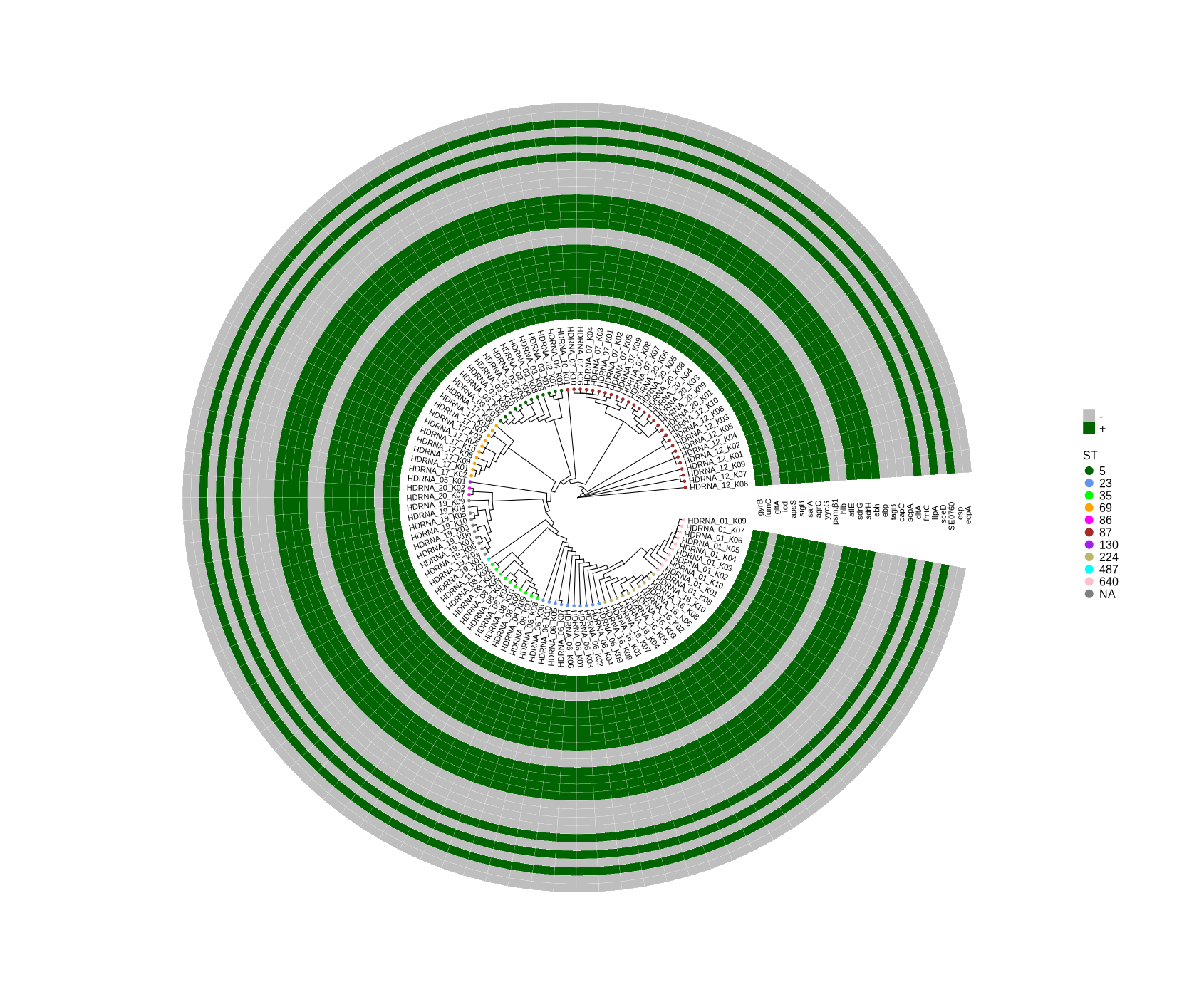
Enhanced Visualization of Gene Presence for the Selected Genes in Bongarts_S.epidermidis_HDRNA
gene_x 0 like s 593 view s
Tags: plot, processing, pipeline
-
preparing gene seqences
(Optional online search) https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=MicrobialGenomes #Title:Refseq prokaryote representative genomes (contains refseq assembly) #Molecule Type:mixed DNA #Update date:2024/10/16 #Number of sequences:1038672 #cat gyrB_revcomp.fasta fumC_revcomp.fasta gltA.fasta icd_revcomp.fasta apsS.fasta sigB_revcomp.fasta sarA_revcomp.fasta ... # SCCmec,agr.typing, # gyrB(House keeper), # fumC,gltA,icd(Metabolic genes), # apsS,sigB,sarA,agrC,yycG(Virulence regulators), # psm.β1,hlb(Toxins), # atlE,sdrG,sdrH,ebh,ebp,tagB(Biofilm formation), # capC,sepA,dltA,fmtC,lipA,sceD,SE0760(Immune evasion & colonization), # esp,ecpA(Serine protease) gene complement(1547773..1549233) /gene="ebp" /locus_tag="SAKG22_13940" CDS complement(1547773..1549233) /gene="ebp" samtools faidx AP019545.1.fasta samtools faidx AP019545.1.fasta "gi|1602080981|dbj|AP019545.1|":1547773-1549233 > ebp.fasta revcomp ebp.fasta > ebp_revcomp.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/gyrB_revcomp.fasta gyrB.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/fumC_revcomp.fasta fumC.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/gltA.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/icd_revcomp.fasta icd.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/apsS.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sigB_revcomp.fasta sigB.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sarA_revcomp.fasta sarA.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/agrC.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/yycG.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/psm-beta1.fasta psm.β1.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/hlb.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/atlE.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sdrG.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sdrH_revcomp.fasta sdrH.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ebh_revcomp.fasta ebh.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ebp_revcomp.fasta ebp.fasta cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/tagB.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/capC.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sepA.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/dltA.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/fmtC.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/lipA.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/sceD.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/SE0760.