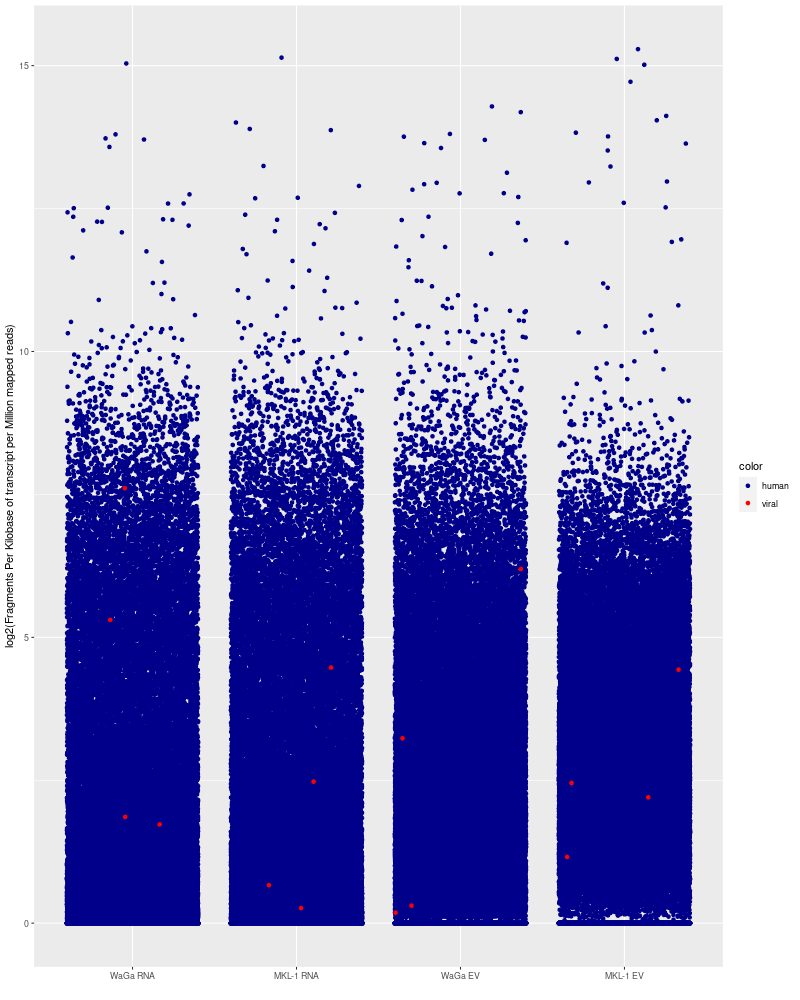
Display viral transcripts found in mRNA-seq MKL-1, WaGa EVs compared to cells
gene_x 0 like s 1107 view s
Tags: python, R, processing, virus, human, genome
Dowload the file fpkm_values_WaGa.csv
Download the file fpkm_values_MKL-1.csv
STEP1: using following code (R_scripts.txt) to generate merged_gene_counts_WaGa.csv and merged_gene_counts_MKL-1.csv
# ---- GENERATE BED-file for WaGa ----
cp WaGa.gff3 WaGa.gff3_backup
sed -i -e "s/\tGenbank\tgene\t/\tGenbank_gene\t/g" WaGa.gff3
grep "Genbank_gene" WaGa.gff3 > WaGa_gene.gff3
sed -i -e "s/\tGenbank_gene\t/\tGenbank\tgene\t/g" WaGa_gene.gff3
grep "gene_biotype=pseudogene" WaGa.gff3_backup >> WaGa_gene.gff3 #-->4
#The whole point of the GTF format was to standardise certain aspects that are left open in GFF. Hence, there are many different valid ways to encode the same information in a valid GFF format, and any parser or converter needs to be written specifically for the choices the author of the GFF file made. For example, a GTF file requires the gene ID attribute to be called "gene_id", while in GFF files, it may be "ID", "Gene", something different, or completely missing.
