-
preparing gene expression matrix: calculate DESeq2 results
#Input: merged_gene_counts.txt setwd("/home/jhuang/DATA/Data_Susanne_Carotis_RNASeq/run_2023_GSVA/") library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") #BiocManager::install("org.Hs.eg.db") library("org.Hs.eg.db") library(DESeq2) library(gplots) d.raw<- read.delim2("merged_gene_counts.txt",sep="\t", header=TRUE, row.names=1) colnames(d.raw)<-c("gene_name", "leer_mock_2h_r2", "Ace2_mock_2h_r2", "leer_inf_24h_r1", "Ace2_inf_2h_r1", "leer_inf_24h_r2", "leer_inf_2h_r1", "leer_mock_2h_r1", "leer_inf_2h_r2", "Ace2_inf_2h_r2", "Ace2_mock_2h_r1", "Ace2_inf_24h_r2", "Ace2_inf_24h_r1") col_order <- c("gene_name", "leer_mock_2h_r1","leer_mock_2h_r2","leer_inf_2h_r1","leer_inf_2h_r2","leer_inf_24h_r1","leer_inf_24h_r2","Ace2_mock_2h_r1","Ace2_mock_2h_r2","Ace2_inf_2h_r1","Ace2_inf_2h_r2","Ace2_inf_24h_r1","Ace2_inf_24h_r2") reordered.raw <- d.raw[,col_order] reordered.raw$gene_name <- NULL #d <- d.raw[rowSums(reordered.raw>3)>2,] condition = as.factor(c("leer_mock_2h","leer_mock_2h","leer_inf_2h","leer_inf_2h","leer_inf_24h","leer_inf_24h","Ace2_mock_2h","Ace2_mock_2h","Ace2_inf_2h","Ace2_inf_2h","Ace2_inf_24h","Ace2_inf_24h")) ids = as.factor(c("leer_mock_2h_r1","leer_mock_2h_r2","leer_inf_2h_r1","leer_inf_2h_r2","leer_inf_24h_r1","leer_inf_24h_r2","Ace2_mock_2h_r1","Ace2_mock_2h_r2","Ace2_inf_2h_r1","Ace2_inf_2h_r2","Ace2_inf_24h_r1","Ace2_inf_24h_r2")) #cData = data.frame(row.names=colnames(reordered.raw), condition=condition, batch=batch, ids=ids) #dds<-DESeqDataSetFromMatrix(countData=reordered.raw, colData=cData, design=~batch+condition) cData = data.frame(row.names=colnames(reordered.raw), condition=condition, ids=ids) dds<-DESeqDataSetFromMatrix(countData=reordered.raw, colData=cData, design=~condition) #----more detailed and specific with the following code!---- dds$condition <- relevel(dds$condition, "Ace2_mock_2h") dds = DESeq(dds, betaPrior=FALSE) # betaPrior default value is FALSE resultsNames(dds) -
preparing selected_geneSets in gsva(exprs, selected_geneSets, method=”gsva”). Note that methods are different than methods for nanoString, here are ENSEMBL listed.
#Input: "Signatures.xls" + "Signatures_additional.xls" library(readxl) library(gridExtra) library(ggplot2) library(GSVA) # Paths to the Excel files file_paths <- list("Signatures.xls", "Signatures_additional.xls") # Get sheet names for each file sheet_names_list <- lapply(file_paths, excel_sheets) # Initialize an empty list to hold gene sets geneSets <- list() # Loop over each file path and its corresponding sheet names for (i in 1:length(file_paths)) { file_path <- file_paths[[i]] sheet_names <- sheet_names_list[[i]] # Loop over each sheet, extract the ENSEMBL IDs, and add to the list for (sheet in sheet_names) { # Read the sheet data <- read_excel(file_path, sheet = sheet) # Process the GeneSet names (replacing spaces with underscores, for example) gene_set_name <- gsub(" ", "_", unique(data$GeneSet)[1]) # Add ENSEMBL IDs to the list geneSets[[gene_set_name]] <- as.character(data$ENSEMBL) } } # Print the result to check print(geneSets) summary(geneSets) #desired_geneSets <- c("Monocytes", "Plasma_cells", "T_regs", "Cyt._act._T_cells", "Neutrophils", "Inflammatory_neutrophils", "Suppressive_neutrophils", "LDG", "CD40_activated") desired_geneSets <- c("IFN", "TNF", "IL-6R_complex", "IL-1_cytokines", "Pro-inflam._IL-1", "Monocyte_secreted", "Apoptosis", "NFkB_complex", "NLRP3_inflammasome") selected_geneSets <- geneSets[desired_geneSets] # Print the selected gene sets print(selected_geneSets) -
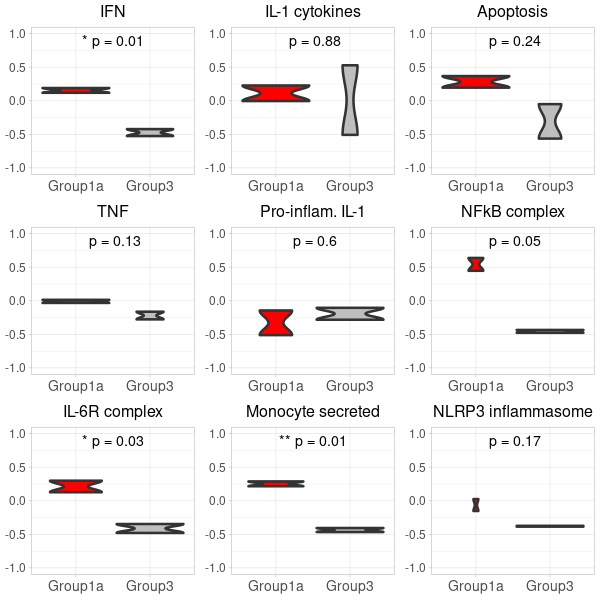
prepare violin plots