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/esp.fasta . cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/ecpA.fasta . #cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/MT880870.fasta . #cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/MT880871.fasta . #cp ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/MT880872.fasta . >gi|1602080981|dbj|AP019545.1|:1547773-1549233 >gi|34980227|gb|AY322150.1|:3091-8793 ./gyrB_revcomp.fasta ./agrC.fasta ./CP133692.fasta ./sceD_mibi1435_revcomp.fasta ./CP133682.fasta ./CP133697.fasta ./sdrG.fasta ./sarA_revcomp.fasta ./CP133679.fasta ./CP133686.fasta ./sepA.fasta ./icd.fasta ./CP133691.fasta ./CP133698.fasta ./yycG.fasta ./CP133701.fasta ./sdrH_revcomp.fasta ./SE0760.fasta ./CP133681.fasta ./CP133677.fasta ./yycG_revcomp.fasta ./CP000029.fasta ./atlE_old.fasta ./ebpS.fasta ./ebpS_revcomp.fasta ./atlE_revcomp.fasta ./sceDAE.fasta ./hlb.fasta >gi|441494790|gb|KC242859.1|:16-1008 ATGGTGAAAAAAACAAAATCCAATTCACTAAAAAAAGTTGCAACACTTGCATTAGCAAAT TTATTATTAGTTGGTGCACTTACTGACAATAGTGCCAAAGCCGAATCTAAGAAAGATGAT ACTGATTTGAAGTTAGTTAGTCATAACGTTTATATGTTATCGACCGTTTTGTATCCAAAC TGGGGGCAATATAAACGCGCTGATTTAATCGGGCAATCTTCTTATATTAAAAATAATGAT GTCGTAATATTCAATGAAGCATTTGATAATGGCGCATCAGATAAGCTATTAAGTAATGTA AAAAAAGAATATCCTTACCAAACACCTGTACTCGGTCGTTCTCAATCAGGGTGGGACAAA ACTGAAGGTAGCTACTCATCAACTGTTGCTGAAGATGGTGGCGTAGCGATTGTAAGTAAA TATCCTATTAAAGAAAAAATCCAGCATGTTTTCAAAAGCGGTTGTGGATTCGACAATGAT AGCAACAAAGGCTTTGTTTATACAAAAATAGAGAAAAATGGGAAGAACGTTCATGTTATC GGTACACATACACAATCTGAAGATTCACGTTGTGGTGCTGGACATGATCGAAAAATTAGA GCTGAACAAATGAAAGAAATCAGTGATTTTGTTAAAAAGAAAAATATCCCTAAAGATGAA ACGGTATATATAGGTGGCGACCTTAATGTAAATAAAGGTACTCCAGAGTTCAAAGATATG CTTAAAAACTTGAATGTAAATGATGTTCTATATGCAGGTCATAATAGCACATGGGACCCT CAATCAAATTCAATTGCGAAATATAATTATCCTAATGGTAAACCAGAACATTTAGACTAT ATATTTACAGATAAAGATCATAAACAACCAAAACAATTAGTCAATGAAGTTGTGACTGAA AAACCTAAGCCATGGGATGTATATGCGTTCCCATATTACTACGTTTACAATGATTTTTCA GATCATTACCCAATCAAAGCCTATAGTAAATAA >hlb_ >gi|441494790|gb|KC242859.1| Staphylococcus aureus strain 226 beta-hemolysin (hlb) gene, complete cds AAAGGAGTGATAATG ATGGTGAAAAAAACAAAATCCAATTCACTAAAAAAAGTTGCAACACTTGCATTAGCAAATTTATTATTAGTTGGTGCACTTACTGACAATAGTGCCAAAGCCGAATCTAAGAAAGATGATACTGA TTTGAAGTTAGTTAGTCATAACGTTTATATGTTATCGACCGTTTTGTATCCAAACTGGGGGCAATATAAA CGCGCTGATTTAATCGGGCAATCTTCTTATATTAAAAATAATGATGTCGTAATATTCAATGAAGCATTTG ATAATGGCGCATCAGATAAGCTATTAAGTAATGTAAAAAAAGAATATCCTTACCAAACACCTGTACTCGG TCGTTCTCAATCAGGGTGGGACAAAACTGAAGGTAGCTACTCATCAACTGTTGCTGAAGATGGTGGCGTA GCGATTGTAAGTAAATATCCTATTAAAGAAAAAATCCAGCATGTTTTCAAAAGCGGTTGTGGATTCGACA ATGATAGCAACAAAGGCTTTGTTTATACAAAAATAGAGAAAAATGGGAAGAACGTTCATGTTATCGGTAC ACATACACAATCTGAAGATTCACGTTGTGGTGCTGGACATGATCGAAAAATTAGAGCTGAACAAATGAAA GAAATCAGTGATTTTGTTAAAAAGAAAAATATCCCTAAAGATGAAACGGTATATATAGGTGGCGACCTTA ATGTAAATAAAGGTACTCCAGAGTTCAAAGATATGCTTAAAAACTTGAATGTAAATGATGTTCTATATGC AGGTCATAATAGCACATGGGACCCTCAATCAAATTCAATTGCGAAATATAATTATCCTAATGGTAAACCA GAACATTTAGACTATATATTTACAGATAAAGATCATAAACAACCAAAACAATTAGTCAATGAAGTTGTGA CTGAAAAACCTAAGCCATGGGATGTATATGCGTTCCCATATTACTACGTTTACAATGATTTTTCAGATCA