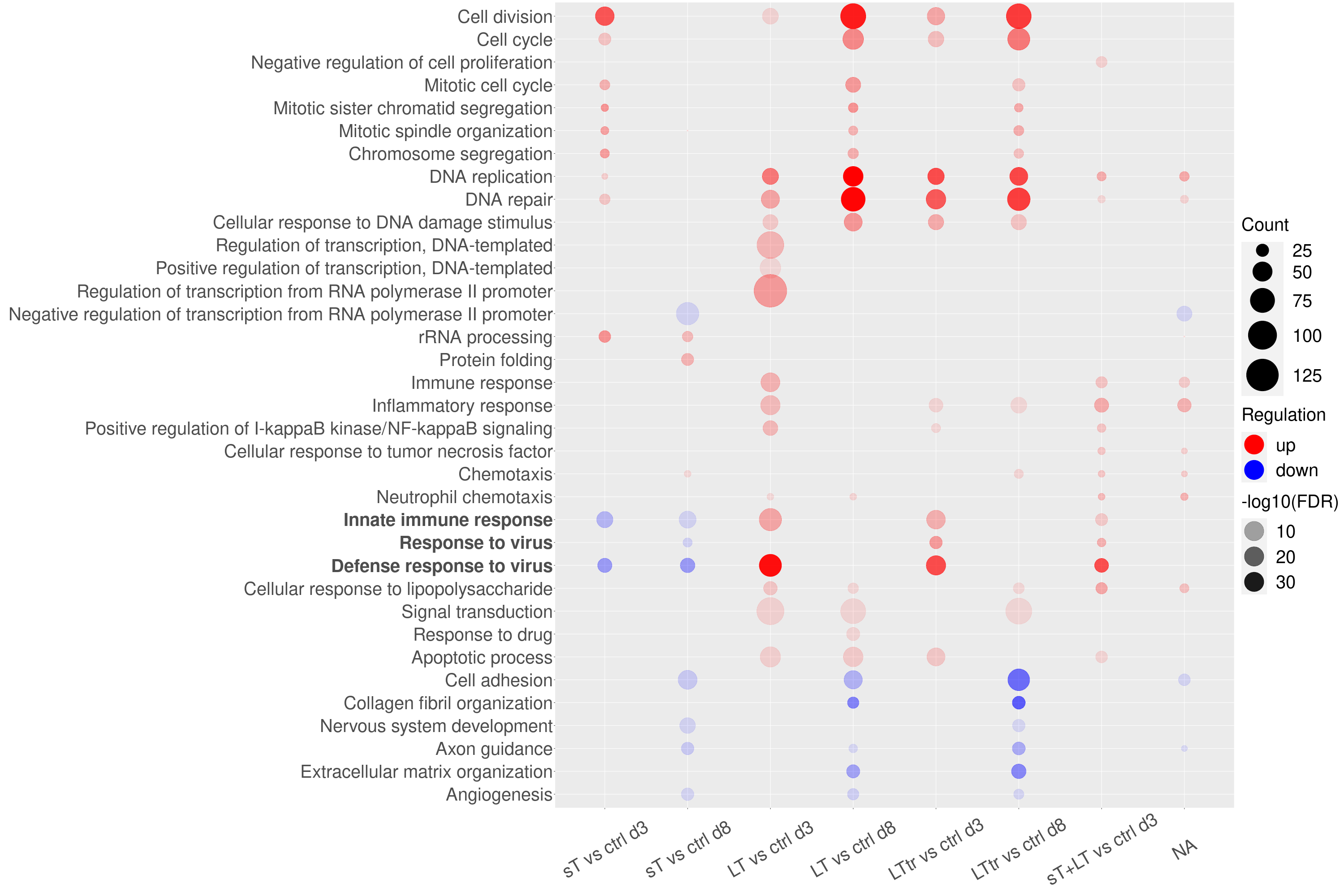
Bubble Plot Visualization of Gene Expression Changes
gene_x 0 like s 650 view s
Tags: plot, R
Download the file Bubble_R_allTantigens.csv
library(ggplot2)
library(dplyr)
library(magrittr)
library(tidyr)
library(forcats)
mydat <- read.csv("Bubble_R_allTantigens.csv", sep=";", header=TRUE)
mydat$GeneRatio <- sapply(mydat$GeneRatio_frac, function(x) eval(parse(text=x)))
#mydat$FoldEnrichment <- as.numeric(mydat$FoldEnrichment)
mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down"))
mydat$Comparison <- factor(mydat$Comparison, levels=c("sT vs ctrl d3","sT vs ctrl d8","LT vs ctrl d3","LT vs ctrl d8","LTtr vs ctrl d3","LTtr vs ctrl d8","sT+LT vs ctrl d3","sT+LTtr vs ctrl d912"))
mydat$Term <- factor(mydat$Term, levels=rev(c("Cell division", "Cell cycle", "Negative regulation of cell proliferation", "Mitotic cell cycle", "Mitotic sister chromatid segregation", "Mitotic spindle organization", "Chromosome segregation", "DNA replication", "DNA repair", "Cellular response to DNA damage stimulus", "Regulation of transcription, DNA-templated", "Positive regulation of transcription, DNA-templated", "Regulation of transcription from RNA polymerase II promoter", "Negative regulation of transcription from RNA polymerase II promoter", "rRNA processing", "Protein folding", "Immune response", "Inflammatory response", "Positive regulation of I-kappaB kinase/NF-kappaB signaling", "Cellular response to tumor necrosis factor", "Chemotaxis", "Neutrophil chemotaxis", "Innate immune response", "Response to virus", "Defense response to virus", "Cellular response to lipopolysaccharide", "Signal transduction", "Response to drug", "Apoptotic process", "Cell adhesion", "Collagen fibril organization", "Nervous system development", "Axon guidance", "Extracellular matrix organization", "Angiogenesis")))
tiff("Bubble_all-Tantigens_big.tiff", units = "in", width = 35, height = 50, res=500)
#png("bubble.png", width=1400, height=600)
xl <- factor(rev(c("Cell division", "Cell cycle", "Negative regulation of cell proliferation", "Mitotic cell cycle", "Mitotic sister chromatid segregation", "Mitotic spindle organization", "Chromosome segregation", "DNA replication", "DNA repair", "Cellular response to DNA damage stimulus", "Regulation of transcription, DNA-templated", "Positive regulation of