine Medium)
- MH2B(MHB):Mueller–Hinton Broth 第二配方
- EPI:外排泵抑制剂(Efflux Pump Inhibitor)
1. 研究背景与问题定位
1.1 临床与公共卫生动因
- MDR 革兰阴性菌威胁升级,A. baumannii 在 ICU/院感中占比高,治疗选择有限。
- 最后线药物(替加环素、碳青霉烯等)敏感率下滑,且地区波动大,提示环境/宿主体内因素的重要性。
- 亚MIC 暴露在临床(药代/组织渗透/生物膜)与环境(废水/污泥/水体)里普遍存在,能驱动适应、耐受、异质耐药等演化过程。
1.2 科学空白
- RND 外排泵被公认为 A. baumannii 耐药关键,但基因表达与表型耐药不总一致;
- 异质性(heteroresistance/耐受/持留)与宿主相关培养基(AUM/尿液)的影响在常规 AST 中难以显现;
- 缺少将亚MIC → 表型异质 → 宿主环境转录重编程串联起来的系统证据链。
2. 科学问题、核心假说与研究目标
2.1 科学问题(What)
- 亚MIC 替加环素如何诱发/放大 A. baumannii 的表型异质性与耐受/适应亚群?
- RND 外排泵(AdeABC/AdeIJK)在这一过程中扮演何种动力学与调控角色?
- 尿液/AUM等宿主相关环境如何重编程转录网络、改变药敏表型与毒力?
2.2 核心假说(Why + How)
- H1:反复亚MIC 暴露通过应激响应/膜稳健性/代谢重分配,选择或诱导“在膜/外排应激下仍能成殖”的 R 亚群上升;RND 缺失株(ΔadeIJ)这一现象尤显著。
- H2:RND 泵不仅通过外排直接降低胞内药物,还通过变构/能量耦联影响膜稳健、代谢与应激路径,放大表型异质。
- H3:尿液/AUM汇聚多因素(渗透压、尿素、金属离子、碳源限制),系统性重编程外排泵与代谢网络,导致MIC 变化与毒力改变。
2.3 研究目标(Deliverables)
- 建立亚MIC → R/S 亚群的定量框架(PAP、MDK、脉冲生存)并定位分子机制;
- 解析 RND 在异质性与宿主环境适应中的作用;
- 构建UTI 相关药敏解读范式与EPI 靶点清单。
3. 实验总体设计与样本体系
3.1 株系与处理
- 菌株:WT(野生型)与 ΔadeIJ(RND 缺失),必要时扩展至 ΔadeIJK。
- 处理与命名逻辑:
- No(未暴露):WT_17/24、deltaIJ_17/24
- One exposure(一次亚MIC):pre_WT_17/24、pre_deltaIJ_17/24