TTACCCAATCAAAGCCTATAGTAAATAA ./dltA.fasta ./atlE.fasta ./gyrB.fasta ./ORF123_sepA_ORF5.fasta ./ebh.fasta ./CP133696.fasta ./yycG_old.fasta ./CP133700.fasta ./CP133695.fasta ./ecpA.fasta ./capBCA_ywtC.fasta ./CP133690.fasta ./gltA.fasta ./esp.fasta ./sigB.fasta ./CP133699.fasta ./CP133685.fasta ./CP133680.fasta ./MT880872.fasta ./icd_revcomp.fasta ./agrC_old.fasta ./ecpA_.fasta ./agrC_revcomp.fasta ./MT880870.fasta ./CP133687.fasta ./CP133693.fasta ./psm-beta1.fasta >gi|380448412|gb|JQ066320.1| Staphylococcus aureus strain JP1 Psm beta1 (psm beta1) and Psm beta2 (psm beta2) genes, complete cds ACACGTTTAACAACACAAGAATTATTATATCTAAATGAACTAAAATTAGCAATACCTTGTAAATAAAAAA TGTTTATATTTTTCACTATTATAGAGCTATTTATCTAAAAAGGTTCAATAAGACTTAAATACGAATTCAG GCAACTTAATTGTGTTAAATACAGTTTTGAATGCCTAACTGTATTTCTTTTCTCTTTAAAATACAGTTAA GTACATTATAAGATGTTGTGCGGATAAACAAACTAATTGCATCAAATTTATTTTAAAATAACAACAACAA AACGTTAAGAGAATAACATTTCGGTGATTTAAAAGCTACGCACGTTTTTGTTATCTTCAAATTTAAATTT TAAGGAGTGTTTTCA ATGGAAGGTTTATTTAACGCAATTAAAGATACCGTAACTGCAGCAATTAATAATGATGGC GCAAAATTAGGCACAAGCATTGTGAGCATCGTTGAAAATGGCGTAGGTTTATTAGGTAAA TTATTCGGATTCTAA TTTCAATATGTTATGTAAGTAATCAGTATTATTTCAAAGGTGAGGGAGAGATTTAAATGA CTGGACTAGCAGAAGCAATCGCAAATACTGTGCAAGCTGCACAACAACATGATAGTGTGAAATTAGGCAC AAGTATCGTAGACATCGTTGCTAACGGTGTGGGTTTACTAGGTAAATTATTTGGATTCTAATATAATAAC TAATATTCTTTAAAATAAACTGGGTGAGCATACTTTAATGTTATGCACTCAGTTTATTTTATTTGCAGAA ATTTGAGCCTCTGTTAAGATTTAGATACATAGACAATATAGGAGATGGGGAAATTGGGATATAAAAATAT TTTGATAGACTTTGATGATACAATTGTTGATTTTTATGATGCAGA ATGGAAGGTTTATTTAACGCAATTAAAGATACCGTAACTGCAGCAATTAATAATGATGGC GCAAAATTAGGCACAAGCATTGTGAGCATCGTTGAAAATGGCGTAGGTTTATTAGGTAAA TTATTCGGATTCTAA ./psm-beta.fasta ./lipA.fasta ./capC.fasta ./fumC_revcomp.fasta ./CP133689.fasta ./sepA_mibi1435.fasta ./sigB_revcomp.fasta ./sceD.fasta ./fumC.fasta ./Enterococcus_faecium_isolate_E300_pathogenicity_island.fasta ./agrABCD_hld.fasta ./CP133688.fasta ./ecpA_mibi1435_revcomp.fasta ./hlb_.fasta ./CP133684.fasta ./tagB_mibi1435_revcomp.fasta ./CP133678.fasta ./tagB.fasta ./MT880871.fasta ./CP133676.fasta ./sdrH.fasta ./Staphylococcus_epidermidis_RP62A.fasta ./ebh_revcomp.fasta ./CP133683.fasta ./apsS.fasta ./Staphylococcus_aureus_MRSA252.fasta ./fmtC.fasta ./CP133694.fasta ./sarA.fasta -
prepare the long fasta if we use only the long sequencing
ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_01_K01_conservative.fasta HDRNA_01_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_03_K01_bold_bandage.fasta HDRNA_03_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_06_K01_conservative.fasta HDRNA_06_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_07_K01_conservative.fasta HDRNA_07_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_08_K01_conservative.fasta HDRNA_08_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_12_K01_bold.fasta HDRNA_12_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_16_K01_conservative.fasta HDRNA_16_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_17_K01_conservative.fasta HDRNA_17_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_19_K01_bold.fasta HDRNA_19_K01.fasta ln -s ../Data_Holger_S.epidermidis_long/finished_assemblies/HDRNA_20_K01_conservative.fasta HDRNA_20_K01.fasta -
