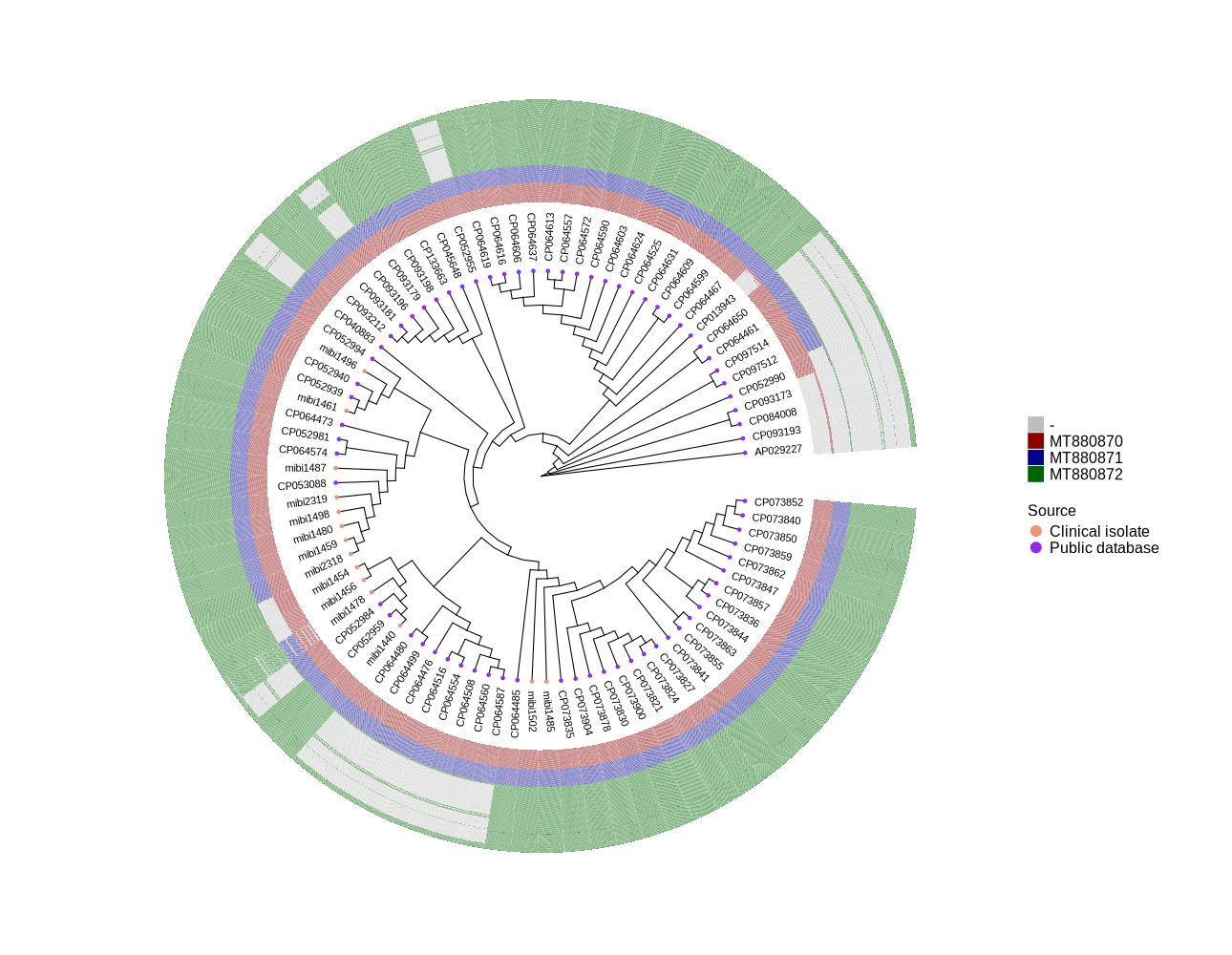
Identify all occurrences of Phages MT880870, MT880871 and MT880872 in S. epidermidis ST2 genomes from public and clinical isolates
gene_x 0 like s 518 view s
Tags: plot, pipeline
-
prepare typing_ST2_until_QPB07622.csv under ~/DATA/Data_Luise_Sepi_STKN/presence_absence_ST2
#grep ">" MT880870.fasta | cut -d' ' -f1 > headers.txt # -- under presence_absence_ST2 -- samtools faidx MT880870.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07572.1_1" > QPB07572.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07573.1_2" > QPB07573.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07574.1_3" > QPB07574.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07575.1_4" > QPB07575.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07576.1_5" > QPB07576.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07577.1_6" > QPB07577.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07578.1_7" > QPB07578.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07579.1_8" > QPB07579.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07580.1_9" > QPB07580.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07581.1_10" > QPB07581.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07582.1_11" > QPB07582.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07583.1_12" > QPB07583.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07584.1_13" > QPB07584.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07585.1_14" > QPB07585.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07586.1_15" > QPB07586.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07587.1_16" > QPB07587.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07588.1_17" > QPB07588.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07589.1_18" > QPB07589.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07590.1_19" > QPB07590.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07591.1_20" > QPB07591.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07592.1_21" > QPB07592.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07593.1_22" > QPB07593.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07594.1_23" > QPB07594.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07595.1_24" > QPB07595.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07596.1_25" > QPB07596.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07597.1_26" > QPB07597.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07598.1_27" > QPB07598.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07599.1_28" > QPB07599.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07600.1_29" > QPB07600.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07601.1_30" > QPB07601.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07602.1_31" > QPB07602.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07603.1_32" > QPB07603.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07604.1_33" > QPB07604.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07605.1_34" > QPB07605.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07606.1_35" > QPB07606.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07607.1_36" > QPB07607.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07608.1_37" > QPB07608.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07609.1_38" > QPB07609.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07610.1_39" > QPB07610.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07611.1_40" > QPB07611.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07612.1_41" > QPB07612.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07613.1_42" > QPB07613.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07614.1_43" > QPB07614.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07615.1_44" > QPB07615.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07616.1_45" > QPB07616.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07617.1_46" > QPB07617.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07618.1_47" > QPB07618.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07619.1_48" > QPB07619.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07620.1_49" > QPB07620.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07621.1_50" > QPB07621.fasta samtools faidx MT880870.fasta "lcl|MT880870.1_cds_QPB07622.1_51" > QPB07622.fasta for i in {572..622}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" #under plotTreeHeatmap_ST2 for i in {572..622}; do echo "python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/${id} /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/typing_ST2_until_${id_1}.csv /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/typing_ST2_until_${id}.csv" done #reprace all '+' with 'MT880870' in typing_ST2_until_QPB07622.csv cp ../presence_absence_ST2/typing_ST2_until_QPB07622.csv ./ sed -i 's/+/MT880870/g' typing_ST2_until_QPB07622.csv -
prepare typing_ST2_until_QPB07667.csv under ~/DATA/Data_Luise_Sepi_STKN/presence_absence_ST2
