How to run PICRUSt2 (v2)?
gene_x 0 like s 849 view s
Tags: processing, tool
https://github.com/picrust/picrust2/wiki/Infer-pathway-abundances
-
Difference between unstratified and stratified
In the context of PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States), unstratified and stratified outputs refer to different ways of presenting the predicted functional profiles of microbial communities. 1. Unstratified Output: Definition: Unstratified output provides the overall predicted abundance of each function (e.g., gene families, metabolic pathways) across the entire microbial community. Characteristics: Summarized Data: It aggregates the functional predictions for all taxa in a sample, giving a single abundance value for each function. No Taxonomic Information: Does not break down the contribution of each specific taxon to the predicted function. It only provides the total abundance of each function without detailing which taxa are contributing to those functions. Use Case: Useful when the overall functional potential of a microbial community is of interest without needing to know the contribution of individual taxa. It simplifies the data and reduces complexity. Example: If you are interested in the total predicted abundance of a specific gene across all microbes in a sample, you would use the unstratified output. 2. Stratified Output: Definition: Stratified output provides the predicted abundance of each function, but also stratifies (or breaks down) this data by the taxonomic origin of the microbes contributing to each function. Characteristics: Detailed Data: It provides more granular information by showing the predicted abundance of each function for each taxon in the community. Taxonomic Breakdown: This output allows you to see how much each taxon (e.g., a specific species or genus) contributes to the predicted abundance of each function. Use Case: Useful for understanding the functional contributions of specific taxa within a microbial community. It provides insight into which organisms are potentially driving certain functions within a community. Example: If you want to know which specific microbes are contributing to the abundance of a certain gene, the stratified output will give you this information by listing the abundance of that gene for each taxon. Key Differences: Level of Detail: Unstratified output provides a high-level summary, whereas stratified output offers a detailed breakdown by taxon. Data Granularity: Stratified output is more granular and complex, while unstratified output is simpler and more straightforward. Purpose: The choice between unstratified and stratified depends on whether you are interested in the total functional potential of the community (unstratified) or in understanding the functional roles of specific taxa (stratified). Summary: Unstratified: Overall predicted functional abundance without taxonomic breakdown. Stratified: Predicted functional abundance with detailed taxonomic breakdown for each function. 