Virus Genome Analysis Pipeline: Hybrid Capture, DAMIAN Blastn, and VRAP Mapping for Measles (麻疹) Sample
gene_x 0 like s 575 view s
Tags: pipeline
-
DAMIAN calculations
cd /mnt/nvme0n1p1/blast # -- Measles -- #I think it would be good to first analyse the sample in DAMIAN Blastn and then map it to the closest related genome resulting from DAMIAN to see if we cover the entire sequence. damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_measles/p20578/N24_539_1A_measles_S1_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_measles/p20578/N24_539_1A_measles_S1_R2_001.fastq.gz --sample N24_539_1A_measles_S1_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 40 --force damian_report.rb #Please find enclosed two links to further samples that require analysis with DAMIAN. In each case, the host is a human. The samples comprise three liver/serum samples (upper link below) and 12 samples from the ring trial (lower link below). #This is merely an initial evaluation of samples with DAMIAN, using Blastn. # -- Leber/Serum -- #Bei den Lexogen CORALL Libraries muss bei der Analyse darauf geachtet werden, dass diese in Read 1 standardmäßig einen UMI (12 nt) haben und die ersten 9 nt von Read 2 haben eine erhöhte Fehlerrate beim mapping (durch das Priming-Prinzip laut Hersteller) – die könnte man vor der weiteren Analyse trimmen. #cutadapt -u 12 -U 9 -o output_R1_trimmed.fastq -p output_R2_trimmed.fastq input_R1.fastq input_R2.fastq # #Explanation: # # -u 12: Trim 12 bases from the start of Read 1 (this is for the UMI). # -U 9: Trim 9 bases from the start of Read 2 (this is to deal with the higher error rate in the first 9 bases of Read 2). # -o output_R1_trimmed.fastq: Output file for Read 1 after trimming. # -p output_R2_trimmed.fastq: Output file for Read 2 after trimming. # #Why this works: # # The -u option applies trimming to Read 1, while the -U option applies trimming to Read 2. This ensures that the correct bases are trimmed from each read, without affecting the other. # #Now when you run this command, Read 1 will have 12 bases trimmed, and Read 2 will have 9 bases trimmed, as expected. cutadapt -u 12 -U -9 -o Leber1_Corall_wo_Dnase_S5_R1_001.fastq -p Leber1_Corall_wo_Dnase_S5_R2_001.fastq p20575_L3/Leber1_Corall_wo_Dnase_S5_R1_001.fastq.gz p20575_L3/Leber1_Corall_wo_Dnase_S5_R2_001.fastq.gz cutadapt -u 12 -U -9 -o Leber2_Corall_wo_Dnase_S6_R1_001.fastq -p Leber2_Corall_wo_Dnase_S6_R2_001.fastq ./p20576_L3/Leber2_Corall_wo_Dnase_S6_R1_001.fastq.gz ./p20576_L3/Leber2_Corall_wo_Dnase_S6_R2_001.fastq.gz cutadapt -u 12 -U -9 -o Serum_Corall_wo_Dnase_S7_R1_001.fastq -p Serum_Corall_wo_Dnase_S7_R2_001.fastq ./p20577_L3/Serum_Corall_wo_Dnase_S7_R1_001.fastq.gz ./p20577_L3/Serum_Corall_wo_Dnase_S7_R2_001.fastq.gz for sample in Leber1_Corall_wo_Dnase_S5 Leber2_Corall_wo_Dnase_S6 Serum_Corall_wo_Dnase_S7; do java -jar /home/jhuang/Tools/Trimmomatic-0.36/trimmomatic-0.36.jar PE -threads 16 ${sample}_R1_001.fastq ${sample}_R2_001.fastq trimmed/${sample}_R1.fastq.gz trimmed/${sample}_unpaired_R1.fastq.gz trimmed/${sample}_R2.fastq.gz trimmed/${sample}_unpaired_R2.fastq.gz ILLUMINACLIP:/home/jhuang/Tools/Trimmomatic-0.36/adapters/TruSeq3-PE-2.fa:2:30:10:8:TRUE LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 AVGQUAL:20; done fastqc ./trimmed/Leber1_Corall_wo_Dnase_S5_R1.fastq.gz fastqc ./trimmed/Leber1_Corall_wo_Dnase_S5_R2.fastq.gz damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Liver/trimmed/Leber1_Corall_wo_Dnase_S5_R1.