perform blastn searching for long-sequencing
# -- makeblastdb -- for sample in HDRNA_01_K01 HDRNA_03_K01 HDRNA_06_K01 HDRNA_07_K01 HDRNA_08_K01 HDRNA_12_K01 HDRNA_16_K01 HDRNA_17_K01 HDRNA_19_K01 HDRNA_20_K01; do makeblastdb -in ../assembly/${sample}.fasta -dbtype nucl done for id in gyrB fumC gltA icd apsS sigB sarA agrC yycG psm.β1 hlb atlE sdrG sdrH ebh ebp tagB capC sepA dltA fmtC lipA sceD SE0760 esp ecpA; do echo "mkdir ${id}" echo "for sample in in HDRNA_01_K01 HDRNA_03_K01 HDRNA_06_K01 HDRNA_07_K01 HDRNA_08_K01 HDRNA_12_K01 HDRNA_16_K01 HDRNA_17_K01 HDRNA_19_K01 HDRNA_20_K01; do" echo "blastn -db ../assembly/${sample}.fasta -query ${id}.fasta -evalue 1e-40 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done #gyrB fumC gltA icd apsS sigB sarA agrC yycG psm.β1 hlb atlE sdrG sdrH ebh ebp tagB capC sepA dltA fmtC lipA sceD SE0760 esp ecpA python ~/Scripts/process_directory.py gyrB typing.csv typing_until_gyrB.csv python ~/Scripts/process_directory.py fumC typing_until_gyrB.csv typing_until_fumC.csv python ~/Scripts/process_directory.py gltA typing_until_fumC.csv typing_until_gltA.csv python ~/Scripts/process_directory.py icd typing_until_gltA.csv typing_until_icd.csv python ~/Scripts/process_directory.py apsS typing_until_icd.csv typing_until_apsS.csv python ~/Scripts/process_directory.py sigB typing_until_apsS.csv typing_until_sigB.csv python ~/Scripts/process_directory.py sarA typing_until_sigB.csv typing_until_sarA.csv python ~/Scripts/process_directory.py agrC typing_until_sarA.csv typing_until_agrC.csv python ~/Scripts/process_directory.py yycG typing_until_agrC.csv typing_until_yycG.csv python ~/Scripts/process_directory.py psm.β1 typing_until_yycG.csv typing_until_psm.β1.csv python ~/Scripts/process_directory.py hlb typing_until_psm.β1.csv typing_until_hlb.csv python ~/Scripts/process_directory.py atlE typing_until_hlb.csv typing_until_atlE.csv python ~/Scripts/process_directory.py sdrG typing_until_atlE.csv typing_until_sdrG.csv python ~/Scripts/process_directory.py sdrH typing_until_sdrG.csv typing_until_sdrH.csv python ~/Scripts/process_directory.py ebh typing_until_sdrH.csv typing_until_ebh.csv python ~/Scripts/process_directory.py ebp typing_until_ebh.csv typing_until_ebp.csv python ~/Scripts/process_directory.py tagB typing_until_ebp.csv typing_until_tagB.csv python ~/Scripts/process_directory.py capC typing_until_tagB.csv typing_until_capC.csv python ~/Scripts/process_directory.py sepA typing_until_capC.csv typing_until_sepA.csv python ~/Scripts/process_directory.py dltA typing_until_sepA.csv