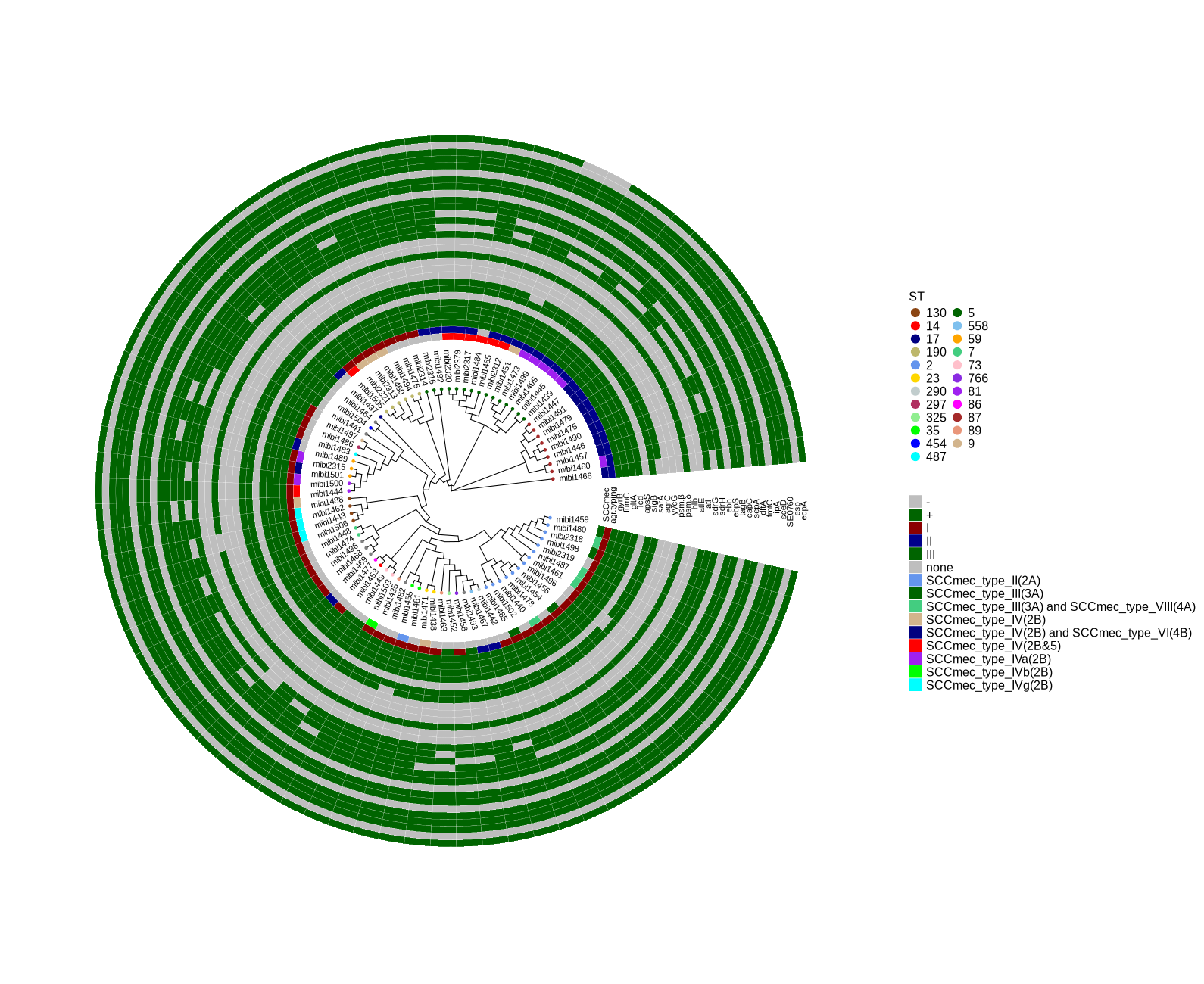
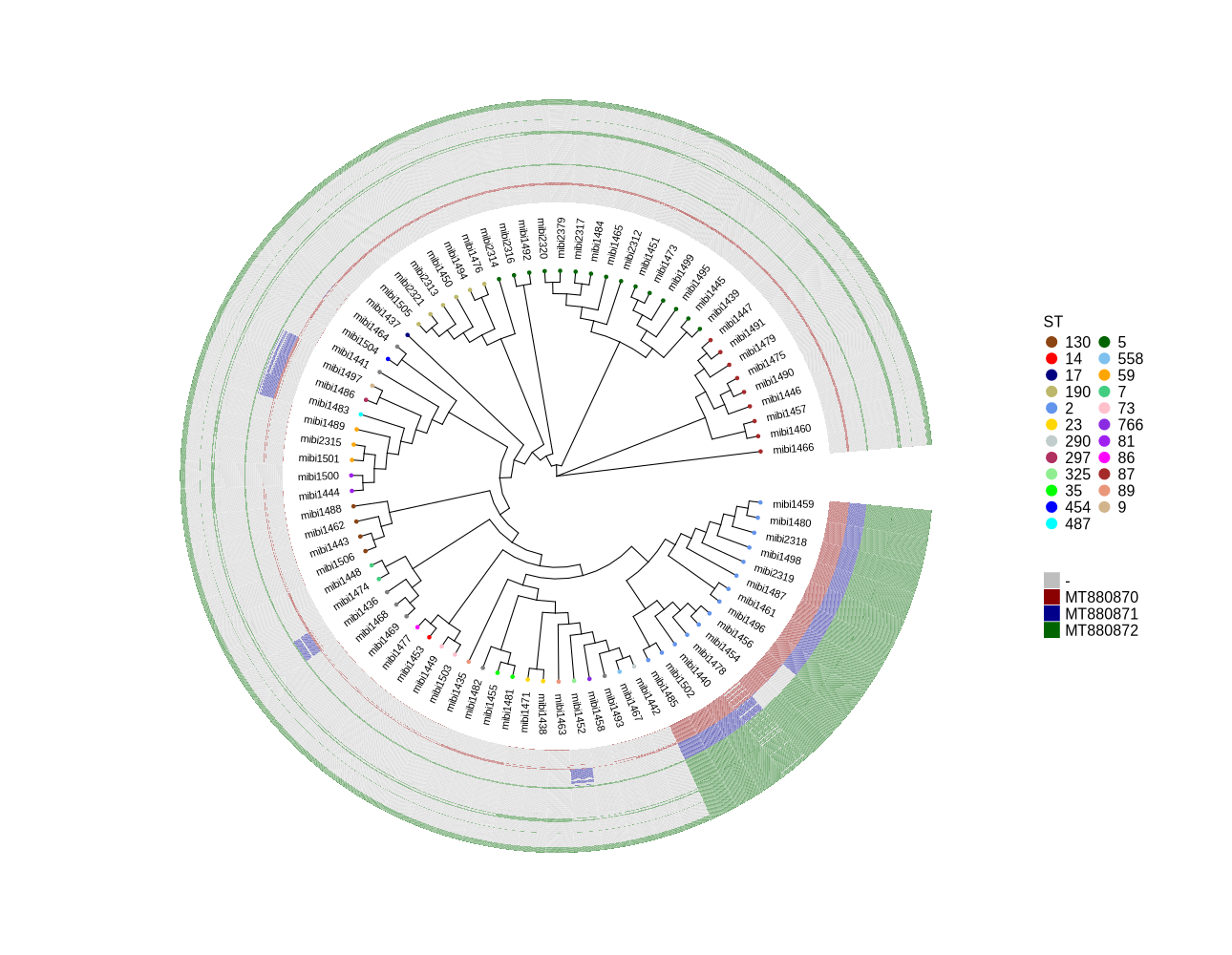
Enhanced Visualization of Gene Presence in Luise_Sepi_STKN
gene_x 0 like s 432 view s
Tags: pipeline
-
prepare typing_ST2_until_QPB07837.csv (under DIR ~/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates and ENV r_env)
cp ../presence_absence_ST2/QPB*.fasta ./ cp ../plotTreeHeatmap/typing.csv ./typing_until_QPB07571.csv # -- makeblastdb -- #for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do # makeblastdb -in ../shovill/${sample}/contigs.fa -dbtype nucl #done for i in {572..622}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do" echo "blastn -db ../shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-40 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {572..622}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done #reprace all '+' with 'MT880870' in typing_until_QPB07622.csv sed -i 's/+/MT880870/g' typing_until_QPB07622.csv -
prepare typing_until_QPB07667.csv
for i in {623..667}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do" echo "blastn -db ../shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-40 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {623..667}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done sed -i 's/+/MT880871/g' typing_until_QPB07667.csv -
prepare typing_until_QPB07667.csv
for i in {668..750}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do" echo "blastn -db ../shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-40 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {751..837}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1435 mibi1436 mibi1437 mibi1438 mibi1439 mibi1440 mibi1441 mibi1442 mibi1443 mibi1444 mibi1445 mibi1446 mibi1447 mibi1448 mibi1449 mibi1450 mibi1451 mibi1452 mibi1453 mibi1454 mibi1455 mibi1456 mibi1457 mibi1458 mibi1459 mibi1460 mibi1461 mibi1462 mibi1463 mibi1464 mibi1465 mibi1466 mibi1467 mibi1468 mibi1469 mibi1471 mibi1473 mibi1474 mibi1475 mibi1476 mibi1477 mibi1478 mibi1479 mibi1480 mibi1481 mibi1482 mibi1483 mibi1484 mibi1485 mibi1486 mibi1487 mibi1488 mibi1489 mibi1490 mibi1491 mibi1492 mibi1493 mibi1494 mibi1495 mibi1496 mibi1497 mibi1498 mibi1499 mibi1500 mibi1501 mibi1502 mibi1503 mibi1504 mibi1505 mibi1506 mibi2312 