# from gff3 to gtf
cp WaGa_gene.gff3 WaGa_gene.gtf
sed -i -e "s/\tID=gene-/\tgene_id \"/g" WaGa_gene.gtf
sed -i -e "s/;/\"; /g" WaGa_gene.gtf
sed -i -e "s/=/=\"/g" WaGa_gene.gtf
#sed -i -e $'s/\n/\"\n/g' WaGa_gene.gtf
gffread -E -F --bed WaGa.gff3 -o WaGa.bed #-->4
# ---- GENERATE BED-file for MKL-1 ----
cp MKL-1.gff3 MKL-1.gff3_backup
sed -i -e "s/\tGenbank\tgene\t/\tGenbank_gene\t/g" MKL-1.gff3
grep "Genbank_gene" MKL-1.gff3 > MKL-1_gene.gff3
sed -i -e "s/\tGenbank_gene\t/\tGenbank\tgene\t/g" MKL-1_gene.gff3
grep "gene_biotype=pseudogene" MKL-1.gff3_backup >> MKL-1_gene.gff3 #-->4
# from gff3 to gtf
cp MKL-1_gene.gff3 MKL-1_gene.gtf
sed -i -e "s/\tID=gene-/\tgene_id \"/g" MKL-1_gene.gtf
sed -i -e "s/;/\"; /g" MKL-1_gene.gtf
sed -i -e "s/=/=\"/g" MKL-1_gene.gtf
#sed -i -e $'s/\n/\"\n/g' MKL-1_gene.gtf
gffread -E -F --bed MKL-1.gff3 -o MKL-1.bed #-->4
# ---- GENERATE GFF3-file for WaGa ----
cp WaGa.gff3_backup WaGa.gff3
grep "^##" WaGa.gff3 > WaGa_gene.gff3
grep "ID=gene" WaGa.gff3 >> WaGa_gene.gff3
#!!!!VERY_IMPORTANT!!!!: change type '\tCDS\t' to '\texon\t'!
sed -i -e "s/\tgene\t/\texon\t/g" WaGa_gene.gff3
#INPUT are 3796 genes (pseudogene, rRNA, tRNA, gene), why output are only 3726 (-pseudogene-rRNA)
# ---- GENERATE GFF3-file for MKL-1 ----
cp MKL-1.gff3_backup MKL-1.gff3
grep "^##" MKL-1.gff3 > MKL-1_gene.gff3
grep "ID=gene" MKL-1.gff3 >> MKL-1_gene.gff3
#!!!!VERY_IMPORTANT!!!!: change type '\tCDS\t' to '\texon\t'!
sed -i -e "s/\tgene\t/\texon\t/g" MKL-1_gene.gff3
#INPUT are 3796 genes (pseudogene, rRNA, tRNA, gene), why output are only 3726 (-pseudogene-rRNA)
#MODIFY LTtr1 to LT1, LTtr2 to LT2, since the gff3 files contains only the records of LT1 and LT2, not LTtr1 and LTtr2.
#KJ128379.1 Genbank exon 196 429 . + . ID=gene-LTtr1;Name=LTtr1;gbkey=Gene;gene=LTtr1;gene_biotype=protein_coding
#KJ128379.1 Genbank exon 861 1463 . + . ID=gene-LTtr2;Name=LTtr2;gbkey=Gene;gene=LTtr2;gene_biotype=protein_coding
# generate ./results_virus/featureCounts/merged_gene_counts_MKL-1.txt
nextflow run rnaseq/main_Fraction_O_for_virus.nf --reads '/home/jhuang/DATA/Data_Ute_RNA-Seq_MKL-1/Raw_Data/*.fastq.gz' --fasta /home/jhuang/DATA/MCPyV_refs/MKL-1.fasta --gtf /home/jhuang/DATA/MCPyV_refs/MKL-1_gene.gff3 --bed12 /home/jhuang/DATA/MCPyV_refs/MKL-1.bed --singleEnd -profile standard --aligner hisat2 --fcGroupFeatures Name --fcGroupFeaturesType gene_biotype --saveReference -resume --saveAlignedIntermediates --skip_rseqc --skip_dupradar --skip_genebody_coverage --skip_preseq --skip_edger #-work-dir
# generate ./results_virus/featureCounts/merged_gene_counts_WaGa.txt
nextflow run rnaseq/main_Fraction_O_for_virus.nf --reads '/home/jhuang/DATA/Data_Ute_RNA-Seq_WaGa/Raw_Data/*.fastq.gz' --fasta /home/jhuang/DATA/MCPyV_refs/WaGa.fasta --gtf /home/jhuang/DATA/MCPyV_refs/WaGa_gene.gff3 --bed12 /home/jhuang/DATA/MCPyV_refs/WaGa.bed --singleEnd -profile standard --aligner hisat2 --fcGroupFeatures Name --fcGroupFeaturesType gene_biotype --saveReference -resume --saveAlignedIntermediates --skip_rseqc --skip_dupradar --skip_genebody_coverage --skip_preseq --skip_edger