typing_until_dltA.csv python ~/Scripts/process_directory.py fmtC typing_until_dltA.csv typing_until_fmtC.csv python ~/Scripts/process_directory.py lipA typing_until_fmtC.csv typing_until_lipA.csv python ~/Scripts/process_directory.py sceD typing_until_lipA.csv typing_until_sceD.csv python ~/Scripts/process_directory.py SE0760 typing_until_sceD.csv typing_until_SE0760.csv python ~/Scripts/process_directory.py esp typing_until_SE0760.csv typing_until_esp.csv python ~/Scripts/process_directory.py ecpA typing_until_esp.csv typing_until_ecpA.csv -
perform blastn searching for short-sequencing
# -- makeblastdb -- for sample in HDRNA_01_K01 HDRNA_01_K02 HDRNA_01_K03 HDRNA_01_K04 HDRNA_01_K05 HDRNA_01_K06 HDRNA_01_K07 HDRNA_01_K08 HDRNA_01_K09 HDRNA_01_K10 HDRNA_02_K01 HDRNA_03_K01 HDRNA_03_K02 HDRNA_03_K03 HDRNA_03_K04 HDRNA_03_K05 HDRNA_03_K06 HDRNA_03_K07 HDRNA_03_K08 HDRNA_03_K09 HDRNA_03_K10 HDRNA_04_K01 HDRNA_05_K01 HDRNA_06_K01 HDRNA_06_K02 HDRNA_06_K03 HDRNA_06_K04 HDRNA_06_K05 HDRNA_06_K06 HDRNA_06_K07 HDRNA_06_K08 HDRNA_06_K09 HDRNA_06_K10 HDRNA_07_K01 HDRNA_07_K02 HDRNA_07_K03 HDRNA_07_K04 HDRNA_07_K05 HDRNA_07_K06 HDRNA_07_K07 HDRNA_07_K08 HDRNA_07_K09 HDRNA_07_K10 HDRNA_08_K01 HDRNA_08_K02 HDRNA_08_K03 HDRNA_08_K04 HDRNA_08_K05 HDRNA_08_K06 HDRNA_08_K07 HDRNA_08_K08 HDRNA_08_K09 HDRNA_08_K10 HDRNA_10_K01 HDRNA_11_K01 HDRNA_12_K01 HDRNA_12_K02 HDRNA_12_K03 HDRNA_12_K04 HDRNA_12_K05 HDRNA_12_K06 HDRNA_12_K07 HDRNA_12_K08 HDRNA_12_K09 HDRNA_12_K10 HDRNA_16_K01 HDRNA_16_K02 HDRNA_16_K03 HDRNA_16_K04 HDRNA_16_K05 HDRNA_16_K06 HDRNA_16_K07 HDRNA_16_K08 HDRNA_16_K09 HDRNA_16_K10 HDRNA_17_K01 HDRNA_17_K02 HDRNA_17_K03 HDRNA_17_K04 HDRNA_17_K05 HDRNA_17_K06 HDRNA_17_K07 HDRNA_17_K08 HDRNA_17_K09 HDRNA_17_K10 HDRNA_19_K01 HDRNA_19_K02 HDRNA_19_K03 HDRNA_19_K04 HDRNA_19_K05 HDRNA_19_K06 HDRNA_19_K07 HDRNA_19_K08 HDRNA_19_K09 HDRNA_19_K10 HDRNA_20_K01 HDRNA_20_K02 HDRNA_20_K03 HDRNA_20_K04 HDRNA_20_K05 HDRNA_20_K06 HDRNA_20_K07 HDRNA_20_K08 HDRNA_20_K09; do makeblastdb -in ../Data_Holger_S.epidermidis_short/shovill/${sample}/contigs.fa -dbtype nucl done #Note: The current -evalue is set to 1e-1; it might need to be made more stringent. for id in gyrB fumC gltA icd apsS sigB sarA agrC yycG psm.β1 hlb atlE sdrG sdrH ebh ebp tagB capC sepA dltA fmtC lipA sceD SE0760 esp ecpA; do echo "mkdir ${id}" echo "for sample in HDRNA_01_K01 HDRNA_01_K02 HDRNA_01_K03 HDRNA_01_K04 HDRNA_01_K05 HDRNA_01_K06 HDRNA_01_K07 HDRNA_01_K08 HDRNA_01_K09 HDRNA_01_K10 HDRNA_02_K01 HDRNA_03_K01 HDRNA_03_K02 HDRNA_03_K03 HDRNA_03_K04 HDRNA_03_K05 HDRNA_03_K06 HDRNA_03_K07 HDRNA_03_K08 HDRNA_03_K09 HDRNA_03_K10 HDRNA_04_K01 HDRNA_05_K01 HDRNA_06_K01 HDRNA_06_K02 HDRNA_06_K03 HDRNA_06_K04 HDRNA_06_K05 HDRNA_06_K06 HDRNA_06_K07 HDRNA_06_K08 HDRNA_06_K09 HDRNA_06_K10 HDRNA_07_K01 HDRNA_07_K02 HDRNA_07_K03 HDRNA_07_K04 HDRNA_07_K05 HDRNA_07_K06 HDRNA_07_K07 HDRNA_07_K08 HDRNA_07_K09 