- Two exposures(两次亚MIC,0.5×MIC ×2):0_5_WT_17/24、0_5_deltaIJ_17/24
- 时间点:17 h、24 h(或 8/16/24 h 批次采样)
- Rationale:“05”表示每次 0.5×MIC 的两次脉冲,强调sub-MIC 叠加效应。
3.2 培养基与应用场景
- LB(液体/平板):生长曲线(液体)、总 CFU 与母板(平板)。
- MacConkey agar(胆盐+结晶紫):膜/外排应激选择,用于显化 R/S 亚群、计算 CFU_Mac/CFU_LB。
- MH2B(液体):标准药敏/动力学参照。
- AUM(液体)/Urine(液体):模拟尿路体内环境,用于生长、MIC 与 RNA-seq。
4. 指标体系与统计学(统一口径)
4.1 关键指标定义
- R 比例(3c 指标):
R% = CFU_Mac / CFU_LB × 100% - R/S 比值(可选报告):
R/S = CFU_Mac / (CFU_LB − CFU_Mac) - PAP 尾部比例:
p_tail = ΣCFU(≥MIC) / CFU(无药) - AUC(PAP 曲线下面积):衡量整体耐受度
- MDK99 / MDK99.99:达到 99%/99.99% 杀灭所需时间
- 脉冲生存率:
survival = CFU_pulse / CFU_start - RNA-seq:DEGs(|log2FC| 与 FDR),通路富集(KEGG/GO),主成分贡献(PC1/PC2)
4.2 统计检验与多重校正
- 组间比较:t 检验/曼–惠特尼;多组用 ANOVA/Kruskal–Wallis + FDR/Bonferroni。
- 比例/计数:二项置信区间、Fisher 精确检验、GLM(logit)。
- 时间–生存类:分段回归/非线性拟合;MDK 的置信区间或 bootstrap。
- RNA-seq:DESeq2(FDR < 0.05;|log2FC| 阈值预先注册)。
4.3 生物学重复与效应量
- 每组 ≥3 生物学重复;报告 效应量(Cohen’s d 或 Cliff’s delta)与 95% CI,避免只看 p 值。
5. SOP:核心实验流程(可直接贴进方法学)
5.1 并行铺板(LB vs MacConkey)与 R 比例
- 标准化 OD/CFU 接种;
- 在目标时点(8/16/24 h)取样,并行涂布到 LB agar 与 MacConkey agar;