rsync -a -P jhuang@hamm:~/DATA/Data_Ute_RNA-Seq_WaGa/results_human/featureCounts/merged_gene_counts.txt ./results_human/featureCounts/merged_gene_counts_WaGa.txt
rsync -a -P jhuang@hamm:~/DATA/Data_Ute_RNA-Seq_WaGa/results_virus/featureCounts/merged_gene_counts.txt ./results_virus/featureCounts/merged_gene_counts_WaGa.txt
rsync -a -P jhuang@hamm:~/DATA/Data_Ute_RNA-Seq_MKL-1/results_human/featureCounts/merged_gene_counts.txt ./results_human/featureCounts/merged_gene_counts_MKL-1.txt
rsync -a -P jhuang@hamm:~/DATA/Data_Ute_RNA-Seq_MKL-1/results_virus/featureCounts/merged_gene_counts.txt ./results_virus/featureCounts/merged_gene_counts_MKL-1.txt
sed 's/Aligned.sortedByCoord.out.bam//g' results_human/featureCounts/merged_gene_counts_WaGa.txt > merged_gene_counts_WaGa_human.txt
sed 's/.sorted.bam//g' results_virus/featureCounts/merged_gene_counts_WaGa.txt > merged_gene_counts_WaGa_virus.txt
sed 's/Aligned.sortedByCoord.out.bam//g' results_human/featureCounts/merged_gene_counts_MKL-1.txt > merged_gene_counts_MKL-1_human.txt
sed 's/.sorted.bam//g' results_virus/featureCounts/merged_gene_counts_MKL-1.txt > merged_gene_counts_MKL-1_virus.txt
python3 concat_files_WaGa.py merged_gene_counts_WaGa_human.txt merged_gene_counts_WaGa_virus.txt merged_gene_counts_WaGa.csv #(23 samples)
python3 concat_files_MKL-1.py merged_gene_counts_MKL-1_human.txt merged_gene_counts_MKL-1_virus.txt merged_gene_counts_MKL-1.csv #(18 samples)
The code snippet of the file concat_files_MKL-1.py
#!/usr/bin/env python3
#"MKL-1 RNA","MKL-1 RNA 118","MKL-1 RNA 147",
#"MKL-1 EV","MKL-1 EV 2","MKL-1 EV 118","MKL-1 EV 87","MKL-1 EV 27","MKL-1 EV 042","MKL-1 EV 0505",
#"MKL-1 EV sT DMSO 042","MKL-1 EV sT DMSO 0505", "MKL-1 EV scr DMSO 042","MKL-1 EV scr DMSO 0505", "MKL-1 EV sT Dox 042","MKL-1 EV sT Dox 0505", "MKL-1 EV scr Dox 042","MKL-1 EV scr Dox 0505",
#"WaGa RNA","WaGa RNA 118","WaGa RNA 147",
#"WaGa EV","WaGa EV 2","WaGa EV 118","WaGa EV 147","WaGa EV 226","WaGa EV 1107","WaGa EV 1605","WaGa EV 2706",
#"WaGa EV sT DMSO 1107","WaGa EV sT DMSO 1605","WaGa EV sT DMSO 2706", "WaGa EV scr DMSO 1107","WaGa EV scr DMSO 1605","WaGa EV scr DMSO 2706", "WaGa EV sT Dox 1107","WaGa EV sT Dox 1605","WaGa EV sT Dox 2706", "WaGa EV scr Dox 1107","WaGa EV scr Dox 1605","WaGa EV scr Dox 2706"
#Geneid gene_name WaGa_RNA 1605_WaGa_wt_EV 2706_WaGa_scr_DMSO_EV WaGa_EV-RNA_226 1107_WaGa_scr_Dox_EV 1107_WaGa_sT_Dox_EV 1605_WaGa_sT_Dox_EV 1107_WaGa_scr_DMSO_EV 1107_WaGa_wt_EV 1605_WaGa_sT_DMSO_EV WaGa_EV-RNA_2 2706_WaGa_sT_Dox_EV WaGa_EV-RNA_147 2706_WaGa_sT_DMSO_EV 1107_WaGa_sT_DMSO_EV 2706_WaGa_scr_Dox_EV WaGa_RNA_147 1605_WaGa_scr_DMSO_EV 2706_WaGa_wt_EV WaGa_RNA_118 1605_WaGa_scr_Dox_EV WaGa_EV-RNA_118 WaGa_EV-RNA
#Geneid gene_name
import sys
import pandas as pd
# Check if the correct number of arguments is provided
if len(sys.argv) != 4:
print("Usage: python merge_files.py <file1.csv> <file2.csv> <output.csv>")