transcription, DNA-templated", "Regulation of transcription from RNA polymerase II promoter", "Negative regulation of transcription from RNA polymerase II promoter", "rRNA processing", "Protein folding", "Immune response", "Inflammatory response", "Positive regulation of I-kappaB kinase/NF-kappaB signaling", "Cellular response to tumor necrosis factor", "Chemotaxis", "Neutrophil chemotaxis", "Innate immune response", "Response to virus", "Defense response to virus", "Cellular response to lipopolysaccharide", "Signal transduction", "Response to drug", "Apoptotic process", "Cell adhesion", "Collagen fibril organization", "Nervous system development", "Axon guidance", "Extracellular matrix organization", "Angiogenesis")))
bold.terms <- c("Innate immune response", "Response to virus", "Defense response to virus")
bold.labels <- ifelse((xl) %in% bold.terms, yes = "bold", no = "plain")
#-log10(FDR) can be renamed as 'Significance'
png("bubble_plot.png", 3000, 2000)
ggplot(mydat, aes(y = Term, x = Comparison)) + geom_point(aes(color = Regulation, size = Count, alpha = abs(log10(FDR)))) + scale_color_manual(values = c("up" = "red", "down" = "blue")) + scale_size_continuous(range = c(1, 34)) + labs(x = "", y = "", color="Regulation", size="Count", alpha="-log10(FDR)") + theme(axis.text.y = element_text(face = bold.labels))+ theme(axis.text.x = element_text(angle = 30, vjust = 0.5)) + theme(axis.text = element_text(size = 40)) + theme(legend.text = element_text(size = 40)) + theme(legend.title = element_text(size = 40))+
guides(color = guide_legend(override.aes = list(size = 20)), alpha = guide_legend(override.aes = list(size = 20)))
dev.off()
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Should the inputs for GSVA be normalized or raw?
- File format for single channel analysis of Agilent microarray data with Limma?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Kraken2 Installation and Usage Guide
最新文章
- DAMIAN Post-processing for Flavivirus and FSME
- All tools and services of BV-BRC
- Variant calling (inter-host + intra-host) for Data_Pietschmann_229ECoronavirus_Mutations_2025 (via docker own_viral_ngs) v2
- How to correlate RNA-seq Data with Mass Spectrometry Proteomics Data?
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
MicrobiotaProcess Group2 vs Group6 (v1)
Bubble plot for 1457∆atlE vs 1457-M10 vs 1457 vs mock