# -- under presence_absence_ST2 -- samtools faidx MT880871.fasta #grep ">" MT880871.fasta | cut -d' ' -f1 > headers.txt samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07623.1_1" > QPB07623.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07624.1_2" > QPB07624.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07625.1_3" > QPB07625.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07626.1_4" > QPB07626.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07627.1_5" > QPB07627.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07628.1_6" > QPB07628.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07629.1_7" > QPB07629.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07630.1_8" > QPB07630.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07631.1_9" > QPB07631.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07632.1_10" > QPB07632.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07633.1_11" > QPB07633.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07634.1_12" > QPB07634.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07635.1_13" > QPB07635.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07636.1_14" > QPB07636.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07637.1_15" > QPB07637.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07638.1_16" > QPB07638.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07639.1_17" > QPB07639.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07640.1_18" > QPB07640.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07641.1_19" > QPB07641.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07642.1_20" > QPB07642.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07643.1_21" > QPB07643.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07644.1_22" > QPB07644.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07645.1_23" > QPB07645.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07646.1_24" > QPB07646.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07647.1_25" > QPB07647.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07648.1_26" > QPB07648.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07649.1_27" > QPB07649.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07650.1_28" > QPB07650.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07651.1_29" > QPB07651.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07652.1_30" > QPB07652.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07653.1_31" > QPB07653.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07654.1_32" > QPB07654.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07655.1_33" > QPB07655.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07656.1_34" > QPB07656.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07657.1_35" > QPB07657.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07658.1_36" > QPB07658.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07659.1_37" > QPB07659.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07660.1_38" > QPB07660.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07661.1_39" > QPB07661.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07662.1_40" > QPB07662.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07663.1_41" > QPB07663.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07664.1_42" > QPB07664.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07665.1_43" > QPB07665.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07666.1_44" > QPB07666.fasta samtools faidx MT880871.fasta "lcl|MT880871.1_cds_QPB07667.1_45" > QPB07667.fasta for i in {623..667}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {660..660}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done # -- under plotTreeHeatmap_ST2 -- for i in {623..667}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/${id} /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/typing_ST2_until_${id_1}.csv /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/typing_ST2_until_${id}.csv" done sed -i 's/+/MT880871/g' typing_ST2_until_QPB07667.csv -
prepare typing_ST2_until_QPB07667.csv under ~/DATA/Data_Luise_Sepi_STKN/presence_absence_ST2
# -- under presence_absence_ST2 -- samtools faidx MT880872.fasta #grep ">" MT880872.fasta | cut -d' ' -f1 > headers.txt samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07668.1_1" > QPB07668.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07669.1_2" > QPB07669.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07670.1_3" > QPB07670.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07671.1_4" > QPB07671.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07672.1_5" > QPB07672.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07673.1_6" > QPB07673.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07674.1_7" > QPB07674.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07675.1_8" > QPB07675.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07676.1_9" > QPB07676.