sys.exit()
# Get the file names from command line arguments
file1 = sys.argv[1]
file2 = sys.argv[2]
output_file = sys.argv[3]
# Read the files
df1 = pd.read_csv(file1, sep='\t')
df2 = pd.read_csv(file2, sep='\t')
# Rename columns in both DataFrames
#df1.rename(columns=column_mapping, inplace=True)
#df2.rename(columns=column_mapping, inplace=True)
# Define the desired column order
#"MKL-1 RNA","MKL-1 RNA 118","MKL-1 RNA 147",
#"MKL-1 EV","MKL-1 EV 2","MKL-1 EV 118","MKL-1 EV 87","MKL-1 EV 27","MKL-1 EV 042","MKL-1 EV 0505",
#"MKL-1 EV sT DMSO 042","MKL-1 EV sT DMSO 0505", "MKL-1 EV scr DMSO 042","MKL-1 EV scr DMSO 0505", "MKL-1 EV sT Dox 042","MKL-1 EV sT Dox 0505", "MKL-1 EV scr Dox 042","MKL-1 EV scr Dox 0505",
#Geneid gene_name 0505_MKL-1_scr_Dox_EV MKL-1_EV-RNA 0505_MKL-1_scr_DMSO_EV 0505_MKL-1_wt_EV MKL-1_RNA_147 MKL-1_EV-RNA_2 MKL-1_RNA MKL-1_EV-RNA_87 042_MKL-1_wt_EV 042_MKL-1_scr_Dox_EV MKL-1_EV-RNA_27 042_MKL-1_sT_Dox 042_MKL-1_sT_DMSO MKL-1_EV-RNA_118 0505_MKL-1_sT_Dox_EV 0505_MKL-1_sT_DMSO_EV MKL-1_RNA_118 042_MKL-1_scr_DMSO_EV
column_order = ['Geneid','gene_name', 'MKL-1_RNA','MKL-1_RNA_118','MKL-1_RNA_147', 'MKL-1_EV-RNA','MKL-1_EV-RNA_2','MKL-1_EV-RNA_118','MKL-1_EV-RNA_87','MKL-1_EV-RNA_27','042_MKL-1_wt_EV','0505_MKL-1_wt_EV', '042_MKL-1_sT_DMSO','0505_MKL-1_sT_DMSO_EV', '042_MKL-1_scr_DMSO_EV','0505_MKL-1_scr_DMSO_EV', '042_MKL-1_sT_Dox','0505_MKL-1_sT_Dox_EV', '042_MKL-1_scr_Dox_EV','0505_MKL-1_scr_Dox_EV']
# Reorder columns in both DataFrames
df1 = df1[column_order]
df2 = df2[column_order]
# Concatenate the dataframes
result = pd.concat([df1, df2], axis=0, ignore_index=True)
# Save the result to a new file
result.to_csv(output_file, index=False)
The code snippet of the file concat_files_WaGa.py
#!/usr/bin/env python3
#"WaGa RNA","WaGa RNA 118","WaGa RNA 147",
#"WaGa EV","WaGa EV 2","WaGa EV 118","WaGa EV 147","WaGa EV 226","WaGa EV 1107","WaGa EV 1605","WaGa EV 2706",
#"WaGa EV sT DMSO 1107","WaGa EV sT DMSO 1605","WaGa EV sT DMSO 2706", "WaGa EV scr DMSO 1107","WaGa EV scr DMSO 1605","WaGa EV scr DMSO 2706", "WaGa EV sT Dox 1107","WaGa EV sT Dox 1605","WaGa EV sT Dox 2706", "WaGa EV scr Dox 1107","WaGa EV scr Dox 1605","WaGa EV scr Dox 2706"
#Geneid gene_name WaGa_RNA 1605_WaGa_wt_EV 2706_WaGa_scr_DMSO_EV WaGa_EV-RNA_226 1107_WaGa_scr_Dox_EV 1107_WaGa_sT_Dox_EV 1605_WaGa_sT_Dox_EV 1107_WaGa_scr_DMSO_EV 1107_WaGa_wt_EV 1605_WaGa_sT_DMSO_EV WaGa_EV-RNA_2 2706_WaGa_sT_Dox_EV WaGa_EV-RNA_147 2706_WaGa_sT_DMSO_EV 1107_WaGa_sT_DMSO_EV 2706_WaGa_scr_Dox_EV WaGa_RNA_147 1605_WaGa_scr_DMSO_EV 2706_WaGa_wt_EV WaGa_RNA_118 1605_WaGa_scr_Dox_EV WaGa_EV-RNA_118 WaGa_EV-RNA
#Geneid gene_name
import sys
import pandas as pd
# Check if the correct number of arguments is provided
if len(sys.argv) != 4:
print("Usage: python merge_files.py <file1.csv> <file2.csv> <output.csv>")
sys.exit()
# Get the file names from command line arguments
file1 = sys.argv[1]
file2 = sys.argv[2]
output_file = sys.argv[3]
# Read the files
df1 = pd.read_csv(file1, sep='\t')