# 0. for Nanostring, the GSVA input requires a gene expression matrix 'exprs' where rows are genes and columns are samples. This matrix must be in non-log space. #exprs <- exprs(filtered_or_neg_target_m666Data) # 0. for RNAseq, the GSVA input requires a gene expression matrix where rows are genes and columns are samples. This matrix must be in non-log space. exprs <- counts(dds, normalized=TRUE) # 1. Compute GSVA scores: gsva_scores <- gsva(exprs, selected_geneSets, method="gsva") # 2. Convert to data.frame for ggplot: gsva_df <- as.data.frame(t(gsva_scores)) # 3. Add conditions to gsva_df: gsva_df$Condition <- dds$condition # 4. Filter the gsva_df to retain only the desired conditions: #group 1 vs. group 3 in the nanostring data gsva_df_filtered <- gsva_df[gsva_df$Condition %in% c("Ace2_mock_2h", "Ace2_inf_24h"), ] # 5. Define a function to plot violin plots: # Update the condition levels in gsva_df_filtered to ensure the desired order on x-axis: gsva_df_filtered$Condition <- gsub("Ace2_mock_2h", "Group3", gsva_df_filtered$Condition) #group3=mock gsva_df_filtered$Condition <- gsub("Ace2_inf_24h", "Group1a", gsva_df_filtered$Condition) #group1a=infection gsva_df_filtered$Condition <- factor(gsva_df_filtered$Condition, levels = c("Group1a", "Group3")) plot_violin <- function(data, gene_name) { # Calculate the t-test p-value for the two conditions condition1_data <- data[data$Condition == "Group1a", gene_name] condition2_data <- data[data$Condition == "Group3", gene_name] p_value <- t.test(condition1_data, condition2_data)$p.value # Convert p-value to annotation p_annotation <- ifelse(p_value < 0.01, "**", ifelse(p_value < 0.05, "*", "")) rounded_p_value <- paste0("p = ", round(p_value, 2)) plot_title <- gsub("_", " ", gene_name) p <- ggplot(data, aes(x=Condition, y=!!sym(gene_name), fill=Condition)) + geom_violin(linewidth=1.2) + scale_fill_manual(values = custom_colors) + labs(title=plot_title, y="GSVA Score") + ylim(-1, 1) + theme_light() + theme( axis.title.x = element_text(size=12), axis.title.y = element_text(size=12), axis.text.x = element_text(size=10), axis.text.y = element_text(size=10), plot.title = element_text(size=12, hjust=0.5), legend.position = "none" # Hide legend since the colors are self-explanatory ) # Add p-value annotation to the plot p <- p + annotate("text", x=1.5, y=0.9, label=paste0(p_annotation, " ", rounded_p_value), size=5, hjust=0.5) return(p) } # 6. Generate the list of plots in a predefined order: genes <- colnames(gsva_df_filtered)[!colnames(gsva_df_filtered) %in% "Condition"] genes <- genes[match(desired_order, genes)] genes <- genes[!is.na(genes)] first_row_plots <- lapply(genes, function(gene) plot_violin(gsva_df_filtered, gene)) -
generating two 3×3 grid plots
# Start by extracting the 1-9 plots from each list first_row_plots <- first_row_plots[1:9] # Function to modify the individual plots based on position in the grid modify_plot <- function(p, row, col) { p <- p + theme(axis.title.y = element_blank()) # remove y-axis title if not the first plot p <- p + theme( axis.title.x = element_blank(), # remove x-axis title for all plots axis.title.y = element_blank(), # remove x-axis title for all plots axis.text = element_text(size = 14), # Increase axis text size axis.title = element_text(size = 16), # Increase axis title size plot.title = element_text(size = 16) # Increase plot title size ) return(p) } # Apply the modifications to the plots for (i in 1:9) { first_row_plots[[i]] <- modify_plot(first_row_plots[[i]], 1, i) } # Increase the font size of x-axis labels for each plot for (i in 1:9) { first_row_plots[[i]] <- first_row_plots[[i]] + theme(axis.text.x = element_text(size = 14), axis.text.y = element_text(size = 12)) } # Pad first_row_plots to have 9 plots remaining_plots <- 9 - length(first_row_plots) if (remaining_plots > 0) { first_row_plots <- c(first_row_plots, rep(list(NULL), remaining_plots)) } # Convert the first_row_plots to a matrix and draw plots_matrix_1 <- matrix(first_row_plots, ncol=3, byrow=TRUE) png("Carotis_RNA-seq_grid_1.png", width=600, height=600) do.call("grid.arrange", c(plots_matrix_1, list(ncol=3))) dev.off()
GSVA-plot for carotis RNA-seq data
Leave a reply