HDRNA_07_K10 HDRNA_08_K01 HDRNA_08_K02 HDRNA_08_K03 HDRNA_08_K04 HDRNA_08_K05 HDRNA_08_K06 HDRNA_08_K07 HDRNA_08_K08 HDRNA_08_K09 HDRNA_08_K10 HDRNA_10_K01 HDRNA_11_K01 HDRNA_12_K01 HDRNA_12_K02 HDRNA_12_K03 HDRNA_12_K04 HDRNA_12_K05 HDRNA_12_K06 HDRNA_12_K07 HDRNA_12_K08 HDRNA_12_K09 HDRNA_12_K10 HDRNA_16_K01 HDRNA_16_K02 HDRNA_16_K03 HDRNA_16_K04 HDRNA_16_K05 HDRNA_16_K06 HDRNA_16_K07 HDRNA_16_K08 HDRNA_16_K09 HDRNA_16_K10 HDRNA_17_K01 HDRNA_17_K02 HDRNA_17_K03 HDRNA_17_K04 HDRNA_17_K05 HDRNA_17_K06 HDRNA_17_K07 HDRNA_17_K08 HDRNA_17_K09 HDRNA_17_K10 HDRNA_19_K01 HDRNA_19_K02 HDRNA_19_K03 HDRNA_19_K04 HDRNA_19_K05 HDRNA_19_K06 HDRNA_19_K07 HDRNA_19_K08 HDRNA_19_K09 HDRNA_19_K10 HDRNA_20_K01 HDRNA_20_K02 HDRNA_20_K03 HDRNA_20_K04 HDRNA_20_K05 HDRNA_20_K06 HDRNA_20_K07 HDRNA_20_K08 HDRNA_20_K09; do" echo "blastn -db ../Data_Holger_S.epidermidis_short/shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-1 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done #gyrB fumC gltA icd apsS sigB sarA agrC yycG psm.β1 hlb atlE sdrG sdrH ebh ebp tagB capC sepA dltA fmtC lipA sceD SE0760 esp ecpA python ~/Scripts/process_directory.py gyrB typing_104.csv typing_until_gyrB.csv python ~/Scripts/process_directory.py fumC typing_until_gyrB.csv typing_until_fumC.csv python ~/Scripts/process_directory.py gltA typing_until_fumC.csv typing_until_gltA.csv python ~/Scripts/process_directory.py icd typing_until_gltA.csv typing_until_icd.csv python ~/Scripts/process_directory.py apsS typing_until_icd.csv typing_until_apsS.csv python ~/Scripts/process_directory.py sigB typing_until_apsS.csv typing_until_sigB.csv python ~/Scripts/process_directory.py sarA typing_until_sigB.csv typing_until_sarA.csv python ~/Scripts/process_directory.py agrC typing_until_sarA.csv typing_until_agrC.csv python ~/Scripts/process_directory.py yycG typing_until_agrC.csv typing_until_yycG.csv python ~/Scripts/process_directory.py psm.β1 typing_until_yycG.csv typing_until_psm.β1.csv python ~/Scripts/process_directory.py hlb typing_until_psm.β1.csv typing_until_hlb.csv python ~/Scripts/process_directory.py atlE typing_until_hlb.csv typing_until_atlE.csv python ~/Scripts/process_directory.py sdrG typing_until_atlE.csv typing_until_sdrG.csv python ~/Scripts/process_directory.py sdrH typing_until_sdrG.csv typing_until_sdrH.csv python ~/Scripts/process_directory.py ebh typing_until_sdrH.csv typing_until_ebh.csv python ~/Scripts/process_directory.py ebp typing_until_ebh.csv typing_until_ebp.csv python ~/Scripts/process_directory.py tagB typing_until_ebp.csv typing_until_tagB.csv python ~/Scripts/process_directory.py capC typing_until_tagB.csv typing_until_capC.csv python ~/Scripts/process_directory.py sepA typing_until_capC.csv typing_until_sepA.csv python ~/Scripts/process_directory.py dltA typing_until_sepA.csv typing_until_dltA.csv python ~/Scripts/process_directory.py fmtC typing_until_dltA.csv