- 过夜培养计数 CFU_LB / CFU_Mac;
- 计算 R% = CFU_Mac/CFU_LB,记录 ±95% CI;
- 统计 No vs One vs Two、WT vs ΔadeIJ 的差异。
5.2 复制平板分离 S(易感)
- 先在 LB 无药制备母板(100–300 个离散菌落);
- 用天鹅绒/膜 整体复制到 MacConkey(±药物板);
- 选择板不长/显著受损的坐标 = S 候选;
- 回到 LB 母板对应坐标挑取 S,纯化 2–3 轮,建库;
- 验证:MIC(不升/接近原始群体)、Time-kill(无长尾)、PAP 尾部低。
5.3 PAP(Population Analysis Profiling)
- 配置药物阶梯:0、0.25×、0.5×、1×、2×MIC(可扩展);
- 等量涂布 → 计数 CFU;
- 作图(log10 CFU vs 浓度),计算 AUC 与 p_tail;
- 统计处理间差异与效应量。
5.4 Time-kill / MDK
- 固定高浓度药(杀菌剂常 ≥10×MIC;四环素类可选联合或耐受表征方案);
- 0–24 h 分时取样计 CFU;
- 拟合曲线、估 MDK99/MDK99.99;
- MDK↑/长尾加重=耐受增强。
5.5 脉冲生存(Pulse Survival)
- 给予短时高浓度脉冲(10–20×MIC,30–60 min);
- 终止药效、复苏、计 CFU;
- 生存比例比较处理差异。
5.6 RNA-seq(尿液/AUM/MH2B & 暴露对照)
- 管线:FastQC → HISAT2/STAR → featureCounts → DESeq2;
- 批次设计:WT/ΔadeIJ ×(尿液/AUM/MH2B)×(No/One/Two)× 时间点;
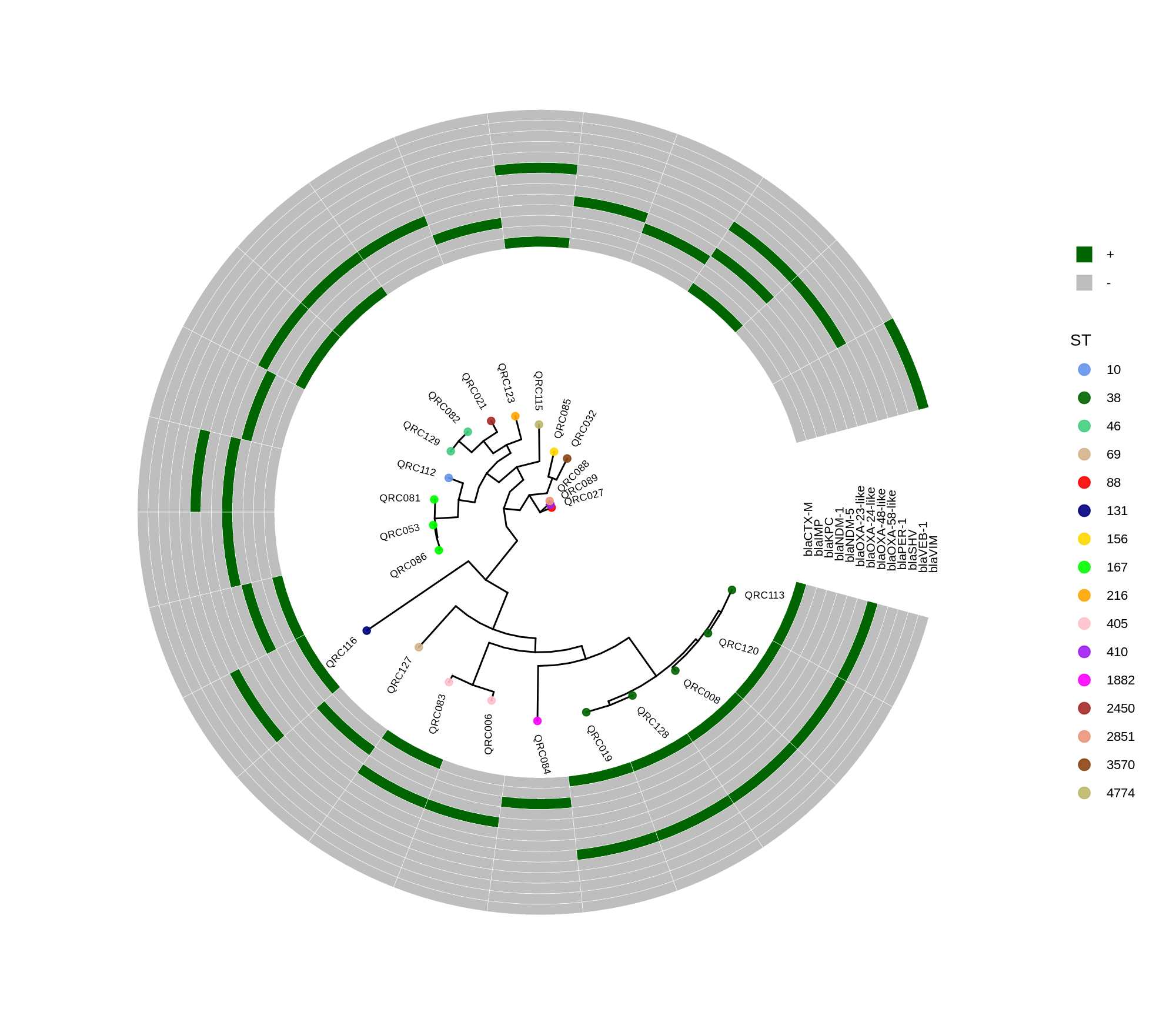
- 输出:PCA(PC1/PC2)、DEGs(FDR<0.05)、KEGG/GO 富集;重点关注 外排泵(adeA/B/C、adeI/J/K)、膜/代谢/应激通路。
6. 初步结果(与前述一致的“证据链”)
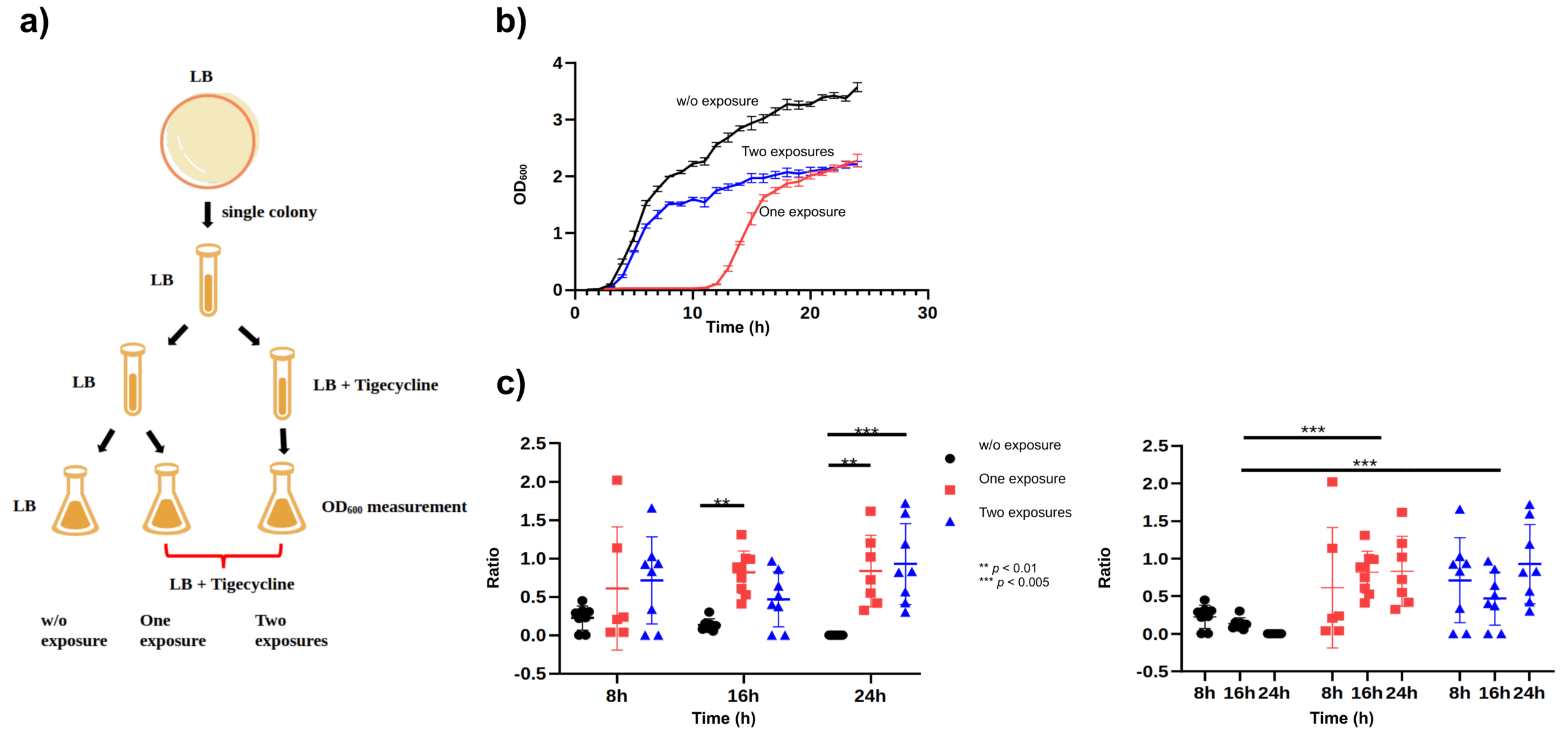
6.1 Fig.3 — 亚MIC 暴露 → 适应与 R 亚群上升
- 3a:流程示意(未画基质)。
- 3b(LB 液体):Two > One > None 的生长恢复/存活。