在PICRUSt2(通过重建未观测状态进行群落的系统发育调查)的背景下,**未分层(unstratified)和分层(stratified)**输出是指呈现微生物群落的预测功能特征的不同方式。 1. 未分层(Unstratified)输出: 定义:未分层输出提供了整个微生物群落中每个功能(例如,基因家族、代谢途径)的总体预测丰度。 特点: 汇总数据:它汇总了样本中所有分类单元的功能预测,为每个功能提供一个总的丰度值。 无分类信息:不显示每个具体分类单元对预测功能的贡献,仅提供每个功能的总丰度,而不细分哪些分类单元在贡献这些功能。 适用场景:当对微生物群落的总体功能潜力感兴趣,而不需要知道单个分类单元的贡献时,未分层输出是有用的。它简化了数据,减少了复杂性。 示例:如果你感兴趣的是一个样本中所有微生物的特定基因的总预测丰度,你可以使用未分层输出。 2. 分层(Stratified)输出: 定义:分层输出提供了每个功能的预测丰度,同时按贡献这些功能的微生物的分类来源进行了分层。 特点: 详细数据:通过显示群落中每个分类单元的每个功能的预测丰度,提供了更详细的信息。 分类细分:这种输出方式可以让你看到每个分类单元(例如,具体的物种或属)对每个功能的预测丰度的贡献。 适用场景:当需要了解特定分类单元在微生物群落中的功能贡献时,分层输出是有用的。它提供了哪些微生物可能在群落中驱动特定功能的见解。 示例:如果你想知道哪些具体的微生物在贡献某个基因的丰度,分层输出将提供此信息,列出每个分类单元的该基因丰度。 关键区别: 细节层次:未分层输出提供的是一个高级概述,而分层输出则提供按分类单元的详细细分。 数据粒度:分层输出更为细化和复杂,而未分层输出更为简单和直接。 用途:选择未分层还是分层,取决于你是对群落的总体功能潜力感兴趣(未分层),还是希望了解特定分类单元的功能作用(分层)。 总结: 未分层(Unstratified):总体的预测功能丰度,不包含分类细分。 分层(Stratified):包含详细分类细分的预测功能丰度。 For analyzing differential pathways expressed between two sample groups, you should use the unstratified input in PICRUSt2. Reason for Choosing Unstratified Input: Focus on Overall Functional Differences: When comparing the functional profiles of two groups of samples, the primary interest is often in identifying which pathways are differentially abundant overall between the groups, regardless of which specific taxa are contributing to these differences. Unstratified input provides a summary of the total abundance of each function or pathway across the entire microbial community in each sample group, making it easier to compare the overall functional profiles. Simpler and More Direct Comparison: Unstratified data aggregates the functional predictions for all taxa within each sample. This aggregation simplifies the comparison between groups because it provides a single value per function or pathway for each sample, allowing for straightforward statistical testing of differential abundance. Reduces Complexity: Stratified input, which breaks down functional contributions by taxa, adds a layer of complexity that is not necessary for identifying overall differential pathways between groups. The unstratified output eliminates this complexity and focuses purely on the functions themselves, rather than on which specific taxa are contributing to these functions. When to Use Stratified Input: If you are interested in which specific taxa are responsible for the differences in pathway abundances between the two groups, then stratified input would be useful. It allows you to see not only which pathways are differentially expressed but also how the contribution of these pathways varies across different taxa. Summary: For identifying differential pathways expressed between two sample groups, use unstratified input to focus on the overall differences in functional profiles without considering the taxonomic breakdown. Use stratified input if you need to understand the taxonomic origins of these functional differences. -
Pathway inference