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07677.1_10" > QPB07677.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07678.1_11" > QPB07678.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07679.1_12" > QPB07679.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07680.1_13" > QPB07680.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07681.1_14" > QPB07681.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07682.1_15" > QPB07682.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07683.1_16" > QPB07683.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07684.1_17" > QPB07684.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07685.1_18" > QPB07685.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07686.1_19" > QPB07686.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07687.1_20" > QPB07687.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07688.1_21" > QPB07688.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07689.1_22" > QPB07689.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07690.1_23" > QPB07690.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07691.1_24" > QPB07691.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07692.1_25" > QPB07692.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07693.1_26" > QPB07693.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07694.1_27" > QPB07694.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07695.1_28" > QPB07695.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07696.1_29" > QPB07696.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07697.1_30" > QPB07697.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07698.1_31" > QPB07698.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07699.1_32" > QPB07699.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07700.1_33" > QPB07700.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07701.1_34" > QPB07701.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07702.1_35" > QPB07702.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07703.1_36" > QPB07703.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07704.1_37" > QPB07704.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07705.1_38" > QPB07705.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07706.1_39" > QPB07706.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07707.1_40" > QPB07707.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07708.1_41" > QPB07708.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07709.1_42" > QPB07709.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07710.1_43" > QPB07710.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07711.1_44" > QPB07711.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07712.1_45" > QPB07712.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07713.1_46" > QPB07713.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07714.1_47" > QPB07714.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07715.1_48" > QPB07715.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07716.1_49" > QPB07716.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07717.1_50" > QPB07717.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07718.1_51" > QPB07718.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07719.1_52" > QPB07719.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07720.1_53" > QPB07720.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07721.1_54" > QPB07721.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07722.1_55" > QPB07722.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07723.1_56" > QPB07723.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07724.1_57" > QPB07724.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07725.1_58" > QPB07725.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07726.1_59" > QPB07726.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07727.1_60" > QPB07727.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07728.1_61" > QPB07728.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07729.1_62" > QPB07729.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07730.1_63" > QPB07730.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07731.1_64" > QPB07731.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07732.1_65" > QPB07732.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07733.1_66" > QPB07733.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07734.1_67" > QPB07734.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07735.1_68" > QPB07735.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07736.1_69" > QPB07736.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07737.1_70" > QPB07737.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07738.1_71" > QPB07738.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07739.1_72" > QPB07739.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07740.1_73" > QPB07740.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07741.1_74" > QPB07741.