- 3c(R%):R% = CFU_Mac/CFU_LB;在 ΔadeIJ 中 Two > One > None(16–24 h 最显著;*p<0.005, p<0.01**),WT 变化小。
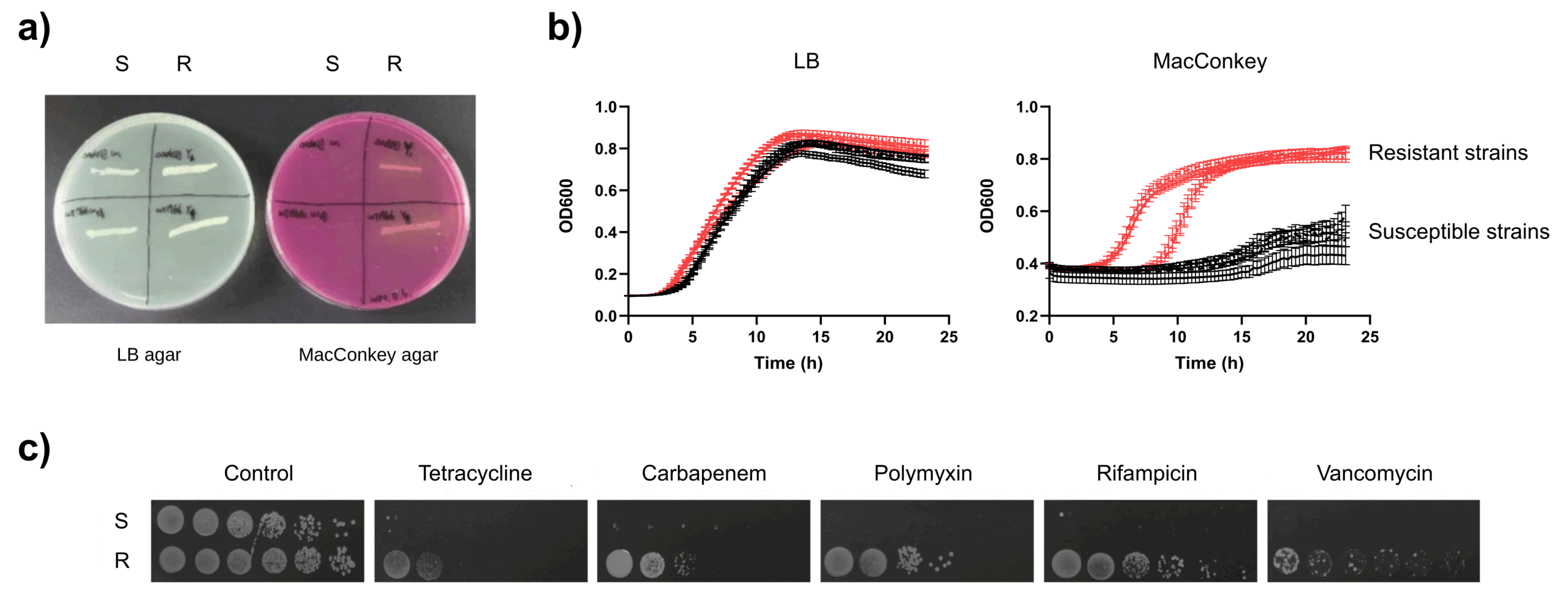
6.2 Fig.4 — 预暴露后 R/S 表型分化与药板验证
- LB vs MacConkey:R 能长,S 不能长/受损;WT 在两板均可生长。
- 药板(四环素、碳青霉烯、利福平、多黏菌素、万古霉素):S 不可成殖,R 表现更耐受。
6.3 Fig.5 — 培养基效应(AUM/Urine/MH2B)
- 生长:MH2B 最快;Urine 适中;AUM 较慢。
- MIC 转变:AUM 中 四环素类 ↓~4×、碳青霉烯 ↓~8×(vs MH2B),提示介质效应。
6.4 Fig.6 — 尿液/AUM 的全转录重编程
- PCA:Urine 沿 PC1(~75%)聚类;AUM 与 MH2B 沿 PC2(~20%)分离。
- DEGs/通路:两者各自独特且部分重叠;脲酶上调;Urine 中 adeB 上调、AUM 中 adeA/adeB 下调;富集 氨基酸/碳代谢、TCA 等。
7. 质量控制(QC)与混杂控制
- 接种量与生长期:统一 OD600/CFU 与生长阶段,避免接种效应。
- 时间点:固定 8/16/24 h(或 17/24 h)窗口;保证重复一致。
- 平板并行:同一稀释度并行涂布 LB vs MacConkey;同批操作。
- 批次效应:RNA-seq 采用阻断设计/ComBat 校正;PCA 检查批次漂移。
- 药物效力:MIC 校准、现配现用、稳定性检查;AUM/尿液配方与 pH/渗透压监控。
- 统计注册:预注册阈值(FDR、|log2FC|、效应量),报告完整。
8. 风险点与备选方案(Mitigation)
- R/S 不显著:提高重复数;增加 MacConkey 胆盐/结晶紫梯度;延长/调整暴露方案。
- RNA-seq 变异大:加大样本量或聚焦关键时间点;加入 RT-qPCR 验证。
- WT 也出现明显 R 上升:进一步引入 ΔadeABC / ΔadeIJK 与互补株,拆解特异性。
- MIC 与耐受定义混淆:采用 “MIC(稳态)+ MDK(动力学)”双标准区分。
9. 项目管理:时间线与里程碑
| 时间 | 核心任务 | 可交付物 |
|---|---|---|
| 2026 Q1–Q2 | 生长曲线/MIC/PAP/并行铺板体系搭建 | QC 报告、首版 SOP、基线数据 |