df2 = pd.read_csv(file2, sep='\t')
# Define the column mapping for renaming columns
column_mapping = {
'WaGa_RNA': 'WaGa RNA',
'WaGa_RNA_118': 'WaGa RNA 118',
'WaGa_RNA_147': 'WaGa RNA 147',
'WaGa_EV-RNA': 'WaGa EV',
'WaGa_EV-RNA_2': 'WaGa EV 2',
'WaGa_EV-RNA_118': 'WaGa EV 118',
'WaGa_EV-RNA_147': 'WaGa EV 147',
'WaGa_EV-RNA_226': 'WaGa EV 226',
'1107_WaGa_wt_EV': 'WaGa EV 1107',
'1605_WaGa_wt_EV': 'WaGa EV 1605',
'2706_WaGa_wt_EV': 'WaGa EV 2706',
'1107_WaGa_sT_DMSO_EV': 'WaGa EV sT DMSO 1107',
'1605_WaGa_sT_DMSO_EV': 'WaGa EV sT DMSO 1605',
'2706_WaGa_sT_DMSO_EV': 'WaGa EV sT DMSO 2706',
'1107_WaGa_scr_DMSO_EV': 'WaGa EV scr DMSO 1107',
'1605_WaGa_scr_DMSO_EV': 'WaGa EV scr DMSO 1605',
'2706_WaGa_scr_DMSO_EV': 'WaGa EV scr DMSO 2706',
'1107_WaGa_sT_Dox_EV': 'WaGa EV sT Dox 1107',
'1605_WaGa_sT_Dox_EV': 'WaGa EV sT Dox 1605',
'2706_WaGa_sT_Dox_EV': 'WaGa EV sT Dox 2706',
'1107_WaGa_scr_Dox_EV': 'WaGa EV scr Dox 1107',
'1605_WaGa_scr_Dox_EV': 'WaGa EV scr Dox 1605',
'2706_WaGa_scr_Dox_EV': 'WaGa EV scr Dox 2706',
} # Replace with your desired column names
# Rename columns in both DataFrames
#df1.rename(columns=column_mapping, inplace=True)
#df2.rename(columns=column_mapping, inplace=True)
# Define the desired column order
#column_order = ['Geneid', 'gene_name', 'WaGa RNA','WaGa RNA 118','WaGa RNA 147','WaGa EV','WaGa EV 2','WaGa EV 118','WaGa EV 147','WaGa EV 226','WaGa EV 1107','WaGa EV 1605','WaGa EV 2706','WaGa EV sT DMSO 1107','WaGa EV sT DMSO 1605','WaGa EV sT DMSO 2706','WaGa EV scr DMSO 1107','WaGa EV scr DMSO 1605','WaGa EV scr DMSO 2706','WaGa EV sT Dox 1107','WaGa EV sT Dox 1605','WaGa EV sT Dox 2706','WaGa EV scr Dox 1107','WaGa EV scr Dox 1605','WaGa EV scr Dox 2706'] # Replace with your desired column order
column_order = ['Geneid','gene_name','WaGa_RNA','WaGa_RNA_118','WaGa_RNA_147','WaGa_EV-RNA','WaGa_EV-RNA_2','WaGa_EV-RNA_118','WaGa_EV-RNA_147','WaGa_EV-RNA_226','1107_WaGa_wt_EV','1605_WaGa_wt_EV','2706_WaGa_wt_EV','1107_WaGa_sT_DMSO_EV','1605_WaGa_sT_DMSO_EV','2706_WaGa_sT_DMSO_EV','1107_WaGa_scr_DMSO_EV','1605_WaGa_scr_DMSO_EV','2706_WaGa_scr_DMSO_EV','1107_WaGa_sT_Dox_EV','1605_WaGa_sT_Dox_EV','2706_WaGa_sT_Dox_EV','1107_WaGa_scr_Dox_EV','1605_WaGa_scr_Dox_EV','2706_WaGa_scr_Dox_EV']
# Reorder columns in both DataFrames
df1 = df1[column_order]
df2 = df2[column_order]
# Concatenate the dataframes
result = pd.concat([df1, df2], axis=0, ignore_index=True)
# Save the result to a new file
result.to_csv(output_file, index=False)
STEP2: using following code (R_scripts.txt) to generate fpkm_values_WaGa.csv and fpkm_values_MKL-1.csv
#if (!requireNamespace("BiocManager", quietly = TRUE))
# install.packages("BiocManager")
#BiocManager::install("biomaRt")
library(biomaRt)
library(dplyr)
# Replace 'input_file.csv' with the name of your CSV file containing Ensembl gene IDs and counts
input_data <- read.csv("merged_gene_counts_WaGa.csv")
#input_data <- read.csv("merged_gene_counts_MKL-1.csv")