Input files: *_metagenome_out/*unstrat.tsv.gz Mapfiles: KEGG_pathways_to_KO.tsv KEGG_modules_to_KO.tsv * ec_level4_to_metacyc_rxn.tsv * metacyc_path2rxn_struc_filt_pro.txt metacyc_path2rxn_struc_filt_euk.txt metacyc_pathways_structured_filtered metacyc_path2rxn_struc_filt_fungi.txt metacyc_path2rxn_struc_filt_fungi_present.txt metacyc_rxn_to_level4ec.tsv Output files: ./MetaCyc_pathways_out/path_abun_unstrat.tsv ./KEGG_pathways_out/path_abun_unstrat.tsv #The default is to map the EC numbers to Metacyc reactions and then to Metacyc Pathways. ERROR: runtime is too long! #pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_contrib.tsv.gz -o pathways_out -p 80 #FILE_GENERATED_FOR_DOWNSTREAM: Map EC numbers to MetaCyc pathways and get stratified output corresponding to contribution of predicted gene family abundances within each predicted genome: pathway_pipeline.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -o MetaCyc_pathways_out_per_seq_contrib -p 80 --per_sequence_contrib --per_sequence_abun EC_metagenome_out/seqtab_norm.tsv.gz --per_sequence_function EC_predicted.tsv.gz ##ERROR: pred_metagenome_strat.tsv.gz does not exist. Mapping predicted KO abundances to legacy KEGG pathways (with stratified output that represents contributions to community-wide abundances): ##Why use '--no_gregroup'? no rows remain after regrouping input table. The default pathway and regroup mapfiles are meant for EC numbers. Note that KEGG pathways are not supported since KEGG is a closed-source database, but you can input custom pathway mapfiles if you have access. If you are using a custom function database did you mean to set the --no-regroup flag and/or change the default pathways mapfile used? #pathway_pipeline.py -i KO_metagenome_out/pred_metagenome_strat.tsv.gz -o KEGG_pathways_out -p 80 --no_regroup --map /home/jhuang/Tools/picrust2/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv #FILE_GENERATED_FOR_DOWNSTREAM pathway_pipeline.py -i KO_metagenome_out/pred_metagenome_unstrat.tsv.gz -o KEGG_pathways_out -p 80 --no_regroup --map /home/jhuang/Tools/picrust2/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv pathway_pipeline.py -i KO_metagenome_out/pred_metagenome_unstrat.tsv.gz -o KEGG_pathways_out_per_seq_contrib -p 80 --per_sequence_contrib --per_sequence_abun KO_metagenome_out/seqtab_norm.tsv.gz --per_sequence_function KO_predicted.tsv.gz --no_regroup --map /home/jhuang/Tools/picrust2/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv #Note that the path of map files is under /home/jhuang/Tools/picrust2/picrust2/default_files/pathway_mapfiles #ERROR: COG does not fit the pathway_mapfiles KEGG_pathways_to_KO.tsv?? #pathway_pipeline.py -i COG_metagenome_out/pred_metagenome_contrib.tsv.gz -o COG_pathways_out -p 80 --no_regroup --map /home/jhuang/Tools/picrust2/picrust2/default_files/pathway_mapfiles/KEGG_pathways_to_KO.tsv # The files KEGG_pathways_out/path_abun_unstrat.tsv KEGG_pathways_out_per_seq_contrib/path_abun_unstrat.tsv are the same!!!!!!! diff KEGG_pathways_out/path_abun_unstrat.tsv KEGG_pathways_out_per_seq_contrib/path_abun_unstrat.tsv -