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Liver/trimmed/Leber1_Corall_wo_Dnase_S5_R2.fastq.gz --sample Leber1_Corall_wo_Dnase_S5_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 60 --force damian_report.rb -a 37 -n Leber1_Corall_wo_Dnase_S5_blastn damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Liver/trimmed/Leber2_Corall_wo_Dnase_S6_R1.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Liver/trimmed/Leber2_Corall_wo_Dnase_S6_R2.fastq.gz --sample Leber2_Corall_wo_Dnase_S6_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 60 --force damian_report.rb -a 39 -n Leber2_Corall_wo_Dnase_S6_blastn damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Liver/trimmed/Serum_Corall_wo_Dnase_S7_R1.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Liver/trimmed/Serum_Corall_wo_Dnase_S7_R2.fastq.gz --sample Serum_Corall_wo_Dnase_S7_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 60 --force damian_report.rb -a 41 -n Serum_Corall_wo_Dnase_S7_blastn zip -r Serum_Corall_wo_Dnase_S7_blastn.zip Serum_Corall_wo_Dnase_S7_blastn # -- Ringversuch -- #Hier waren die DNA-Libraries der Samples ungewöhnlich niedrig konzentriert (das Kit ist normalerweise sehr robust für niedrigen Input, von ChIP-Proben auch z.B.) und die Read-Anzahl ist auch niedriger als angepeilt. Vielleicht klärt die bioinformatische Analyse das ja etwas auf – bzw. wie viel DNA würdet ihr aus dem Ausgangsmaterial erwarten (waren es Zell-Spikeins in der RNA?).. Die RNA-Libraries aus den gleichen Proben sehen jeweils ok aus, Readanzahl wurde in etwa erreicht, die Duplikationsrate ist relativ hoch, was bei low input aber auch normal sein kann. damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20585/07_RV1_RNA_S7_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20585/07_RV1_RNA_S7_R2_001.fastq.gz --sample 07_RV1_RNA_S7_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 100 --force damian_report.rb damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20586/08_RV2_RNA_S8_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20586/08_RV2_RNA_S8_R2_001.fastq.gz --sample 08_RV2_RNA_S8_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 100 --force damian_report.rb damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20587/09_RV3_RNA_S9_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20587/09_RV3_RNA_S9_R2_001.fastq.gz --sample 09_RV3_RNA_S9_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 100 --force damian_report.rb damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20588/10_RV4_RNA_S10_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20588/10_RV4_RNA_S10_R2_001.fastq.gz --sample 10_RV4_RNA_S10_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 100 --force damian_report.rb damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20589/11_RV5_RNA_S11_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20589/11_RV5_RNA_S11_R2_001.fastq.gz --sample 11_RV5_RNA_S11_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 100 --force damian_report.rb damian.rb --host human3 --type rna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20590/12_RV6_RNA_S12_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20590/12_RV6_RNA_S12_R2_001.fastq.gz --sample 12_RV6_RNA_S12_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 100 --force damian_report.rb #END cd jhuang@hamm:/mnt/h1/jhuang/blast damian.rb --host human3 --type dna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20579/01_RV1_DNA_S1_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20579/01_RV1_DNA_S1_R2_001.fastq.gz --sample 01_RV1_DNA_S1_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 55 --force damian_report.rb damian.rb --host human3 --type dna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20580/02_RV2_DNA_S2_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20580/02_RV2_DNA_S2_R2_001.fastq.gz --sample 02_RV2_DNA_S2_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 55 --force damian_report.rb damian.rb --host human3 --type dna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20581/03_RV3_DNA_S3_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20581/03_RV3_DNA_S3_R2_001.fastq.gz --sample 03_RV3_DNA_S3_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 55 --force damian_report.rb damian.rb --host human3 --type dna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20582/04_RV4_DNA_S4_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20582/04_RV4_DNA_S4_R2_001.fastq.gz --sample 04_RV4_DNA_S4_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 55 --force damian_report.rb damian.rb --host human3 --type dna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20583/05_RV5_DNA_S5_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20583/05_RV5_DNA_S5_R2_001.fastq.gz --sample 05_RV5_DNA_S5_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 55 --force damian_report.rb damian.rb --host human3 --type dna -1 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20584/06_RV6_DNA_S6_R1_001.fastq.gz -2 /home/jhuang/DATA/Data_Damian/241213_VH00358_120_AAG523FM5_Ringversuch/p20584/06_RV6_DNA_S6_R2_001.fastq.gz --sample 06_RV6_DNA_S6_blastn --blastn progressive --blastp never --min_contiglength 500 --threads 55 --force damian_report.rb #END #Sent: DNA_S1, S2, S3, RNA_S7, S8, S9, S10. #TODOs: DNA_S4, S5, S6, RNA_S11, S12. -
Trimming
mkdir trimmed for sample in N24_539_1A_measles_S1; do java -jar /home/jhuang/Tools/Trimmomatic-0.36/trimmomatic-0.36.jar PE -threads 16 ${sample}_R1_001.fastq.gz ${sample}_R2_001.fastq.gz trimmed/${sample}_R1.fastq.gz trimmed/${sample}_unpaired_R1.fastq.gz trimmed/${sample}_R2.fastq.gz trimmed/${sample}_unpaired_R2.fastq.gz ILLUMINACLIP:/home/jhuang/Tools/Trimmomatic-0.36/adapters/TruSeq3-PE-2.fa:2:30:10:8:TRUE LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 AVGQUAL:20; done -
Download all virus genomes (18644)
mv datasets /usr/local/bin/ chmod +x /usr/local/bin/datasets #datasets download virus genome --complete-only --assembly-source refseq under ~/REFs datasets download virus genome taxon "Viruses" --complete-only --refseq #To check for RefSeq data only, look for NC_, NM_, or similar prefixes in sequence headers and identifiers. wget -r -np -nH --cut-dirs=3 ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses/ [Option1] sudo apt install ncbi-entrez-direct [Option1] esearch -db nucleotide -query "txid3052345[Organism:exp]" | efetch -format fasta > virus_genome_3052345.fasta # 23788 ~/Scripts/filter_fasta.py virus_genome_3052345.fasta virus_complete_genome_3052345.fasta import sys from Bio import SeqIO # Get input and output file paths from command line arguments input_file = sys.argv[1] output_file = sys.argv[2] # Open the output file to write filtered sequences with open(output_file, "w") as out_handle: # Parse the multi-line FASTA file for record in SeqIO.parse(input_file, "fasta"): # Check if 'complete genome' is in the header (description) if "complete genome" in record.description: # Write the entire record (header + multi-line sequence) to the output file SeqIO.write(record, out_handle, "fasta") print(f"Complete genome sequences saved to {output_file}") [Option1] grep "complete genome" virus_complete_genome_3052345.fasta | wc -l #917 [Option2] https://www.ebi.ac.uk/ena/browser/view/3052345 [Option2] grep "complete genome" ena_3052345_sequence.fasta | wc -l #820 -
The commends for more comprehensive blast annotation, in order to get the closest related genome.