input_data <- input_data %>% rename(ensembl_gene_id = Geneid)
# Specify the rows you want to sum
row1 <- 60665
row2 <- 60666
# Create a new row with the sums of the two specified rows, except for the first two columns
new_row <- input_data[c(row1, row2), -c(1, 2)] %>%
summarise(across(everything(), sum))
# Name the first two columns
new_row$ensembl_gene_id <- "LT"
new_row$gene_name <- ""
# Add the new row to the original data frame
input_data <- bind_rows(input_data, new_row)
# Delete row1 and row2
input_data <- input_data[-c(row1, row2),]
# Assuming your CSV file has columns named 'ensembl_gene_id' and 'counts'
gene_ids <- input_data$ensembl_gene_id
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl")
ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="104")
#geness <- getBM(
# attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
# filters = 'ensembl_gene_id',
# values = rownames(res),
# mart = ensembl)
#geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE)
# Get transcript lengths for each Ensembl gene ID
transcript_lengths <- getBM(
attributes = c("ensembl_gene_id", "transcript_length"),
filters = "ensembl_gene_id",
values = gene_ids,
mart = ensembl
)
# Calculate the median transcript length for each gene
median_transcript_lengths <- transcript_lengths %>%
group_by(ensembl_gene_id) %>%
summarize(median_length = median(transcript_length, na.rm = TRUE))
#add to median_transcript_lengths WaGa
#KJ128379.1 Genbank exon 196 429 . + . ID=gene-LT1;Name=LT1;gbkey=Gene;gene=LT1;gene_biotype=protein_coding --> 234
#KJ128379.1 Genbank exon 861 1463 . + . ID=gene-LT2;Name=LT2;gbkey=Gene;gene=LT2;gene_biotype=protein_coding --> 603
#KJ128379.1 Genbank exon 196 756 . + . ID=gene-ST;Name=ST;gbkey=Gene;gene=ST;gene_biotype=protein_coding --> 561
#KJ128379.1 Genbank exon 3156 4427 . - . ID=gene-VP1;Name=VP1;gbkey=Gene;gene=VP1;gene_biotype=protein_coding --> 1272
#KJ128379.1 Genbank exon 4393 5118 . - . ID=gene-VP2;Name=VP2;gbkey=Gene;gene=VP2;gene_biotype=protein_coding --> 726
#MKL-1
#FJ173815.1 Genbank exon 196 429 . + . ID=gene-LT1;Name=LT1;gbkey=Gene;gene=LT1;gene_biotype=protein_coding --> 234
#FJ173815.1 Genbank exon 861 1619 . + . ID=gene-LT2;Name=LT2;gbkey=Gene;gene=LT2;gene_biotype=protein_coding --> 759
#FJ173815.1 Genbank exon 196 756 . + . ID=gene-ST;Name=ST;gbkey=Gene;gene=ST;gene_biotype=protein_coding --> 561
#FJ173815.1 Genbank exon 3110 4381 . - . ID=gene-VP1;Name=VP1;gbkey=Gene;gene=VP1;gene_biotype=protein_coding --> 1272
#FJ173815.1 Genbank exon 4347 5072 . - . ID=gene-VP2;Name=VP2;gbkey=Gene;gene=VP2;gene_biotype=protein_coding --> 726
# Add four lengths for LT, ST, VP1, and VP2, WaGa
additional_lengths <- data.frame(
ensembl_gene_id = c("LT", "ST", "VP1", "VP2"),
median_length = c(837, 561, 1272, 726)
)
#additional_lengths <- data.frame(
# ensembl_gene_id = c("LT", "ST", "VP1", "VP2"),
# median_length = c(993, 561, 1272, 726)
#)
# Combine the original median_transcript_lengths data frame with the additional lengths
median_transcript_lengths <- rbind(median_transcript_lengths, additional_lengths)