Add descriptions to 5(gene_family)+2(pathway) tables
#description_mapfiles KEGG_pathways_info.tsv.gz KEGG_modules_info.tsv.gz metacyc_pathways_info.txt.gz ec_level4_info.tsv.gz cog_info.tsv.gz tigrfam_info.tsv.gz pfam_info.tsv.gz ko_info.tsv.gz #--6.1. Add descriptions in gene family tables # EC and METACYC is a pair, EC for gene_annotation and METACYC for pathway_annotation, therefore we have 5 m-options for gene family tables, 1 m-option for pathway abundance table, for KEGG a custom description_mapfile is needed. add_descriptions.py -i COG_metagenome_out/pred_metagenome_unstrat.tsv.gz -m COG -o COG_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i EC_metagenome_out/pred_metagenome_unstrat.tsv.gz -m EC -o EC_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i KO_metagenome_out/pred_metagenome_unstrat.tsv.gz -m KO -o KO_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i PFAM_metagenome_out/pred_metagenome_unstrat.tsv.gz -m PFAM -o PFAM_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz add_descriptions.py -i TIGRFAM_metagenome_out/pred_metagenome_unstrat.tsv.gz -m TIGRFAM -o TIGRFAM_metagenome_out/pred_metagenome_unstrat_descrip.tsv.gz #--6.2. Add descriptions in pathway abundance tables -m {METACYC,COG,EC,KO,PFAM,TIGRFAM} cd MetaCyc_pathways_out_per_seq_contrib add_descriptions.py -i path_abun_unstrat.tsv.gz -m METACYC -o path_abun_unstrat_descrip.tsv.gz gunzip path_abun_unstrat_descrip.tsv.gz cd .. cd KEGG_pathways_out_per_seq_contrib add_descriptions.py -i path_abun_unstrat.tsv.gz -o path_abun_unstrat_descrip.tsv.gz --custom_map_table /home/jhuang/Tools/picrust2/picrust2/default_files/description_mapfiles/KEGG_pathways_info.tsv.gz gunzip path_abun_unstrat_descrip.tsv.gz cd .. -
Difference between Kxxxxxxx (gene or protein) and koxxxxxxx (pathway)
The terms "ORTHOLOGY: K10989" and "ko00001" refer to different concepts and components within the KEGG (Kyoto Encyclopedia of Genes and Genomes) database, which is used for understanding high-level functions and utilities of biological systems. 1. ORTHOLOGY: K10989 Definition: K10989 refers to a specific KEGG Orthology (KO) identifier. What It Represents: This identifier is assigned to a specific group of orthologous genes or proteins that perform the same function across different species. For example, K10989 might correspond to a particular enzyme or protein that is conserved across multiple organisms. Usage: K10989 is used to refer to a specific function at the gene/protein level. When you see "ORTHOLOGY: K10989," it indicates that this specific gene or protein in a genome has been classified under this orthology group. 