ln -s ~/Tools/vrap/ . conda activate /home/jhuang/miniconda3/envs/vrap vrap/vrap.py -1 trimmed/N24_539_1A_measles_S1_R1.fastq.gz -2 trimmed/N24_539_1A_measles_S1_R2.fastq.gz -o N24_539_1A_measles_S1 --bt2idx=/home/jhuang/REFs/genome --host=/home/jhuang/REFs/genome.fa --virus=/home/jhuang/REFs/ncbi_dataset/data/genomic.fna --nt=/mnt/nvme0n1p1/blast/nt --nr=/mnt/nvme0n1p1/blast/nr -t 100 -l 200 -g [Optional] vrap/vrap.py -1 trimmed/N24_539_1A_measles_S1_R1.fastq.gz -2 trimmed/N24_539_1A_measles_S1_R2.fastq.gz -o N24_539_1A_measles_S1_3052345 --bt2idx=/home/jhuang/REFs/genome --host=/home/jhuang/REFs/genome.fa --virus=/home/jhuang/REFs/virus_complete_genome_3052345.fasta --nt=/mnt/nvme0n1p1/blast/nt --nr=/mnt/nvme0n1p1/blast/nr -t 100 -l 200 -g # * 4 nt_dbs (--virus, --host, download_db.py(nucleotide, currently alphaherpesvirus), nt), 2 prot_db (download_db.py(protein, currently alphaherpesvirus), nr) for blast, save under ./blast/db/virus, ./blast/db/host, vrap/database/viral_db/viral_nucleotide, vrap/database/viral_db/viral_protein # * 1 bowtie_database for host removal (--host), save under ./bowtie/host. # * bowtie run before assembly # * blast run after assembly for the contigs, therefore it does not exist the taxfilt step in vrap. # * checking the order of the databases for annotation step, namely which database will be taken firstly for annotionn after setting --virus? # * If --host is for both bowtie and blastn. # * if only --bt2idx define, only bowtie, no blastn! == no ""--host=/home/jhuang/REFs/genome.fa"" still has the host-removal step! -
Using the bowtie of vrap to map the reads on ref_genome/reference.fasta (The reference refers to the closest related genome found from the list generated by vrap)
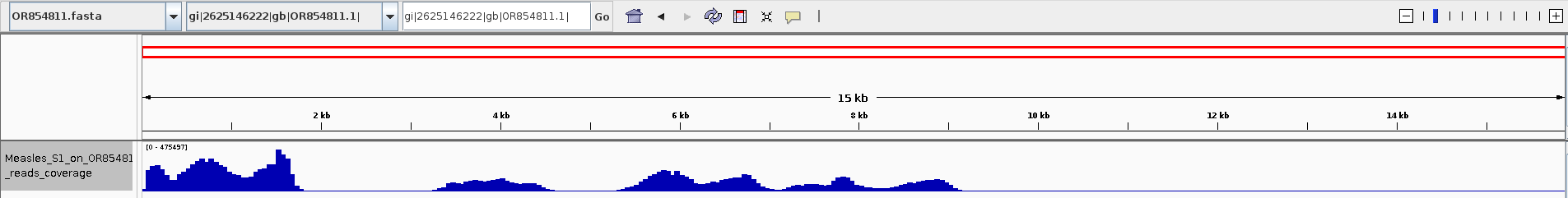
vrap/vrap.py -1 trimmed/N24_539_1A_measles_S1_R1.fastq.gz -2 trimmed/N24_539_1A_measles_S1_R2.fastq.gz -o N24_539_1A_measles_S1_on_OR854811 --host /home/jhuang/Downloads/OR854811.fasta -t 100 -l 200 -g cd bowtie mv mapped mapped.sam samtools view -S -b mapped.sam > mapped.bam samtools sort mapped.bam -o mapped_sorted.bam samtools index mapped_sorted.bam samtools view -H mapped_sorted.bam samtools flagstat mapped_sorted.bam #5755885 + 0 in total (QC-passed reads + QC-failed reads) 0 + 0 secondary 0 + 0 supplementary 0 + 0 duplicates 4457643 + 0 mapped (77.44% : N/A) 1300648 + 0 paired in sequencing 650324 + 0 read1 650324 + 0 read2 637608 + 0 properly paired (49.02% : N/A) 901044 + 0 with itself and mate mapped 77271 + 0 singletons (5.94% : N/A) 0 + 0 with mate mapped to a different chr 0 + 0 with mate mapped to a different chr (mapQ>=5) #4457643 of 5755885 (77.44%) can map the reference. bamCoverage -b mapped_sorted.bam -o ../../Measles_S1_on_OR854811_reads_coverage.bw -
Show the bw on IGV
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
- MCV病毒中的LT与sT蛋白功能
- Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)
- Somatic Variation Detection
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)