mibi2313 mibi2314 mibi2315 mibi2316 mibi2317 mibi2318 mibi2319 mibi2320 mibi2321 mibi2379; do" echo "blastn -db ../shovill/\${sample}/contigs.fa -query ${id}.fasta -evalue 1e-40 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {668..837}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py ${id} typing_until_${id_1}.csv typing_until_${id}.csv" done sed -i 's/+/MT880872/g' typing_until_QPB07837.csv cut -d$'\t' -f1-43 typing_until_QPB07837.csv > temp1 cut -d$'\t' -f44- typing_until_QPB07837.csv > temp2 sed -i 's/MT880870/+/g' temp1 paste -d$'\t' temp1 temp2 > ggtree_and_gheatmap_mibi_phages.csv -
plot tree heatmap under /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates
cp ../plotTreeHeatmap/990_backup.tree ./ library(ggtree) library(ggplot2) library(dplyr) setwd("/home/jhuang/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_mibi_isolates/") # -- edit tree -- #https://icytree.org/ #0.000780 # -- for the figure ggtree_and_gheatmap_mibi_phages.png -- info <- read.csv("ggtree_and_gheatmap_mibi.csv", sep="\t") info$name <- info$Isolate tree <- read.tree("990_backup.tree") cols <- c("2"="cornflowerblue","5"="darkgreen","7"="seagreen3","9"="tan","14"="red", "17"="navyblue", "23"="gold", "35"="green","59"="orange","73"="pink","81"="purple","86"="magenta","87"="brown", "89"="darksalmon","130"="chocolate4","190"="darkkhaki", "290"="azure3", "297"="maroon","325"="lightgreen", "454"="blue","487"="cyan", "558"="skyblue2", "766"="blueviolet") #cols <- c("2"='purple2', "other"='darksalmon') #purple2 skyblue2 heatmapData2 <- info %>% select(Isolate, QPB07572, QPB07573, QPB07574, QPB07575, QPB07576, QPB07577, QPB07578, QPB07579, QPB07580, QPB07581, QPB07582, QPB07583, QPB07584, QPB07585, QPB07586, QPB07587, QPB07588, QPB07589, QPB07590, QPB07591, QPB07592, QPB07593, QPB07594, QPB07595, QPB07596, QPB07597, QPB07598, QPB07599, QPB07600, QPB07601, QPB07602, QPB07603, QPB07604, QPB07605, QPB07606, QPB07607, QPB07608, QPB07609, QPB07610, QPB07611, QPB07612, QPB07613, QPB07614, QPB07615, QPB07616, QPB07617, QPB07618, QPB07619, QPB07620, QPB07621, QPB07622, QPB07623, QPB07624, QPB07625, QPB07626, QPB07627, QPB07628, QPB07629, QPB07630, QPB07631, QPB07632, QPB07633, QPB07634, QPB07635, QPB07636, QPB07637, QPB07638, QPB07