2. ko00001 Definition: ko00001 refers to a specific KEGG pathway map identifier. What It Represents: This identifier is associated with a KEGG pathway, which is a collection of manually drawn pathway maps representing molecular interaction and reaction networks, such as metabolic pathways, signaling pathways, and more. Usage: ko00001 typically refers to a high-level map, like the KEGG pathway overview, which includes an entire collection of pathways or a very broad view of metabolism or other cellular processes. The "ko" prefix indicates that it is a KEGG Orthology-based pathway map. Summary of Differences: Scope: K10989 is specific to a particular orthologous group of genes/proteins. ko00001 refers to a broad KEGG pathway or map. Focus: K10989 focuses on the function of specific genes/proteins across species. ko00001 provides a visual representation of biological processes or pathways. Level of Detail: K10989 is detailed at the molecular or functional level of specific proteins/genes. ko00001 covers a broader, more comprehensive overview of biological systems or networks. These identifiers help researchers navigate between specific gene functions and broader biological processes within the KEGG database. -
Preparing the input files for STAMP, e.g. path_abun_unstrat_descrip.tsv.gz and metadata.tsv
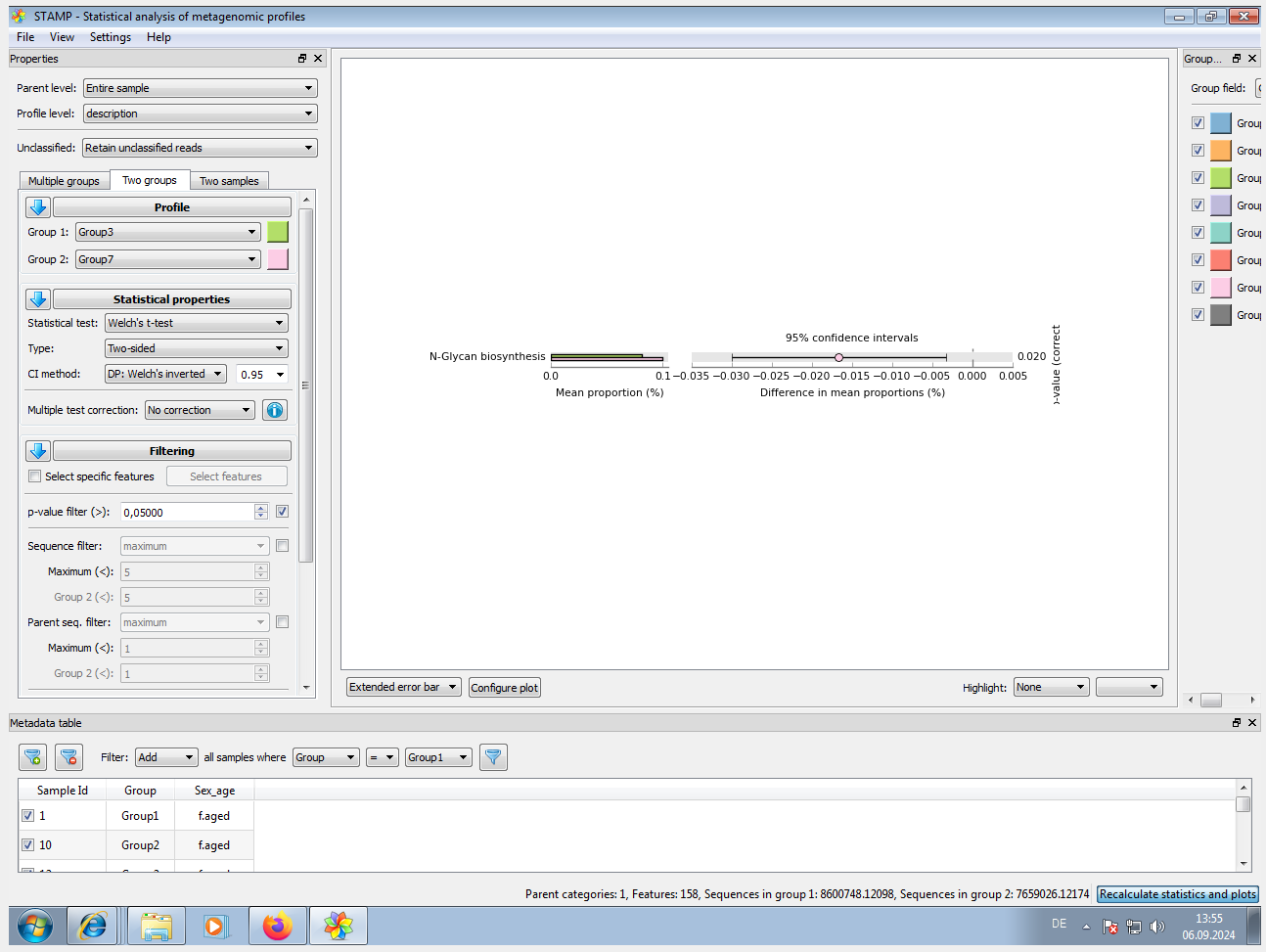
Input files needed for STAMP are: * pred_metagenome_unstrat_descrip.tsv.gz / path_abun_unstrat_descrip.tsv.gz (from STEP 3) * metadata.tsv (see below) cut -d$'\t' -f1 map_corrected.txt > 1 cut -d$'\t' -f5 map_corrected.txt > 5 cut -d$'\t' -f6 map_corrected.txt > 6 paste -d$'\t' 1 5 > 1_5 paste -d$'\t' 1_5 6 > metadata.tsv # NOTE_1: Modify '#SampleID' to 'SampleID' !! SampleID Group Sex_age 1 Group1 f.aged 2 Group1 f.aged 5 Group1 f.aged ... # NOTE_2: for loading of EC[COG|KO|PFAM|TIGRFAM]_metagenome_out/pred_metagenome_unstrat_descrip.tsv, it doesn't work since 'Data does not form a strict hierarchy. Child FAD binding domain has multiple parents (e.g., PF00667, PF00890)'. # NOTE_3: for each pathway type (e.g. KEGG or MetaCyc), we need to restart the program. An example setting see STAMP_Screenshot.png. -
Install STAMP
#https://github.com/picrust/picrust2/wiki/STAMP-example conda activate base conda install mamba # -- Install method 1 (Failed) -- #https://beikolab.cs.dal.ca/software/Quick_installation_instructions_for_STAMP mamba create -n stamp_py2 python=2 pyqt=4 numpy scipy matplotlib biom-format stamp #pip install matplotlib pip install STAMP #Alternative: mamba create -n stamp bioconda::stamp # -- Install method 2 (Failed) -- cd ~/Tools/STAMP-2.1.3 python setup.py install #byte-compiling /home/jhuang/miniconda3/envs/stamp_py2/lib/python2.7/site-packages/stamp/metagenomics/StringHelper.py to StringHelper.pyc #running install_scripts #copying build/scripts-2.7/checkHierarchy.py -> /home/jhuang/miniconda3/envs/stamp_py2/bin #copying build/scripts-2.7/STAMP -> /home/jhuang/miniconda3/envs/stamp_py2/bin #changing mode of /home/jhuang/miniconda3/envs/stamp_py2/bin/checkHierarchy.py to 775 #changing mode of /home/jhuang/miniconda3/envs/stamp_py2/bin/STAMP to 775 #running install_data #copying LICENSE.txt -> /home/jhuang/miniconda3/envs/stamp_py2/. #creating /home/jhuang/miniconda3/envs/stamp_py2/manual #copying ./manual/STAMP_Users_Guide.pdf -> /home/jhuang/miniconda3/envs/stamp_py2/./manual #copying README.md -> /home/jhuang/miniconda3/envs/stamp_py2/. #running install_egg_info #Writing /home/jhuang/miniconda3/envs/stamp_py2/lib/python2.7/site-packages/STAMP-2.1.3-py2.7.egg-info python STAMP_test.py -v python STAMP.py #BUG: The two methods above could successfully install STAMP successfully, however, it stalls if starts? Try to install it on notebook! curl -L -O "https://github.com/conda-forge/miniforge/releases/latest/download/Miniforge3-$(uname)-$(uname -m).sh" bash Miniforge3-$(uname)-$(uname -m).sh mamba install bioconda::stamp pyqt=4 # -- Install method 3 (Failed) -- #--Quick installation instructions for STAMP-- #SUCCESSFUL install on Virtualbox 14 or 16 sudo apt-get install libblas-dev liblapack-dev gfortran sudo apt-get install freetype* python-pip python-dev python-numpy python-scipy python-matplotlib sudo pip install STAMP #pip could not find, manually download the pip-package and install with the following command sudo python setup.py install #in the STAMP-pip-library. #ImportError: No module named biom.parse sudo pip install --upgrade biom-format conda remove -n stamp --all #conda create -n stamp pyqt=4 #conda activate stamp #conda install -c bioconda stamp conda config --show channels mamba create -n stamp_py2 pip python=2 pyqt=4 numpy scipy biom-format mamba activate stamp_py2 #pip install matplotlib pip install stamp # -- Install method 4 (Failed) -- conda remove stamp_pyqt4 mamba install pyqt=4 stamp #conda install icu=56 # -- Install method 5: Windows system on Virtualbox (Failed) -- sudo apt update sudo apt install virtualbox sudo apt install virtualbox-ext-pack virtualbox #http://www.winwin7.com/Win7QiJianBan/XTZJWin7QiJianBan-116517.html #http://win.hgyji.com/fanqiexp.html #https://eprebys.faculty.ucdavis.edu/2020/04/08/installing-windows-xp-in-virtualbox-or-other-vm/ https://jingyan.baidu.com/article/a17d52851540e08098c8f219.html https://msdn.cyanlemon.net/%E6%93%8D%E4%BD%9C%E7%B3%BB%E7%BB%9F/Windows%20XP/%E4%B8%AD%E6%96%87-%E7%AE%80%E4%BD%93/ MRX3F-47B9T-2487J-KWKMF-RPWBY https://blog.51cto.com/u_16213618/11137698 https://msdn.itellyou.cn/ # -- Install method 6: STAMP_2_1_3.exe on Windows 7 in VirtualBox (Successful) -- -
ALDEx2 (Not_Used!)
https://bioconductor.org/packages/release/bioc/html/ALDEx2.html -
Convert png to svg and pdf
inkscape error_bar.png --export-plain-svg=error_bar.svg (embbed) sudo apt update sudo apt install autotrace sudo apt-get install -y libpng-dev libtiff-dev imagemagick git clone https://github.com/autotrace/autotrace.git cd autotrace #sudo apt install intltool #sudo apt install gettext libglib2.0-dev #sudo apt install libtool libtool-bin #sudo apt install automake sudo apt-get install libxml-parser-perl ./autogen.sh ./configure make autotrace -output-format svg -output-file error_bar.svg error_bar.png
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
- MCV病毒中的LT与sT蛋白功能
- Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
Workflow and Tools for Integrating ChIP-seq and RNA-seq Data Analysis