typing_until_fmtC.csv python ~/Scripts/process_directory.py lipA typing_until_fmtC.csv typing_until_lipA.csv python ~/Scripts/process_directory.py sceD typing_until_lipA.csv typing_until_sceD.csv python ~/Scripts/process_directory.py SE0760 typing_until_sceD.csv typing_until_SE0760.csv python ~/Scripts/process_directory.py esp typing_until_SE0760.csv typing_until_esp.csv python ~/Scripts/process_directory.py ecpA typing_until_esp.csv typing_until_ecpA.csv -
plotTreeHeatmap
~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/plotTreeHeatmap #FastTree -gtr -nt variants/snippy.core_without_reference.aln > plotTreeHeatmap/snippy.core.tree #cp ../Data_Holger_S.epidermidis_short/plotTreeHeatmap/typing_104.csv . #cp ../Data_Holger_S.epidermidis_short/results_HDRNA_01-20/variants/snippy.core.aln_104.tree . #Run step 4 library(ggtree) library(ggplot2) library(dplyr) setwd("/home/jhuang/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/plotTreeHeatmap_short/") # -- edit tree -- info <- read.csv("typing_until_ecpA.csv", sep="\t") # Convert 'ST' column to factor with levels in the desired order info$ST <- factor(info$ST, levels = c("5", "23", "35", "69", "86", "87", "130", "224", "487", "640", "none")) info$name <- info$Isolate tree <- read.tree("snippy.core.aln_104.tree") cols <- c("5"="darkgreen", "23"="cornflowerblue", "35"="green","69"="orange","86"="magenta","87"="brown", "130"="purple","224"="darkkhaki", "487"="cyan", "640"="pink","none"="grey") #"2"="", #"7"="seagreen3", #"9"="tan","14"="red", "17"="navyblue", #"73"="pink","81"="purple", #"89"="darksalmon", #"454"="blue","487"="cyan", "558"="skyblue2", "766"="blueviolet" #"5", "23", "35", "69", "86", "87", "130", "224", "487", "640", "none" heatmapData2 <- info %>% select(Isolate, gyrB, fumC, gltA, icd, apsS, sigB, sarA, agrC, yycG, psm.β1, hlb, atlE, sdrG, sdrH, ebh, ebp, tagB, capC, sepA, dltA, fmtC, lipA, sceD, SE0760, esp, ecpA) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn #heatmap.colours <- c("darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("MT880870","MT880871","MT880872","-") #heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "gold", "green","orange","pink","purple","magenta","brown", "darksalmon","chocolate4","darkkhaki", "azure3", "maroon","lightgreen", "blue","cyan", "skyblue2", "blueviolet", "darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("2","5","7","9","14", "17","23", "35","59","73", "81","86","87","89","130","190","290", "297","325", "454","487","558","766", "MT880870","MT880871","MT880872","-") #TEMP_DEACTIVATED! heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "purple", "green","cyan", "darkred", "darkblue", "darkgreen", "grey", "darkgreen", "grey") #TEMP_DEACTIVATED! names(heatmap.colours) <- c("SCCmec_type_II(2A)", "SCCmec_type_III(3A)", "SCCmec_type_III(3A) and SCCmec_type_VIII(4A)", "SCCmec_type_IV(2B)", "SCCmec_type_IV(2B&5)", "SCCmec_type_IV(2B) and