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07742.1_75" > QPB07742.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07743.1_76" > QPB07743.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07744.1_77" > QPB07744.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07745.1_78" > QPB07745.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07746.1_79" > QPB07746.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07747.1_80" > QPB07747.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07748.1_81" > QPB07748.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07749.1_82" > QPB07749.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07750.1_83" > QPB07750.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07751.1_84" > QPB07751.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07752.1_85" > QPB07752.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07753.1_86" > QPB07753.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07754.1_87" > QPB07754.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07755.1_88" > QPB07755.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07756.1_89" > QPB07756.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07757.1_90" > QPB07757.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07758.1_91" > QPB07758.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07759.1_92" > QPB07759.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07760.1_93" > QPB07760.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07761.1_94" > QPB07761.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07762.1_95" > QPB07762.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07763.1_96" > QPB07763.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07764.1_97" > QPB07764.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07765.1_98" > QPB07765.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07766.1_99" > QPB07766.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07767.1_100" > QPB07767.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07768.1_101" > QPB07768.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07769.1_102" > QPB07769.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07770.1_103" > QPB07770.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07771.1_104" > QPB07771.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07772.1_105" > QPB07772.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07773.1_106" > QPB07773.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07774.1_107" > QPB07774.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07775.1_108" > QPB07775.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07776.1_109" > QPB07776.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07777.1_110" > QPB07777.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07778.1_111" > QPB07778.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07779.1_112" > QPB07779.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07780.1_113" > QPB07780.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07781.1_114" > QPB07781.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07782.1_115" > QPB07782.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07783.1_116" > QPB07783.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07784.1_117" > QPB07784.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07785.1_118" > QPB07785.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07786.1_119" > QPB07786.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07787.1_120" > QPB07787.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07788.1_121" > QPB07788.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07789.1_122" > QPB07789.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07790.1_123" > QPB07790.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07791.1_124" > QPB07791.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07792.1_125" > QPB07792.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07793.1_126" > QPB07793.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07794.1_127" > QPB07794.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07795.1_128" > QPB07795.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07796.1_129" > QPB07796.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07797.1_130" > QPB07797.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07798.1_131" > QPB07798.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07799.1_132" > QPB07799.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07800.1_133" > QPB07800.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07801.1_134" > QPB07801.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07802.1_135" > QPB07802.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07803.1_136" > QPB07803.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07804.1_137" > QPB07804.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07805.1_138" > QPB07805.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07806.1_139" > QPB07806.