# Merge input_data with median_transcript_lengths
merged_data <- merge(input_data, median_transcript_lengths, by = "ensembl_gene_id")
# [1] ensembl_gene_id gene_name WaGa_RNA
# [4] WaGa_RNA_118 WaGa_RNA_147 WaGa_EV.RNA
# [7] WaGa_EV.RNA_2 WaGa_EV.RNA_118 WaGa_EV.RNA_147
#[10] WaGa_EV.RNA_226 X1107_WaGa_wt_EV X1605_WaGa_wt_EV
#[13] X2706_WaGa_wt_EV X1107_WaGa_sT_DMSO_EV X1605_WaGa_sT_DMSO_EV
#[16] X2706_WaGa_sT_DMSO_EV X1107_WaGa_scr_DMSO_EV X1605_WaGa_scr_DMSO_EV
#[19] X2706_WaGa_scr_DMSO_EV X1107_WaGa_sT_Dox_EV X1605_WaGa_sT_Dox_EV
#[22] X2706_WaGa_sT_Dox_EV X1107_WaGa_scr_Dox_EV X1605_WaGa_scr_Dox_EV
#[25] X2706_WaGa_scr_Dox_EV median_length
#<0 rows> (or 0-length row.names)
# List of column names containing count data
count_columns <- c("WaGa_RNA", "WaGa_RNA_118", "WaGa_RNA_147", "WaGa_EV.RNA", "WaGa_EV.RNA_2", "WaGa_EV.RNA_118", "WaGa_EV.RNA_147", "WaGa_EV.RNA_226", "X1107_WaGa_wt_EV", "X1605_WaGa_wt_EV", "X2706_WaGa_wt_EV", "X1107_WaGa_sT_DMSO_EV", "X1605_WaGa_sT_DMSO_EV", "X2706_WaGa_sT_DMSO_EV", "X1107_WaGa_scr_DMSO_EV", "X1605_WaGa_scr_DMSO_EV", "X2706_WaGa_scr_DMSO_EV", "X1107_WaGa_sT_Dox_EV", "X1605_WaGa_sT_Dox_EV", "X2706_WaGa_sT_Dox_EV", "X1107_WaGa_scr_Dox_EV", "X1605_WaGa_scr_Dox_EV", "X2706_WaGa_scr_Dox_EV")
# [1] ensembl_gene_id gene_name MKL.1_RNA
# [4] MKL.1_RNA_118 MKL.1_RNA_147 MKL.1_EV.RNA
# [7] MKL.1_EV.RNA_2 MKL.1_EV.RNA_118 MKL.1_EV.RNA_87
#[10] MKL.1_EV.RNA_27 X042_MKL.1_wt_EV X0505_MKL.1_wt_EV
#[13] X042_MKL.1_sT_DMSO X0505_MKL.1_sT_DMSO_EV X042_MKL.1_scr_DMSO_EV
#[16] X0505_MKL.1_scr_DMSO_EV X042_MKL.1_sT_Dox X0505_MKL.1_sT_Dox_EV
#[19] X042_MKL.1_scr_Dox_EV X0505_MKL.1_scr_Dox_EV median_length
#count_columns <- c("MKL.1_RNA","MKL.1_RNA_118","MKL.1_RNA_147","MKL.1_EV.RNA","MKL.1_EV.RNA_2","MKL.1_EV.RNA_118","MKL.1_EV.RNA_87","MKL.1_EV.RNA_27","X042_MKL.1_wt_EV","X0505_MKL.1_wt_EV","X042_MKL.1_sT_DMSO","X0505_MKL.1_sT_DMSO_EV","X042_MKL.1_scr_DMSO_EV","X0505_MKL.1_scr_DMSO_EV","X042_MKL.1_sT_Dox","X0505_MKL.1_sT_Dox_EV","X042_MKL.1_scr_Dox_EV","X0505_MKL.1_scr_Dox_EV")
# Calculate FPKM values for each sample
fpkm_data <- merged_data %>%
mutate(across(all_of(count_columns), ~ .x / median_length * 1e9 / sum(.x), .names = "FPKM_{.col}"))
# # Calculate FPKM values
# total_counts <- sum(merged_data$counts)
# merged_data$FPKM <- (merged_data$counts / (merged_data$median_length / 1000)) / (total_counts / 1e6)
# merged_data$FPKM <- (merged_data$counts * 1e9) / (sum(merged_data$counts) * merged_data$median_length)
# Save the FPKM values to a CSV file
write.csv(fpkm_data, "fpkm_values_WaGa.csv", row.names = FALSE)
STEP3: using following code to draw plots
#For plotting a category against values with randomly assigned x-axis positions for the points within each category, you can use the geom_jitter() function in ggplot2. This function adds a small amount of random noise to the x-axis position of each point, allowing them to be distinguished from each other and preventing overplotting.
#This will produce a scatter plot with the category on the x-axis and the values on the y-axis, with the x-axis positions of the points randomly assigned within each category. You can adjust the amount of jitter by changing the width parameter.
library(ggplot2)
fpkm_data_waga = read.csv("fpkm_values_WaGa.csv")
fpkm_data_waga_selected = fpkm_data_waga[, c('ensembl_gene_id', 'FPKM_WaGa_RNA', 'FPKM_X1107_WaGa_wt_EV')]
fpkm_data_mkl1= read.csv("fpkm_values_MKL-1.csv")
fpkm_data_mkl1_selected = fpkm_data_mkl1[, c('ensembl_gene_id', 'FPKM_MKL.1_RNA', 'FPKM_X042_MKL.1_wt_EV')]
# merge the two data frames by the 'id' column
fpkm_data_selected <- merge(fpkm_data_waga_selected, fpkm_data_mkl1_selected, by = "ensembl_gene_id")
## reorder the columns
#fpkm_data_selected <- select(fpkm_data_selected, ensembl_gene_id, FPKM_WaGa_RNA, FPKM_MKL.1_RNA, FPKM_WaGa_EV.RNA, FPKM_MKL.1_EV.RNA)
# select columns with FPKM values
fpkm_cols <- c('FPKM_WaGa_RNA', 'FPKM_X1107_WaGa_wt_EV', 'FPKM_MKL.1_RNA', 'FPKM_X042_MKL.1_wt_EV')
## take the log2 of selected columns
#fpkm_data_selected_log2 <- fpkm_data_selected %>%
# mutate(across(fpkm_cols, log2, .names = "log2_{.col}"))
#fpkm_data <- fpkm_data %>%
# mutate(across(c(count_columns), ~log2(. + 1), .names = "{.col}_log2")) %>%
# select(-all_of(count_columns))
# subset the data frame to exclude rows with missing values in column "ensembl_gene_id"
#fpkm_data_filtered <- fpkm_data_selected %>% filter(ensembl_gene_id != "")
fpkm_data_selected <- fpkm_data_selected %>%
mutate(across(all_of(fpkm_cols), ~log2(. + 1), .names = "log2_{.col}")) %>% select(-all_of(fpkm_cols)) %>% rename("WaGa RNA" = "log2_FPKM_WaGa_RNA", "MKL-1 RNA" = "log2_FPKM_MKL.1_RNA", "WaGa EV" = "log2_FPKM_X1107_WaGa_wt_EV", "MKL-1 EV" = "log2_FPKM_X042_MKL.1_wt_EV")
df_long <- pivot_longer(fpkm_data_selected, cols = -ensembl_gene_id, names_to = "x", values_to = "y")
# use mutate() and ifelse() to add a color column
df_long <- mutate(df_long, color = ifelse(ensembl_gene_id %in% c("ST", "LT", "VP1", "VP2"),
"viral",
"human"))
df_long$x <- factor(df_long$x, levels=c("WaGa RNA","MKL-1 RNA","WaGa EV","MKL-1 EV"))
# Sample 1000 rows from your data frame
#df_sampled <- df_long %>% sample_n(1000)
# plot using geom_jitter
png("plot_samples_against_FPKM.png", width = 800, height = 1000)
#svg("plot_samples_against_FPKM.svg")
ggplot(df_long, aes(x = x, y = y, color=color)) +
scale_color_manual(values = c("human" = "darkblue", "viral" = "red")) +
geom_jitter(width = 0.4) +
labs(x = "", y = "log2(Fragments Per Kilobase of transcript per Million mapped reads)")
dev.off()
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Workflow using PICRUSt2 for Data_Karoline_16S_2025
- Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences
- 阳光房漏水怎么办?丁基胶带才是最佳密封选择
- DAMIAN Post-processing for Flavivirus and FSME
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Plot phylogenetic tree_heatmap and MSA on yopBDJTEMKOH[NR]
QIIME + Phyloseq + MicrobiotaProcess (v1)