SCCmec_type_VI(4B)", "SCCmec_type_IVa(2B)", "SCCmec_type_IVb(2B)", "SCCmec_type_IVg(2B)", "I", "II", "III", "none", "+","-") ## The heatmap colours should correspond to the factor levels #heatmap.colours <- #setNames(c("cornflowerblue","darkgreen","seagreen3","tan","red","green","orange","pink","brown","magenta","cyan", #"magenta","navyblue","cornflowerblue","gold","orange","darkgreen", "green", "seagreen3", #"chocolate4","brown","purple","pink", "tan","cyan","red"), #c("5", "23", "35", "69", "86", "87", "130", "224", "487", "640", "none", "P01", "P02", "P03", "P04", "P05", "P06", "P07", #"P08", "P10", "P11", "P12", "P16", "P17", "P19", "P20")) #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=ST)) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree.png", width=1260, height=1260) #svg("ggtree.svg", width=1260, height=1260) p dev.off() #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) png("ggtree_and_gheatmap_selected_genes.png", width=1690, height=1400) #svg("ggtree_and_gheatmap_mibi_selected_genes.svg", width=17, height=15) gheatmap(p, heatmapData2, width=2, colnames_position="top", colnames_angle=90, colnames_offset_y = 2.0, hjust=0.7, font.size=4, offset = 17) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() -
Response
As mentioned earlier, the presence-absence matrix of genes was generated using a BLASTn comparison between the genome and the gene sequences. Ensuring the accuracy of these gene sequences is crucial for reliable results. I prepared the sequences by searching GeneBank using the gene names, but there may be some ambiguity where the same gene name corresponds to different sequences. Attached, you will find the sequences for the 11 genes that were not identified in the isolates: gltA, psm-beta1, hlb, ebp, tagB, capA, sepA, fmtC, sceD, esp, and ecpA. I think it would be helpful if you could verify whether these sequences correspond to the genes we are looking for. In addition, I have attached a figure showing all samples with short-read sequencing. As illustrated in the plot, SCCmec and agr typing are still pending. If necessary, I can perform the typing and include the information in the plot. Alternatively, we could focus exclusively on the 10 samples obtained through long-read sequencing. Please let me know how you would prefer to proceed.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- Should the inputs for GSVA be normalized or raw?
- pheatmap vs heatmap.2
- Kraken2 Installation and Usage Guide
- Setup conda environments
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- From Genbank-ID to Phylogenetic Tree Visualization using roary, raxml and ete3
- 📊 Transposon Analysis: Sequencing Depth Requirements
- Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
MicrobiotaProcess Group2 vs Group6 (v1)
RNA-seq Tam on CP059040.1 (Acinetobacter baumannii strain ATCC 19606)