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07807.1_140" > QPB07807.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07808.1_141" > QPB07808.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07809.1_142" > QPB07809.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07810.1_143" > QPB07810.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07811.1_144" > QPB07811.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07812.1_145" > QPB07812.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07813.1_146" > QPB07813.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07814.1_147" > QPB07814.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07815.1_148" > QPB07815.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07816.1_149" > QPB07816.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07817.1_150" > QPB07817.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07818.1_151" > QPB07818.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07819.1_152" > QPB07819.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07820.1_153" > QPB07820.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07821.1_154" > QPB07821.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07822.1_155" > QPB07822.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07823.1_156" > QPB07823.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07824.1_157" > QPB07824.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07825.1_158" > QPB07825.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07826.1_159" > QPB07826.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07827.1_160" > QPB07827.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07828.1_161" > QPB07828.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07829.1_162" > QPB07829.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07830.1_163" > QPB07830.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07831.1_164" > QPB07831.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07832.1_165" > QPB07832.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07833.1_166" > QPB07833.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07834.1_167" > QPB07834.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07835.1_168" > QPB07835.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07836.1_169" > QPB07836.fasta samtools faidx MT880872.fasta "lcl|MT880872.1_cds_QPB07837.1_170" > QPB07837.fasta for i in {668..750}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {751..837}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done for i in {708..708}; do id=$(printf "QPB07%03d" "$i") echo "mkdir ${id}" echo "for sample in mibi1440 mibi1454 mibi1456 mibi1459 mibi1461 mibi1478 mibi1480 mibi1485 mibi1487 mibi1496 mibi1498 mibi1502 mibi2318 mibi2319 AP029227 CP013943 CP040883 CP045648 CP052939 CP052940 CP052955 CP052959 CP052981 CP052984 CP052990 CP052994 CP053088 CP064461 CP064467 CP064473 CP064476 CP064480 CP064485 CP064499 CP064508 CP064516 CP064525 CP064554 CP064557 CP064560 CP064572 CP064574 CP064587 CP064590 CP064599 CP064603 CP064606 CP064609 CP064613 CP064616 CP064619 CP064624 CP064631 CP064637 CP064650 CP073821 CP073824 CP073827 CP073830 CP073835 CP073836 CP073840 CP073841 CP073844 CP073847 CP073850 CP073852 CP073855 CP073857 CP073859 CP073862 CP073863 CP073878 CP073900 CP073904 CP084008 CP093173 CP093179 CP093181 CP093193 CP093196 CP093198 CP093212 CP097512 CP097514 CP120425 CP133663; do" echo "blastn -db \${sample}.fasta -query ${id}.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ./${id}/\${sample}.blastn" echo "done" done # -- under plotTreeHeatmap_ST2 -- for i in {668..837}; do id_1=$(printf "QPB07%03d" "$((i-1))") id=$(printf "QPB07%03d" "$i") echo "python ~/Scripts/process_directory.py /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/presence_absence_ST2/${id} /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/typing_ST2_until_${id_1}.csv /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/typing_ST2_until_${id}.csv" done sed -i 's/+/MT880872/g' typing_ST2_until_QPB07837.csv -
plot tree heatmap under /mnt/md1/DATA_md1/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2_MT880871
library(ggtree) library(ggplot2) library(dplyr) setwd("/home/jhuang/DATA/Data_Luise_Sepi_STKN/plotTreeHeatmap_ST2/") # -- edit tree -- #https://icytree.org/ #0.000780 info <- read.csv("typing_ST2_until_QPB07837.csv", sep="\t") info$name <- info$Isolate tree <- read.tree("core_gene_alignment.aln.treefile") cols <- c("Public database"='purple2', "Clinical isolate"='darksalmon') #purple2 skyblue2 heatmapData2 <- info %>% select(Isolate, QPB07572, QPB07573, QPB07574, QPB07575, QPB07576, QPB07577, QPB07578, QPB07579, QPB07580, QPB07581, QPB07582, QPB07583, QPB07584, QPB07585, QPB07586, QPB07587, QPB07588, QPB07589, QPB07590, QPB07591, QPB07592, QPB07593, QPB07594, QPB07595, QPB07596, QPB07597, QPB07598, QPB07599, QPB07600, QPB07601, QPB07602, QPB07603, QPB07604, QPB07605, QPB07606, QPB07607, QPB07608, QPB07609, QPB07610, QPB07611, QPB07612, QPB07613, QPB07614, QPB07615, QPB07616, QPB07617, QPB07618, QPB07619, QPB07620, QPB07621, QPB07622, QPB07623, QPB07624, QPB07625, QPB07626, QPB07627, QPB07628, QPB07629, QPB07630, QPB07631, QPB07632, QPB07633, QPB07634, QPB07635, QPB07636, QPB07637, QPB07638, QPB07639, QPB07640, QPB07641, QPB07642, QPB07643, QPB07644, QPB07645, QPB07646, QPB07647, QPB07648, QPB07649, QPB07650, QPB07651, QPB07652, QPB07653, QPB07654, QPB07655, QPB07656, QPB07657, QPB07658, QPB07659, QPB07660, QPB07661, QPB07662, QPB07663, QPB07664, QPB07665, QPB07666, QPB07667, QPB07668, QPB07669, QPB07670, QPB07671, QPB07672, QPB07673, QPB07674, QPB07675, QPB07676, QPB07677, QPB07678, QPB07679, QPB07680, QPB07681, QPB07682, QPB07683, QPB07684, QPB07685, QPB07686, QPB07687, QPB07688, QPB07689, QPB07690, QPB07691, QPB07692, QPB07693, QPB07694, QPB07695, QPB07696, QPB07697, QPB07698, QPB07699, QPB07700, QPB07701, QPB07702, QPB07703, QPB07704, QPB07705, QPB07706, QPB07707, QPB07708, QPB07709, QPB07710, QPB07711, QPB07712, QPB07713, QPB07714, QPB07715, QPB07716, QPB07717, QPB07718, QPB07719, QPB07720, QPB07721, QPB07722, QPB07723, QPB07724, QPB07725, QPB07726, QPB07727, QPB07728, QPB07729, QPB07730, QPB07731, QPB07732, QPB07733, QPB07734, QPB07735, QPB07736, QPB07737, QPB07738, QPB07739, QPB07740, QPB07741, QPB07742, QPB07743, QPB07744, QPB07745, QPB07746, QPB07747, QPB07748, QPB07749, QPB07750, QPB07751, QPB07752, QPB07753, QPB07754, QPB07755, QPB07756, QPB07757, QPB07758, QPB07759, QPB07760, QPB07761, QPB07762, QPB07763, QPB07764, QPB07765, QPB07766, QPB07767, QPB07768, QPB07769, QPB07770, QPB07771, QPB07772, QPB07773, QPB07774, QPB07775, QPB07776, QPB07777, QPB07778, QPB07779, QPB07780, QPB07781, QPB07782, QPB07783, QPB07784, QPB07785, QPB07786, QPB07787, QPB07788, QPB07789, QPB07790, QPB07791, QPB07792, QPB07793, QPB07794, QPB07795, QPB07796, QPB07797, QPB07798, QPB07799, QPB07800, QPB07801, QPB07802, QPB07803, QPB07804, QPB07805, QPB07806, QPB07807, QPB07808, QPB07809, QPB07810, QPB07811, QPB07812, QPB07813, QPB07814, QPB07815, QPB07816, QPB07817, QPB07818, QPB07819, QPB07820, QPB07821, QPB07822, QPB07823, QPB07824, QPB07825, QPB07826, QPB07827, QPB07828, QPB07829, QPB07830, QPB07831, QPB07832, QPB07833, QPB07834, QPB07835, QPB07836, QPB07837) #ST, rn <- heatmapData2$Isolate heatmapData2$Isolate <- NULL heatmapData2 <- as.data.frame(sapply(heatmapData2, as.character)) rownames(heatmapData2) <- rn heatmap.colours <- c("darkred", "darkblue", "darkgreen", "grey") names(heatmap.colours) <- c("MT880870","MT880871","MT880872","-") #mydat$Regulation <- factor(mydat$Regulation, levels=c("up","down")) #circular p <- ggtree(tree, layout='circular', branch.length='none') %<+% info + geom_tippoint(aes(color=Source)) + scale_color_manual(values=cols) + geom_tiplab2(aes(label=name), offset=1) png("ggtree.png", width=1260, height=1260) #svg("ggtree.svg", width=1260, height=1260) p dev.off() #png("ggtree_and_gheatmap.png", width=1290, height=1000) #svg("ggtree_and_gheatmap.svg", width=17, height=15) #gheatmap(p, heatmapData2, width=0.1, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=4, offset = 5) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 14)) + theme(legend.title = element_text(size = 14)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) png("ggtree_and_gheatmap.png", width=1290, height=1000) gheatmap(p, heatmapData2, width=0.5, colnames_position="top", colnames_angle=90, colnames_offset_y = 0.1, hjust=0.5, font.size=0, offset = 8) + scale_fill_manual(values=heatmap.colours) + theme(legend.text = element_text(size = 16)) + theme(legend.title = element_text(size = 16)) + guides(fill=guide_legend(title=""), color = guide_legend(override.aes = list(size = 5))) dev.off() -
Report
Explanation for the discrepancy between the last analysis and this one: Note that many partial genes of the phages do not match because, in the previous analysis, only the longest BLASTn hits were considered and recorded in the table.
We have a total of 14 ST2 genomes from Luise's project, and I found 73 ST2 genomes from NCBI.
I examined the presence and absence of all 51 + 45 + 170 genes (details below) across the 87 genomes and visualized the results in a heatmap (attached as ggtree_and_gheatmap_phages.png).
- MT880870: 51 putative genes
- MT880871: 45 putative genes
- MT880872: 170 putative genes
In the heatmap:
Genes not present in the corresponding isolate are shown in grey. Genes present are highlighted in the corresponding colors:
- Dark red for MT880870
- Dark blue for MT880871
- Dark green for MT880872
The graphic is based on the Excel file typing_ST2.xlsx. If you have any suggestions or improvements, please let me know.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
- MCV病毒中的LT与sT蛋白功能
- Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Enhanced Visualization of Gene Presence for the Selected Genes in Bongarts_S.epidermidis_HDRNA