| 2026 Q3 | 演化实验/Time-kill/MDK/脉冲生存 | AUC/MDK/生存率统计与图表 |
| 2026 Q4 | RNA-seq(尿液/AUM/MH2B)、WGS | PCA、DEGs、KEGG/GO、突变谱 |
| 2027 Q1 | 敲除株构建与表型/分子验证 | RT-qPCR/WB/银染/代谢物数据 |
| 2027 Q2 | 综合分析与模型建立 | 机制图、整合图谱(Fig. Model) |
| 2027 Q3–Q4 | 论文/会议/专利 | 3–4 篇论文草稿、会议摘要、EPI 靶点清单 |
10. 预期产出与学术/临床价值
- 机制贡献:完整链路——亚MIC 暴露 → RND/膜/代谢 → R/S 异质 → UTI 环境转录重编程 → 药敏变化。
- 工具与范式:R 比例/PAP/MDK/脉冲生存的统一报告范式;UTI 相关 AST 的培养基效应提示。
- 转化潜力:聚焦 TM 变构/能量耦联位点的 EPI 设计;提示在 UTI 情景下药物/联合方案的优化路径。
11. 附:可直接复用的图注(精炼版,供英文化)
- Fig. 3:Sub-MIC tigecycline exposures (0.5×MIC ×1/×2) promote adaptation in ΔadeIJ; LB growth (OD600) shows Two > One > None; MacConkey tolerance ratio R% = CFU_Mac/CFU_LB increases significantly at 16–24 h (*p<0.005, p<0.01**); WT shows minimal change.
- Fig. 4:Post-exposure ΔadeIJ splits into R (grows on MacConkey) and S (impaired); S fails on drug plates (TET/CARB/RIF/PMB/VAN).
- Fig. 5:MH2B fastest growth; urine moderate; AUM slower; MICs drop in AUM (TET ~4×, CARB ~8× vs MH2B).
- Fig. 6:Urine clusters on PC1 (~75%); AUM separates on PC2 (~20%); urease up; adeB up in urine, adeA/adeB down in AUM; AA/carbon/TCA enriched.