639, QPB07640, QPB07641, QPB07642, QPB07643, QPB07644, QPB07645, QPB07646, QPB07647, QPB07648, QPB07649, QPB07650, QPB07651, QPB07652, QPB07653, QPB07654, QPB07655, QPB07656, QPB07657, QPB07658, QPB07659, QPB07660, QPB07661, QPB07662, QPB07663, QPB07664, QPB07665, QPB07666, QPB07667, QPB07668, QPB07669, QPB07670, QPB07671, QPB07672, QPB07673, QPB07674, QPB07675, QPB07676, QPB07677, QPB07678, QPB07679, QPB07680, QPB07681, QPB07682, QPB07683, QPB07684, QPB07685, QPB07686, QPB07687, QPB07688, QPB07689, QPB07690, QPB07691, QPB07692, QPB07693, QPB07694, QPB07695, QPB07696, QPB07697, QPB07698, QPB07699, QPB07700, QPB07701, QPB07702, QPB07703, QPB07704, QPB07705, QPB07706, QPB07707, QPB07708, QPB07709, QPB07710, QPB07711, QPB07712, QPB07713, QPB07714, QPB07715, QPB07716, QPB07717, QPB07718, QPB07719, QPB07720, QPB07721, QPB07722, QPB07723, QPB07724, QPB07725, QPB07726, QPB07727, QPB07728, QPB07729, QPB07730, QPB07731, QPB07732, QPB07733, QPB07734, QPB07735, QPB07736, QPB07737, QPB07738, QPB07739, QPB07740, QPB07741, QPB07742, QPB07743, QPB07744, QPB07745, QPB07746, QPB07747, QPB07748, QPB07749, QPB07750, QPB07751, QPB07752, QPB07753, QPB07754, QPB07755, QPB07756, QPB07757, QPB07758, QPB07759, QPB07760, QPB07761, QPB07762, QPB07763, QPB07764, QPB07765, QPB07766, QPB07767, QPB07768, QPB07769, QPB07770, QPB07771, QPB07772, QPB07773, QPB07774, QPB07775, QPB07776, QPB07777, QPB07778, QPB07779, QPB07780, QPB07781, QPB07782, QPB07783, QPB07784, QPB07785, QPB07786, QPB07787, QPB07788, QPB07789, QPB07790, QPB07791, QPB07792, QPB07793, QPB07794, QPB07795, QPB07796, QPB07797, QPB07798, QPB07799, QPB07800, QPB07801, QPB07802, QPB07803, QPB07804, QPB07805, QPB07806, QPB07807, QPB07808, QPB07809, QPB07810, QPB07811, QPB07812, QPB07813, QPB07814, QPB07815, QPB07816, QPB07817, QPB07818, QPB07819, QPB07820, QPB07821, QPB07822, QPB07823, QPB07824, QPB07825, QPB07826, QPB07827, QPB07828, QPB07829, QPB07830, QPB07831, QPB07832, QPB07833, QPB07834, QPB07835, QPB07836, QPB07837) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn heatmap.colours <- c("darkred", "darkblue", "darkgreen", "grey") names(heatmap.colours) <- c("MT880870","MT880871","MT880872","-") #heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "gold", "green","orange","pink","purple","magenta","brown", "darksalmon","chocolate4","darkkhaki", "azure3", "maroon","lightgreen", "blue","cyan", "skyblue2", "blueviolet", "darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("2","5","7","9","14", "17","23", "35","59","73", "81","86","87","89","130","190","290", "297","325", "454","487","558","766", "MT880870","MT880871","MT880872","-") #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=ST)) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree.png", width=1260, height=1260) #svg("ggtree.svg", width=1260, height=1260) p dev.off() #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) png("ggtree_and_gheatmap_mibi_phages.png", width=1290, height=1000) #svg("ggtree_and_gheatmap_mibi_phages.svg", width=17, height=15) gheatmap(p, heatmapData2, width=0.5, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=0, offset = 6) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() # -- for the figure ggtree_and_gheatmap_mibi_selected_genes.png -- info <- read.csv("ggtree_and_gheatmap_mibi.csv", sep="\t") info$name <- info$Isolate tree <- read.tree("990_backup.tree") cols <- c("2"="cornflowerblue","5"="darkgreen","7"="seagreen3","9"="tan","14"="red", "17"="navyblue", "23"="gold", "35"="green","59"="orange","73"="pink","81"="purple","86"="magenta","87"="brown", "89"="darksalmon","130"="chocolate4","190"="darkkhaki", "290"="azure3", "297"="maroon","325"="lightgreen", "454"="blue","487"="cyan", "558"="skyblue2", "766"="blueviolet") heatmapData2 <- info %>% select(Isolate, SCCmec, agr.typing, gyrB, fumC, gltA, icd, apsS, sigB, sarA, agrC, yycG, psm.β, psm.δ, hlb, atlE, atl, sdrG, sdrH, ebh, ebpS, tagB, capC, sepA, dltA, fmtC, lipA, sceD, SE0760, esp, ecpA) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn #heatmap.colours <- c("darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("MT880870","MT880871","MT880872","-") #heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "gold", "green","orange","pink","purple","magenta","brown", "darksalmon","chocolate4","darkkhaki", "azure3", "maroon","lightgreen", "blue","cyan", "skyblue2", "blueviolet", "darkred", "darkblue", "darkgreen", "grey") #names(heatmap.colours) <- c("2","5","7","9","14", "17","23", "35","59","73", "81","86","87","89","130","190","290", "297","325", "454","487","558","766", "MT880870","MT880871","MT880872","-") heatmap.colours <- c("cornflowerblue","darkgreen","seagreen3","tan","red", "navyblue", "purple", "green","cyan", "darkred", "darkblue", "darkgreen", "grey", "darkgreen", "grey") names(heatmap.colours) <- c("SCCmec_type_II(2A)", "SCCmec_type_III(3A)", "SCCmec_type_III(3A) and SCCmec_type_VIII(4A)", "SCCmec_type_IV(2B)", "SCCmec_type_IV(2B&5)", "SCCmec_type_IV(2B) and SCCmec_type_VI(4B)", "SCCmec_type_IVa(2B)", "SCCmec_type_IVb(2B)", "SCCmec_type_IVg(2B)", "I", "II", "III", "none", "+","-") #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=ST)) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree.png", width=1260, height=1260) #svg("ggtree.svg", width=1260, height=1260) p dev.off() #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) png("ggtree_and_gheatmap_mibi_selected_genes.png", width=1590, height=1300) #svg("ggtree_and_gheatmap_mibi_selected_genes.svg", width=17, height=15) gheatmap(p, heatmapData2, width=2, colnames_position="top", colnames_angle=90, colnames_offset_y = 2.0, hjust=0.7, font.size=4, offset = 8) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() -
Report
I’ve attached a figure (ggtree_and_gheatmap_mibi_selected_genes.png) that illustrates the presence and absence of the selected genes. This is a visual representation of the table I sent to you earlier. The presence or absence of each gene in the corresponding genomes was determined using a BLASTn comparison between the genome and the gene sequences. Additionally, I’ve updated the figure ggtree_and_gheatmap_mibi_phages.png. In this new version, all ST types are represented in distinct colors. The raw data for both figures can be found in the attached Excel file (ggtree_and_gheatmap_mibi.xlsx).
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Processing Data_Tam_RNAseq_2024_MHB_vs_Urine_ATCC19606
- Workflow using PICRUSt2 for Data_Karoline_16S_2025
- Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences
- 阳光房漏水怎么办?丁基胶带才是最佳密封选择
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing Data_Tam_RNAseq_2024_MHB_vs_Urine_ATCC19606
Workflow using PICRUSt2 for Data_Karoline_16S_2025
Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences