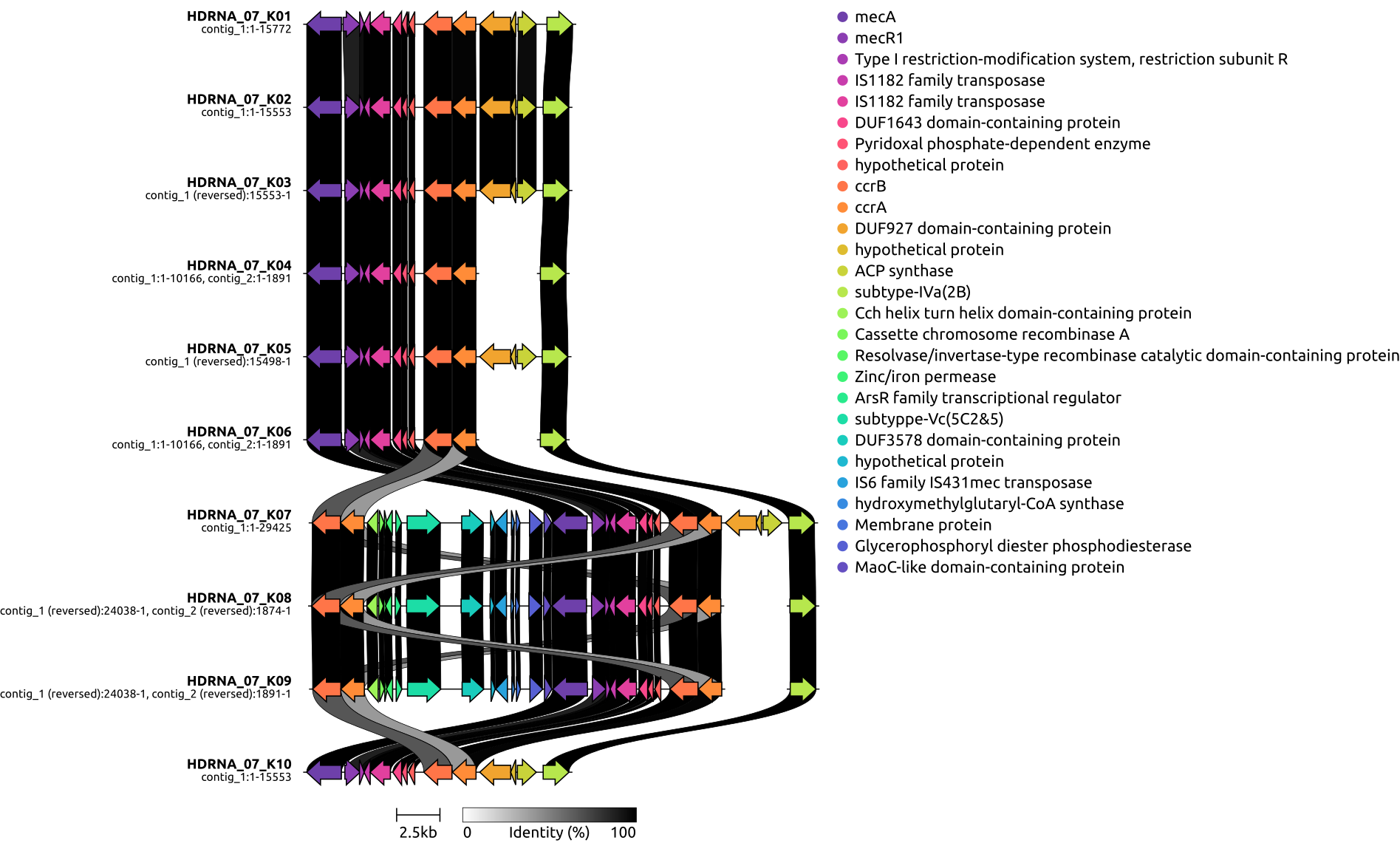
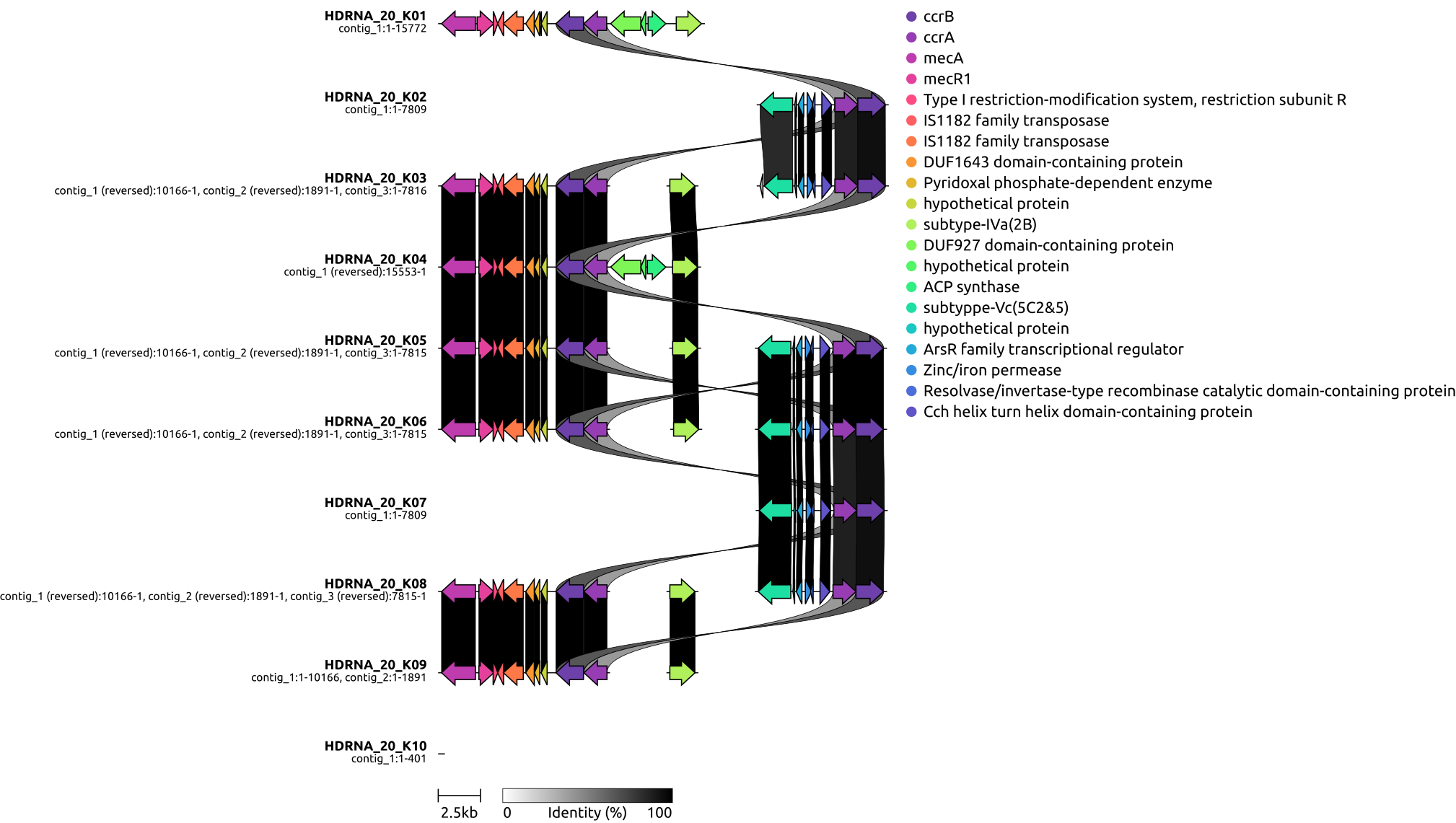
draw graphics on local genetic environments of SCCmec using clinker
gene_x 0 like s 757 view s
Tags: pipeline
TODO: read some papers about recombination and horizontal gene transfer of SCCmec
Identification and characterization of SCCmec typing with psm-mec positivity in staphylococci from patients with coagulase-negative staphylococci peritoneal dialysis-related peritonitis
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10517493/
SCCmec in staphylococci: genes on the move
https://academic.oup.com/femspd/article/46/1/8/598528
https://en.wikipedia.org/wiki/SCCmec
1, SCCmec background
SCCmec is a 21 to 60 kb long genetic element that confers broad-spectrum β-lactam resistance to MRSA.[2] Moreover, additional genetic elements like Tn554, pT181, and pUB110 can be found in SCCmec, which have the capability to render resistance to various non-β-lactam drugs.
The mec complex is divided further into five types (I through V) based on the arrangement of regulatory genetic features such as mecR1, an inducer.[7] The mec gene complex in SCCmec, comprising mec gene, its regulators (mecR1, mecI), and insertion sequences (IS), is categorized into five classes (A to E).
- Class A includes mecA, full mecR1, mecI, and IS431.
- Class B has IS1272, mecA, partial mecR1, and IS431.
- Class C, with two versions (C1, C2), contains mecA, partial mecR1, IS431, differing in IS431 orientation.
- Class D includes IS431, mecA, partial mecR1;
- Class E consists of blaZ, mecC, mecR1, mecI.[7][8][9]
2, prepare files in directory gbs (cd ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/gbs/)
cp /home/jhuang/Tools/bacto/db/CP133676.gb_converted.fna ./
#cp ../snippy_CP133676/shovill/HDRNA_01_K01/contigs.fa HDRNA_01_K01_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K02/contigs.fa HDRNA_01_K02_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K03/contigs.fa HDRNA_01_K03_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K04/contigs.fa HDRNA_01_K04_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K05/contigs.fa HDRNA_01_K05_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K06/contigs.fa HDRNA_01_K06_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K07/contigs.fa HDRNA_01_K07_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K08/contigs.fa HDRNA_01_K08_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K09/contigs.fa HDRNA_01_K09_contigs.fa
cp ../snippy_CP133676/shovill/HDRNA_01_K10/contigs.fa HDRNA_01_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133677.gb_converted.fna ./
#cp ../snippy_CP133677/shovill/HDRNA_03_K01/contigs.fa HDRNA_03_K01_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K02/contigs.fa HDRNA_03_K02_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K03/contigs.fa HDRNA_03_K03_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K04/contigs.fa HDRNA_03_K04_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K05/contigs.fa HDRNA_03_K05_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K06/contigs.fa HDRNA_03_K06_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K07/contigs.fa HDRNA_03_K07_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K08/contigs.fa HDRNA_03_K08_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K09/contigs.fa HDRNA_03_K09_contigs.fa
cp ../snippy_CP133677/shovill/HDRNA_03_K10/contigs.fa HDRNA_03_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133678.gb_converted.fna ./
#cp ../snippy_CP133678/shovill/HDRNA_06_K01/contigs.fa HDRNA_06_K01_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K02/contigs.fa HDRNA_06_K02_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K03/contigs.fa HDRNA_06_K03_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K04/contigs.fa HDRNA_06_K04_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K05/contigs.fa HDRNA_06_K05_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K06/contigs.fa HDRNA_06_K06_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K07/contigs.fa HDRNA_06_K07_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K08/contigs.fa HDRNA_06_K08_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K09/contigs.fa HDRNA_06_K09_contigs.fa
cp ../snippy_CP133678/shovill/HDRNA_06_K10/contigs.fa HDRNA_06_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133680.gb_converted.fna ./
#cp ../snippy_CP133680/shovill/HDRNA_07_K01/contigs.fa HDRNA_07_K01_contigs.fa
#cp ../snippy_CP133680/shovill/HDRNA_07_K01-BB28/contigs.fa HDRNA_07_K01-BB28_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K02/contigs.fa HDRNA_07_K02_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K03/contigs.fa HDRNA_07_K03_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K04/contigs.fa HDRNA_07_K04_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K05/contigs.fa HDRNA_07_K05_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K06/contigs.fa HDRNA_07_K06_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K07/contigs.fa HDRNA_07_K07_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K08/contigs.fa HDRNA_07_K08_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K09/contigs.fa HDRNA_07_K09_contigs.fa
cp ../snippy_CP133680/shovill/HDRNA_07_K10/contigs.fa HDRNA_07_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133682.gb_converted.fna ./
#cp ../snippy_CP133682/shovill/HDRNA_08_K01/contigs.fa HDRNA_08_K01_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K02/contigs.fa HDRNA_08_K02_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K03/contigs.fa HDRNA_08_K03_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K04/contigs.fa HDRNA_08_K04_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K05/contigs.fa HDRNA_08_K05_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K06/contigs.fa HDRNA_08_K06_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K07/contigs.fa HDRNA_08_K07_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K08/contigs.fa HDRNA_08_K08_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K09/contigs.fa HDRNA_08_K09_contigs.fa
cp ../snippy_CP133682/shovill/HDRNA_08_K10/contigs.fa HDRNA_08_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133684.gb_converted.fna ./
#cp ../snippy_CP133684/shovill/HDRNA_12_K01/contigs.fa HDRNA_12_K01_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K02/contigs.fa HDRNA_12_K02_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K03/contigs.fa HDRNA_12_K03_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K04/contigs.fa HDRNA_12_K04_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K05/contigs.fa HDRNA_12_K05_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K06/contigs.fa HDRNA_12_K06_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K07/contigs.fa HDRNA_12_K07_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K08/contigs.fa HDRNA_12_K08_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K09/contigs.fa HDRNA_12_K09_contigs.fa
cp ../snippy_CP133684/shovill/HDRNA_12_K10/contigs.fa HDRNA_12_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133688.gb_converted.fna ./
#cp ../snippy_CP133688/shovill/HDRNA_16_K01/contigs.fa HDRNA_16_K01_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K02/contigs.fa HDRNA_16_K02_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K03/contigs.fa HDRNA_16_K03_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K04/contigs.fa HDRNA_16_K04_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K05/contigs.fa HDRNA_16_K05_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K06/contigs.fa HDRNA_16_K06_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K07/contigs.fa HDRNA_16_K07_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K08/contigs.fa HDRNA_16_K08_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K09/contigs.fa HDRNA_16_K09_contigs.fa
cp ../snippy_CP133688/shovill/HDRNA_16_K10/contigs.fa HDRNA_16_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133693.gb_converted.fna ./
#cp ../snippy_CP133693/shovill/HDRNA_17_K01/contigs.fa HDRNA_17_K01_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K02/contigs.fa HDRNA_17_K02_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K03/contigs.fa HDRNA_17_K03_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K04/contigs.fa HDRNA_17_K04_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K05/contigs.fa HDRNA_17_K05_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K06/contigs.fa HDRNA_17_K06_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K07/contigs.fa HDRNA_17_K07_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K08/contigs.fa HDRNA_17_K08_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K09/contigs.fa HDRNA_17_K09_contigs.fa
cp ../snippy_CP133693/shovill/HDRNA_17_K10/contigs.fa HDRNA_17_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133696.gb_converted.fna ./
#cp ../snippy_CP133696/shovill/HDRNA_19_K01/contigs.fa HDRNA_19_K01_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K02/contigs.fa HDRNA_19_K02_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K03/contigs.fa HDRNA_19_K03_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K04/contigs.fa HDRNA_19_K04_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K05/contigs.fa HDRNA_19_K05_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K06/contigs.fa HDRNA_19_K06_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K07/contigs.fa HDRNA_19_K07_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K08/contigs.fa HDRNA_19_K08_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K09/contigs.fa HDRNA_19_K09_contigs.fa
cp ../snippy_CP133696/shovill/HDRNA_19_K10/contigs.fa HDRNA_19_K10_contigs.fa
cp /home/jhuang/Tools/bacto/db/CP133700.gb_converted.fna ./
#cp ../snippy_CP133700/shovill/HDRNA_20_K01/contigs.fa HDRNA_20_K01_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K02/contigs.fa HDRNA_20_K02_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K03/contigs.fa HDRNA_20_K03_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K04/contigs.fa HDRNA_20_K04_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K05/contigs.fa HDRNA_20_K05_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K06/contigs.fa HDRNA_20_K06_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K07/contigs.fa HDRNA_20_K07_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K08/contigs.fa HDRNA_20_K08_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K09/contigs.fa HDRNA_20_K09_contigs.fa
cp ../snippy_CP133700/shovill/HDRNA_20_K10/contigs.fa HDRNA_20_K10_contigs.fa
https://github.com/gamcil/clinker
3, draw graphics on local genetic environments of SCCmec using clinker
# - draw a graphics for the gene organization of SCCmec regions for 10 groups, using phylogenetic tree for the 100 isolates.
# - regenerate a SNP Indels table for each groups, marked the SNPs and Indels in the regions of SCCmec yellow.
#in snippy_CP133676
cp /home/jhuang/Tools/bacto/db/CP133676.gb_converted.fna ./
#cp ../snippy_CP133676/shovill/HDRNA_01_K01/contigs.fa HDRNA_01_K01_contigs.fa #SCCmec_type_IVa(2B) --> 40444-55810 --> chr:30444-65810
cp ../snippy_CP133676/shovill/HDRNA_01_K02/contigs.fa HDRNA_01_K02_contigs.fa #SCCmec_type_IVa(2B) --> contig00005+contig00011
cp ../snippy_CP133676/shovill/HDRNA_01_K03/contigs.fa HDRNA_01_K03_contigs.fa #SCCmec_type_IVa(2B) --> contig00006+contig00004
cp ../snippy_CP133676/shovill/HDRNA_01_K04/contigs.fa HDRNA_01_K04_contigs.fa #SCCmec_type_IVa(2B) --> contig00002
cp ../snippy_CP133676/shovill/HDRNA_01_K05/contigs.fa HDRNA_01_K05_contigs.fa #SCCmec_type_IVa(2B) --> contig00006+contig00004
cp ../snippy_CP133676/shovill/HDRNA_01_K06/contigs.fa HDRNA_01_K06_contigs.fa #SCCmec_type_IVa(2B) --> contig00002
cp ../snippy_CP133676/shovill/HDRNA_01_K07/contigs.fa HDRNA_01_K07_contigs.fa #SCCmec_type_IVa(2B) --> contig00002
cp ../snippy_CP133676/shovill/HDRNA_01_K08/contigs.fa HDRNA_01_K08_contigs.fa #SCCmec_type_IVa(2B) --> contig00021+contig00011
cp ../snippy_CP133676/shovill/HDRNA_01_K09/contigs.fa HDRNA_01_K09_contigs.fa #SCCmec_type_IVa(2B) --> contig00006+contig00004
cp ../snippy_CP133676/shovill/HDRNA_01_K10/contigs.fa HDRNA_01_K10_contigs.fa #SCCmec_type_IVa(2B) --> contig00008+contig00002
#samtools faidx CP133676.gb_converted.fna CP133676:30444-65810 > CP133676_30444-65810.fasta
#samtools faidx HDRNA_01_K02_contigs.fa contig00005 > HDRNA_01_K02_selected.fasta
##samtools faidx HDRNA_01_K02_contigs.fa contig00011 >> HDRNA_01_K02_selected.fasta
#samtools faidx HDRNA_01_K03_contigs.fa contig00006 > HDRNA_01_K03_selected.fasta
##samtools faidx HDRNA_01_K03_contigs.fa contig00004 >> HDRNA_01_K03_selected.fasta
#samtools faidx HDRNA_01_K04_contigs.fa contig00002 > HDRNA_01_K04_selected.fasta
#samtools faidx HDRNA_01_K05_contigs.fa contig00006 > HDRNA_01_K05_selected.fasta
##samtools faidx HDRNA_01_K05_contigs.fa contig00004 >> HDRNA_01_K05_selected.fasta
#samtools faidx HDRNA_01_K06_contigs.fa contig00002 > HDRNA_01_K06_selected.fasta
#samtools faidx HDRNA_01_K07_contigs.fa contig00002 > HDRNA_01_K07_selected.fasta
#samtools faidx HDRNA_01_K08_contigs.fa contig00021 > HDRNA_01_K08_selected.fasta
##samtools faidx HDRNA_01_K08_contigs.fa contig00011 >> HDRNA_01_K08_selected.fasta
#samtools faidx HDRNA_01_K09_contigs.fa contig00006 > HDRNA_01_K09_selected.fasta
##samtools faidx HDRNA_01_K09_contigs.fa contig00004 >> HDRNA_01_K09_selected.fasta
#samtools faidx HDRNA_01_K10_contigs.fa contig00008 > HDRNA_01_K10_selected.fasta
##samtools faidx HDRNA_01_K10_contigs.fa contig00002 >> HDRNA_01_K10_selected.fasta
# -- install bakta --
conda create --name bakta
conda activate bakta
mamba install -c conda-forge -c bioconda bakta
bakta_db list
bakta_db download --output /mnt/nvme0n1p1/REFs --type full
mv /mnt/nvme0n1p1/REFs/db /mnt/nvme0n1p1/REFs/bakta_db
#Run Bakta using '--db /mnt/nvme0n1p1/REFs/bakta_db' or set a BAKTA_DB environment variable: 'export BAKTA_DB=/mnt/nvme0n1p1/REFs/db'
# -- HDRNA_01_K01=CP133676
#mecA:12:AB505628 100.00 2010/2010 CP133676 40444..42453
#dmecR1:1:AB033763 100.00 987/987 CP133676 42550..43536
#IS1272:3:AM292304 100.00 1843/1843 CP133676 43525..45367
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133676 47209..48858
#ccrA2:7:81108:AB096217 99.63 1350/1350 CP133676 48859..50203
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133676 54320..55810 #subtype-IVa(2B) is always located in another contig --> ignoring this entity!
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133676.gb_converted.fna
#ADAPT the gene_name and positions following the info below in CP133676_adapted.gb, e.g. adding /gene="mecA" and correct the positions.
python3 ~/Scripts/extract_subregion.py CP133676.gb_converted.gbff 40244 56010 CP133676_sub.gbff
#50303-52324 contains DUF927 domain-containing protein
#54320-40244=14076
#14076..15566
#/product="AAA family ATPase"
# -- HDRNA_01_K02
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00005 3825..5169
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00005 5170..6819
#IS1272:3:AM292304 100.00 1843/1843 contig00005 8661..10503
#dmecR1:1:AB033763 100.00 987/987 contig00005 10492..11478
#mecA:12:AB505628 100.00 2010/2010 contig00005 11575..13584
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00011 86471..87961
#python3 extract_gb_from_gbk.py snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.gbk contig00005 HDRNA_01_K02_contig00005.gb
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.fna contig00005:3625-13784 > HDRNA_01_K02_sub.fna
#+4117=92278
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.fna contig00011:86271-88161 >> HDRNA_01_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K02_sub.fna
#python3 ~/Scripts/extract_subregion.py HDRNA_01_K02_contig00005.gbff 3725 13684 HDRNA_01_K02_contig00005_sub.gb
# -- HDRNA_01_K03
#dmecR1:1:AB033763 100.00 987/987 contig00006 10492..11478
#mecA:12:AB505628 100.00 2010/2010 contig00006 11575..13584
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00006 3825..5169
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00006 5170..6819
#IS1272:3:AM292304 100.00 1843/1843 contig00006 8661..10503
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00004 211325..212815
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K03/HDRNA_01_K03.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K03/HDRNA_01_K03.fna contig00006:3625-13784 > HDRNA_01_K03_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K03/HDRNA_01_K03.fna contig00004:211125-213015 >> HDRNA_01_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K03_sub.fna
# -- HDRNA_01_K04
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00002 389566..390910
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 390911..392560
#IS1272:3:AM292304 100.00 1843/1843 contig00002 394402..396244
#dmecR1:1:AB033763 100.00 987/987 contig00002 396233..397219
#mecA:12:AB505628 100.00 2010/2010 contig00002 397316..399325
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 384124..385614
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K04/HDRNA_01_K04.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K04/HDRNA_01_K04.fna contig00002:383924-399525 > HDRNA_01_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K04_sub.fna
# -- HDRNA_01_K05
#dmecR1:1:AB033763 100.00 987/987 contig00006 10657..11643
#mecA:12:AB505628 100.00 2010/2010 contig00006 11740..13749
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00006 3990..5334
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00006 5335..6984
#IS1272:3:AM292304 100.00 1843/1843 contig00006 8826..10668
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00004 212178..213668
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K05/HDRNA_01_K05.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K05/HDRNA_01_K05.fna contig00006:3790-13949 > HDRNA_01_K05_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K05/HDRNA_01_K05.fna contig00004:211978-213868 >> HDRNA_01_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K05_sub.fna
# -- HDRNA_01_K06
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00002 389999..391343
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 391344..392993
#IS1272:3:AM292304 100.00 1843/1843 contig00002 394835..396677
#dmecR1:1:AB033763 100.00 987/987 contig00002 396666..397652
#mecA:12:AB505628 100.00 2010/2010 contig00002 397749..399758
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 384557..386047
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K06/HDRNA_01_K06.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K06/HDRNA_01_K06.fna contig00002:384357-399858 > HDRNA_01_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K06_sub.fna
# -- HDRNA_01_K07
#mecA:12:AB505628 100.00 2010/2010 contig00002 172913..174922
#dmecR1:1:AB033763 100.00 987/987 contig00002 175019..176005
#IS1272:3:AM292304 100.00 1843/1843 contig00002 175994..177836
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 179678..181327
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00002 181328..182672
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 186624..188114
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K07/HDRNA_01_K07.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K07/HDRNA_01_K07.fna contig00002:172813-188314 > HDRNA_01_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K07_sub.fna
# -- HDRNA_01_K08
#dmecR1:1:AB033763 100.00 987/987 contig00021 10445..11431
#mecA:3:AB037671 100.00 2004/2007 contig00021 11535..13538
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00021 3778..5122
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00021 5123..6772
#IS1272:3:AM292304 100.00 1843/1843 contig00021 8614..10456
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00011 72699..74189
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K08/HDRNA_01_K08.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K08/HDRNA_01_K08.fna contig00021:3578-13738 > HDRNA_01_K08_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K08/HDRNA_01_K08.fna contig00011:72499-74389 >> HDRNA_01_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K08_sub.fna
# -- HDRNA_01_K09
#dmecR1:1:AB033763 100.00 987/987 contig00006 10492..11478
#mecA:12:AB505628 100.00 2010/2010 contig00006 11575..13584
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00006 3825..5169
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00006 5170..6819
#IS1272:3:AM292304 100.00 1843/1843 contig00006 8661..10503
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00004 211303..212793
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K09/HDRNA_01_K09.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K09/HDRNA_01_K09.fna contig00006:3625-13784 > HDRNA_01_K09_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K09/HDRNA_01_K09.fna contig00004:211103-212993 >> HDRNA_01_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K09_sub.fna
# -- HDRNA_01_K10
#dmecR1:1:AB033763 100.00 987/987 contig00008 10417..11403
#mecA:12:AB505628 100.00 2010/2010 contig00008 11500..13509
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00008 3750..5094
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00008 5095..6744
#IS1272:3:AM292304 100.00 1843/1843 contig00008 8586..10428
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 382889..384379
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K10/HDRNA_01_K10.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K10/HDRNA_01_K10.fna contig00008:3550-13709 > HDRNA_01_K10_sub.fna
samtools faidx ../snippy_CP133676/prokka/HDRNA_01_K10/HDRNA_01_K10.fna contig00002:382689-384579 >> HDRNA_01_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_01_K10_sub.fna
# -- CP133677
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133677 2112527..2114017
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133677 2117969..2119318
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133677 2119319..2120968
#IS1272:3:AM292304 99.95 1844/1843 CP133677 2122810..2124653
#dmecR1:1:AB033763 100.00 987/987 CP133677 2124642..2125628
#mecA:12:AB505628 100.00 2010/2010 CP133677 2125725..2127734
#bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133677.gb_converted.fna
#python3 ~/Scripts/extract_subregion.py CP133677.gb_converted.gbff 2112327 2127934 HDRNA_03_K01.gbff
revseq ~/Tools/bacto/db/CP133677.gb_converted.fna
#Re-submit cp133677.rev to https://cge.food.dtu.dk/services/SCCmecFinder-1.2/
#mecA:12:AB505628 100.00 2010/2010 CP133677 462542..464551
#dmecR1:1:AB033763 100.00 987/987 CP133677 464648..465634
#IS1272:3:AM292304 99.95 1844/1843 CP133677 465623..467466
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133677 469308..470957
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133677 470958..472307
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133677 476259..477749
bakta --db /mnt/nvme0n1p1/REFs/bakta_db cp133677.rev
python3 ~/Scripts/extract_subregion.py cp133677.gbff 462342 477949 HDRNA_03_K01.gbff
# -- HDRNA_03_K02
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00026 13584..14933
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00026 14934..16583
#IS1272:3:AM292304 99.95 1844/1843 contig00026 18425..20268
#dmecR1:1:AB033763 100.00 987/987 contig00026 20257..21243
#mecA:12:AB505628 100.00 2010/2010 contig00026 21340..23349
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00026 8252..9742
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K02/HDRNA_03_K02.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K02/HDRNA_03_K02.fna contig00026:8052-23549 > HDRNA_03_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K02_sub.fna
# -- HDRNA_03_K03
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00029 13619..14968
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 14969..16618
#IS1272:3:AM292304 99.95 1844/1843 contig00029 18460..20303
#dmecR1:1:AB033763 100.00 987/987 contig00029 20292..21278
#mecA:12:AB505628 100.00 2010/2010 contig00029 21375..23384
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 8232..9722
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K03/HDRNA_03_K03.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K03/HDRNA_03_K03.fna contig00029:8032-23584 > HDRNA_03_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K03_sub.fna
# -- HDRNA_03_K04
#dmecR1:1:AB033763 100.00 987/987 contig00032 10498..11484
#mecA:12:AB505628 100.00 2010/2010 contig00032 11581..13590
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00032 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00032 5175..6824
#IS1272:3:AM292304 99.95 1844/1843 contig00032 8666..10509
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00036 8277..9767
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K04/HDRNA_03_K04.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K04/HDRNA_03_K04.fna contig00032:3625-13790 > HDRNA_03_K04_sub.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K04/HDRNA_03_K04.fna contig00036:8077-9967 >> HDRNA_03_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K04_sub.fna
# -- HDRNA_03_K05
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00029 13774..15123
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 15124..16773
#IS1272:3:AM292304 99.95 1844/1843 contig00029 18615..20458
#dmecR1:1:AB033763 100.00 987/987 contig00029 20447..21433
#mecA:12:AB505628 100.00 2010/2010 contig00029 21530..23539
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 8387..9877
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K05/HDRNA_03_K05.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K05/HDRNA_03_K05.fna contig00029:8187-23739 > HDRNA_03_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K05_sub.fna
# -- HDRNA_03_K06
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00030 13739..15088
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00030 15089..16738
#IS1272:3:AM292304 99.95 1844/1843 contig00030 18580..20423
#dmecR1:1:AB033763 100.00 987/987 contig00030 20412..21398
#mecA:12:AB505628 100.00 2010/2010 contig00030 21495..23504
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00030 8352..9842
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K06/HDRNA_03_K06.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K06/HDRNA_03_K06.fna contig00030:8152-23704 > HDRNA_03_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K06_sub.fna
# -- HDRNA_03_K07
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00027 11087..12436
#mecA:12:AB505628 100.00 2010/2010 contig00027 2671..4680
#dmecR1:1:AB033763 100.00 987/987 contig00027 4777..5763
#IS1272:3:AM292304 99.95 1844/1843 contig00027 5752..7595
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 9437..11086
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00027 16333..17823
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K07/HDRNA_03_K07.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K07/HDRNA_03_K07.fna contig00027:2471-18023 > HDRNA_03_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K07_sub.fna
# -- HDRNA_03_K08
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00029 13631..14980
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 14981..16630
#IS1272:3:AM292304 99.95 1844/1843 contig00029 18472..20315
#dmecR1:1:AB033763 100.00 987/987 contig00029 20304..21290
#mecA:12:AB505628 100.00 2010/2010 contig00029 21387..23396
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 8244..9734
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K08/HDRNA_03_K08.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K08/HDRNA_03_K08.fna contig00029:8044-23596 > HDRNA_03_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K08_sub.fna
# -- HDRNA_03_K09
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00025 25311..26660
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00025 26661..28310
#IS1272:3:AM292304 99.95 1844/1843 contig00025 30152..31995
#dmecR1:1:AB033763 100.00 987/987 contig00025 31984..32970
#mecA:12:AB505628 100.00 2010/2010 contig00025 33067..35076
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00025 19924..21414
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K09/HDRNA_03_K09.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K09/HDRNA_03_K09.fna contig00025:19724-35276 > HDRNA_03_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K09_sub.fna
# -- HDRNA_03_K10
#ccrA2:7:81108:AB096217 99.63 1350/1350 contig00027 13754..15103
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 15104..16753
#IS1272:3:AM292304 99.95 1844/1843 contig00027 18595..20438
#dmecR1:1:AB033763 100.00 987/987 contig00027 20427..21413
#mecA:12:AB505628 100.00 2010/2010 contig00027 21510..23519
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00027 8367..9857
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K10/HDRNA_03_K10.fna
samtools faidx ../snippy_CP133677/prokka/HDRNA_03_K10/HDRNA_03_K10.fna contig00027:8167-23719 > HDRNA_03_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_03_K10_sub.fna
#CP133678
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 CP133678 42063..43217
#ccrA2:7:81108:AB096217 99.93 1350/1350 CP133678 47234..48583
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 CP133678 48584..50233
#IS1272:2:AB033763 100.00 1585/1585 CP133678 52075..53659
#dmecR1:1:AB033763 100.00 987/987 CP133678 55466..56452
#mecA:12:AB505628 100.00 2010/2010 CP133678 56549..58558
#bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133678.gb_converted.fna
#python3 ~/Scripts/extract_subregion.py CP133678.gb_converted.gbff 41863 58758 HDRNA_06_K01.gbff
revseq ~/Tools/bacto/db/CP133678.gb_converted.fna
#Re-submit cp133678.rev to https://cge.food.dtu.dk/services/SCCmecFinder-1.2/
#mecA:12:AB505628 100.00 2010/2010 CP133678 2406703..2408712
#dmecR1:1:AB033763 100.00 987/987 CP133678 2408809..2409795
#IS1272:2:AB033763 100.00 1585/1585 CP133678 2411602..2413186
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 CP133678 2415028..2416677
#ccrA2:7:81108:AB096217 99.93 1350/1350 CP133678 2416678..2418027
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 CP133678 2422044..2423198
bakta --db /mnt/nvme0n1p1/REFs/bakta_db cp133678.rev
python3 ~/Scripts/extract_subregion.py cp133678.gbff 2406503 2423398 HDRNA_06_K01.gbff
#HDRNA_06_K02
##IS1272:2:AB033763 91.00 1523/1585 contig00036 86..1608
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00031 1464..2618
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K02/HDRNA_06_K02.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K02/HDRNA_06_K02.fna contig00031:1264-2818 > HDRNA_06_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K02_sub.fna
#HDRNA_06_K03
##IS1272:2:AB033763 91.00 1523/1585 contig00038 1..1523
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00033 4048..5202
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K03/HDRNA_06_K03.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K03/HDRNA_06_K03.fna contig00033:3848-5402 > HDRNA_06_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K03_sub.fna
#HDRNA_06_K04
##IS1272:2:AB033763 91.00 1523/1585 contig00043 1..1523
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00039 1444..2598
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K04/HDRNA_06_K04.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K04/HDRNA_06_K04.fna contig00039:1244-2798 > HDRNA_06_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K04_sub.fna
#HDRNA_06_K05
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00025 13961..15310
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00025 15311..16960
#IS1272:2:AB033763 100.00 1585/1585 contig00025 18802..20386
#dmecR1:1:AB033763 100.00 987/987 contig00033 357..1343
#mecA:12:AB505628 100.00 2010/2010 contig00033 1440..3449
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00025 8790..9944
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K05/HDRNA_06_K05.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K05/HDRNA_06_K05.fna contig00025:8590-20586 > HDRNA_06_K05_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K05/HDRNA_06_K05.fna contig00033:157-3649 >> HDRNA_06_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K05_sub.fna
#HDRNA_06_K06
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00027 11981..13330
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00027 13331..14980
#IS1272:2:AB033763 100.00 1585/1585 contig00027 16822..18406
#mecA:12:AB505628 100.00 2010/2010 contig00032 1634..3643
#dmecR1:1:AB033763 100.00 987/987 contig00032 3740..4726
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00027 6810..7964
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K06/HDRNA_06_K06.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K06/HDRNA_06_K06.fna contig00027:6610-18606 > HDRNA_06_K06_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K06/HDRNA_06_K06.fna contig00032:1434-4926 >> HDRNA_06_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K06_sub.fna
#HDRNA_06_K07
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00024 13961..15310
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00024 15311..16960
#IS1272:2:AB033763 100.00 1585/1585 contig00024 18802..20386
#mecA:12:AB505628 100.00 2010/2010 contig00032 1633..3642
#dmecR1:1:AB033763 100.00 987/987 contig00032 3739..4725
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00024 8790..9944
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K07/HDRNA_06_K07.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K07/HDRNA_06_K07.fna contig00024:8590-20586 > HDRNA_06_K07_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K07/HDRNA_06_K07.fna contig00032:1433-4925 >> HDRNA_06_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K07_sub.fna
#HDRNA_06_K08
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00026 6635..7984
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00026 7985..9634
#IS1272:2:AB033763 100.00 1585/1585 contig00026 11476..13060
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00026 1464..2618
#dmecR1:1:AB033763 100.00 987/987 contig00029 357..1343
#mecA:12:AB505628 100.00 2010/2010 contig00029 1440..3449
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K08/HDRNA_06_K08.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K08/HDRNA_06_K08.fna contig00026:1264-13260 > HDRNA_06_K08_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K08/HDRNA_06_K08.fna contig00029:157-3649 >> HDRNA_06_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K08_sub.fna
#HDRNA_06_K09
##IS1272:2:AB033763 91.00 1523/1585 contig00045 1..1523
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00039 4048..5202
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K09/HDRNA_06_K09.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K09/HDRNA_06_K09.fna contig00039:3848-5402 > HDRNA_06_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K09_sub.fna
#HDRNA_06_K10
#dmecR1:1:AB033763 100.00 987/987 contig00031 357..1343
#mecA:12:AB505628 100.00 2010/2010 contig00031 1440..3449
#ccrA2:7:81108:AB096217 99.93 1350/1350 contig00026 6635..7984
#ccrB2:9:JCSC4469:AB097677 99.88 1650/1650 contig00026 7985..9634
#IS1272:2:AB033763 100.00 1585/1585 contig00026 11476..13060
#subtype-IVc(2B):3:81108:AB096217 100.00 1155/1155 contig00026 1464..2618
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K10/HDRNA_06_K10.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K10/HDRNA_06_K10.fna contig00026:1264-13260 > HDRNA_06_K10_sub.fna
samtools faidx ../snippy_CP133678/prokka/HDRNA_06_K10/HDRNA_06_K10.fna contig00031:157-3649 >> HDRNA_06_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_06_K10_sub.fna
#CP133680
#mecA:12:AB505628 100.00 2010/2010 CP133680 37611..39620
#dmecR1:1:AB033763 100.00 987/987 CP133680 39717..40703
#IS1272:3:AM292304 99.95 1844/1843 CP133680 40692..42535
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133680 44377..46026
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133680 46027..47376
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133680 51493..52983
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133680.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133680.gb_converted.gbff 37411 53183 CP133680_sub.gbff
#HDRNA_07_K02
#mecA:12:AB505628 100.00 2010/2010 contig00003 137822..139831
#dmecR1:1:AB033763 100.00 987/987 contig00003 139928..140914
#IS1272:3:AM292304 99.95 1844/1843 contig00003 140903..142746
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 144588..146237
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 146238..147587
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 151484..152974
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K02/HDRNA_07_K02.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K02/HDRNA_07_K02.fna contig00003:137622-153174 > HDRNA_07_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K02_sub.fna
#HDRNA_07_K03
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 52228..53718
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 57615..58964
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 58965..60614
#IS1272:3:AM292304 99.95 1844/1843 contig00003 62456..64299
#dmecR1:1:AB033763 100.00 987/987 contig00003 64288..65274
#mecA:12:AB505628 100.00 2010/2010 contig00003 65371..67380
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K03/HDRNA_07_K03.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K03/HDRNA_07_K03.fna contig00003:52028-67580 > HDRNA_07_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K03_sub.fna
#HDRNA_07_K04
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 11087..12436
#mecA:12:AB505628 100.00 2010/2010 contig00027 2671..4680
#dmecR1:1:AB033763 100.00 987/987 contig00027 4777..5763
#IS1272:3:AM292304 99.95 1844/1843 contig00027 5752..7595
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 9437..11086
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 231..1721
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K04/HDRNA_07_K04.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K04/HDRNA_07_K04.fna contig00027:2471-12636 > HDRNA_07_K04_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K04/HDRNA_07_K04.fna contig00016:31-1921 >> HDRNA_07_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K04_sub.fna
#HDRNA_07_K05
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 52208..53698
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00015 57540..58889
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00015 58890..60539
#IS1272:3:AM292304 99.95 1844/1843 contig00015 62381..64224
#dmecR1:1:AB033763 100.00 987/987 contig00015 64213..65199
#mecA:12:AB505628 100.00 2010/2010 contig00015 65296..67305
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K05/HDRNA_07_K05.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K05/HDRNA_07_K05.fna contig00015:52008-67505 > HDRNA_07_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K05_sub.fna
#HDRNA_07_K06
#mecA:12:AB505628 100.00 2010/2010 contig00005 137910..139919
#dmecR1:1:AB033763 100.00 987/987 contig00005 140016..141002
#IS1272:3:AM292304 99.95 1844/1843 contig00005 140991..142834
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00005 144676..146325
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00005 146326..147675
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00017 211..1701
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K06/HDRNA_07_K06.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K06/HDRNA_07_K06.fna contig00005:137710-147875 > HDRNA_07_K06_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K06/HDRNA_07_K06.fna contig00017:11-1901 >> HDRNA_07_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K06_sub.fna
#TODO_1: DESCRIBE it as the fact below, one is ccrB4+ccrA4+subtyppe-Vc(5C2&5), another is ccrB2+ccrA2+subtype-IVa(2B)!
#TODO_2: CONSIDERING whether "#IS1272:2:AB033763 91.00" should be added to the plot?
#HDRNA_07_K07
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00002 142538..144172
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00002 144169..145530
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00002 148018..149952
#mecA:12:AB505628 100.00 2010/2010 contig00002 156410..158419
#dmecR1:1:AB033763 100.00 987/987 contig00002 158516..159502
#IS1272:3:AM292304 99.95 1844/1843 contig00002 159491..161334
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00002 163176..164825
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00002 164826..166175
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00002 170072..171562
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K07/HDRNA_07_K07.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K07/HDRNA_07_K07.fna contig00002:142338-171762 > HDRNA_07_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K07_sub.fna
#mecR1
#IS1182 family transposase
#Cation-transporting P-type ATPase --> subtyppe-Vc(5C2&5)
#AAA family ATPase --> subtype-IVa(2B)
#HDRNA_07_K08
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5175..6824
#IS1272:3:AM292304 99.95 1844/1843 contig00004 8666..10509
#dmecR1:1:AB033763 100.00 987/987 contig00004 10498..11484
#mecA:12:AB505628 100.00 2010/2010 contig00004 11581..13590
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 20048..21982
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 24470..25831
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 25828..27462
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 50618..52108
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K08/HDRNA_07_K08.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K08/HDRNA_07_K08.fna contig00004:3625-27662 > HDRNA_07_K08_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K08/HDRNA_07_K08.fna contig00016:50418-52308 >> HDRNA_07_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K08_sub.fna
#HDRNA_07_K09
#dmecR1:1:AB033763 100.00 987/987 contig00004 10423..11409
#mecA:12:AB505628 100.00 2010/2010 contig00004 11506..13515
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 19973..21907
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 24395..25756
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 25753..27387
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5100..6749
#IS1272:3:AM292304 99.95 1844/1843 contig00004 8591..10434
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 52208..53698
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K09/HDRNA_07_K09.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K09/HDRNA_07_K09.fna contig00004:3550-27587 > HDRNA_07_K09_sub.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K09/HDRNA_07_K09.fna contig00016:52008-53898 >> HDRNA_07_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K09_sub.fna
#HDRNA_07_K10
#mecA:12:AB505628 100.00 2010/2010 contig00003 137894..139903
#dmecR1:1:AB033763 100.00 987/987 contig00003 140000..140986
#IS1272:3:AM292304 99.95 1844/1843 contig00003 140975..142818
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 144660..146309
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 146310..147659
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 151556..153046
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K10/HDRNA_07_K10.fna
samtools faidx ../snippy_CP133680/prokka/HDRNA_07_K10/HDRNA_07_K10.fna contig00003:137694-153246 > HDRNA_07_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_07_K10_sub.fna
#CP133682
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 CP133682 47094..48722
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 CP133682 55568..57439
#mecA:12:AB505628 100.00 2010/2010 CP133682 64626..66635
#mecR1:1:D86934 100.00 1758/1758 CP133682 66732..68489
#mecI:1:D86934 100.00 372/372 CP133682 68489..68860
##IS1272:2:AB033763 91.06 1577/1585 CP133682 865163..866739
##IS1272:2:AB033763 91.06 1577/1585 CP133682 968562..970138
##IS1272:2:AB033763 91.06 1577/1585 CP133682 1659758..1661334
##IS1272:2:AB033763 91.06 1577/1585 CP133682 2357991..2359567
##IS1272:2:AB033763 91.06 1577/1585 CP133682 293604..295180
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133682.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133682.gb_converted.gbff 46894 69060 CP133682_sub.gbff
#Zinc-ribbon domain-containing protein-->
#HDRNA_08_K02
#mecI:1:D86934 100.00 372/372 contig00005 34179..34550
#mecR1:1:D86934 100.00 1758/1758 contig00005 34550..36307
#mecA:12:AB505628 100.00 2010/2010 contig00005 36404..38413
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 45600..47471
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 54317..55945
##IS1272:2:AB033763 91.01 1524/1585 contig00020 66..1589
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K02/HDRNA_08_K02.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K02/HDRNA_08_K02.fna contig00005:33979-56145 > HDRNA_08_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K02_sub.fna
#HDRNA_08_K03
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 111644..113272
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 120118..121989
#mecA:12:AB505628 100.00 2010/2010 contig00005 129176..131185
#mecR1:1:D86934 100.00 1758/1758 contig00005 131282..133039
#mecI:1:D86934 100.00 372/372 contig00005 133039..133410
##IS1272:2:AB033763 91.01 1524/1585 contig00020 66..1589
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K03/HDRNA_08_K03.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K03/HDRNA_08_K03.fna contig00005:111444-133610 > HDRNA_08_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K03_sub.fna
#HDRNA_08_K04
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 111072..112700
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 119546..121417
#mecA:12:AB505628 100.00 2010/2010 contig00005 128604..130613
#mecR1:1:D86934 100.00 1758/1758 contig00005 130710..132467
#mecI:1:D86934 100.00 372/372 contig00005 132467..132838
##IS1272:2:AB033763 91.06 1577/1585 contig00019 7..1583
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K04/HDRNA_08_K04.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K04/HDRNA_08_K04.fna contig00005:110872-133038 > HDRNA_08_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K04_sub.fna
#HDRNA_08_K05
##IS1272:2:AB033763 91.01 1524/1585 contig00021 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 34179..34550
#mecR1:1:D86934 100.00 1758/1758 contig00006 34550..36307
#mecA:12:AB505628 100.00 2010/2010 contig00006 36404..38413
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45600..47471
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 54317..55945
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K05/HDRNA_08_K05.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K05/HDRNA_08_K05.fna contig00006:33979-56145 > HDRNA_08_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K05_sub.fna
#HDRNA_08_K06
##IS1272:2:AB033763 91.01 1524/1585 contig00020 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 34179..34550
#mecR1:1:D86934 100.00 1758/1758 contig00006 34550..36307
#mecA:12:AB505628 100.00 2010/2010 contig00006 36404..38413
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45600..47471
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 54317..55945
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K06/HDRNA_08_K06.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K06/HDRNA_08_K06.fna contig00006:33979-56145 > HDRNA_08_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K06_sub.fna
#HDRNA_08_K07
#mecI:1:D86934 100.00 372/372 contig00005 32299..32670
#mecR1:1:D86934 100.00 1758/1758 contig00005 32670..34427
#mecA:12:AB505628 100.00 2010/2010 contig00005 34524..36533
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 43720..45591
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 52437..54065
##IS1272:2:AB033763 91.01 1524/1585 contig00025 66..1589
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K07/HDRNA_08_K07.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K07/HDRNA_08_K07.fna contig00005:32099-54265 > HDRNA_08_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K07_sub.fna
#HDRNA_08_K08
##IS1272:2:AB033763 91.01 1524/1585 contig00022 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 33701..34072
#mecR1:1:D86934 100.00 1758/1758 contig00006 34072..35829
#mecA:12:AB505628 100.00 2010/2010 contig00006 35926..37935
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45122..46993
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 53839..55467
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K08/HDRNA_08_K08.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K08/HDRNA_08_K08.fna contig00006:33501-55667 > HDRNA_08_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K08_sub.fna
#HDRNA_08_K09
##IS1272:2:AB033763 91.01 1524/1585 contig00021 1..1524
#mecI:1:D86934 100.00 372/372 contig00006 34178..34549
#mecR1:1:D86934 100.00 1758/1758 contig00006 34549..36306
#mecA:12:AB505628 100.00 2010/2010 contig00006 36403..38412
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00006 45599..47470
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00006 54316..55944
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K09/HDRNA_08_K09.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K09/HDRNA_08_K09.fna contig00006:33978-56144 > HDRNA_08_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K09_sub.fna
#HDRNA_08_K10
#ccrB4:2:BK20781:FJ670542 93.06 1629/1629 contig00005 111072..112700
#subtype-IVd(2B):4:JCSC4469:AB097677 97.76 1872/1872 contig00005 119546..121417
#mecA:12:AB505628 100.00 2010/2010 contig00005 128604..130613
#mecR1:1:D86934 100.00 1758/1758 contig00005 130710..132467
#mecI:1:D86934 100.00 372/372 contig00005 132467..132838
##IS1272:2:AB033763 91.06 1577/1585 contig00018 7..1583
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K10/HDRNA_08_K10.fna
samtools faidx ../snippy_CP133682/prokka/HDRNA_08_K10/HDRNA_08_K10.fna contig00005:110872-133038 > HDRNA_08_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_08_K10_sub.fna
#CP133684
#mecA:12:AB505628 100.00 2010/2010 CP133684 37596..39605
#dmecR1:1:AB033763 100.00 987/987 CP133684 39702..40688
#IS1272:3:AM292304 99.95 1844/1843 CP133684 40677..42520
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133684 44362..46011
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133684 46012..47361
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133684 51478..52968
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133684.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133684.gb_converted.gbff 37396 53168 CP133684_sub.gbff
#HDRNA_12_K02
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00012 52208..53698
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00012 57540..58889
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00012 58890..60539
#IS1272:3:AM292304 99.95 1844/1843 contig00012 62381..64224
#dmecR1:1:AB033763 100.00 987/987 contig00012 64213..65199
#mecA:12:AB505628 100.00 2010/2010 contig00012 65296..67305
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K02/HDRNA_12_K02.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K02/HDRNA_12_K02.fna contig00012:52008-67505 > HDRNA_12_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K02_sub.fna
#HDRNA_12_K03
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00029 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00029 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00029 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00029 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 211..1701
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K03/HDRNA_12_K03.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K03/HDRNA_12_K03.fna contig00029:2451-12616 > HDRNA_12_K03_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K03/HDRNA_12_K03.fna contig00015:11-1901 >> HDRNA_12_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K03_sub.fna
#HDRNA_12_K04
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00031 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00031 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00031 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00031 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00031 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 184..1674
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K04/HDRNA_12_K04.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K04/HDRNA_12_K04.fna contig00031:2451-12616 > HDRNA_12_K04_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K04/HDRNA_12_K04.fna contig00014:84-1874 >> HDRNA_12_K04_sub.fna
bakta --skip-crispr --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K04_sub.fna
#HDRNA_12_K05
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00030 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00030 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00030 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00030 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00030 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 211..1701
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K05/HDRNA_12_K05.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K05/HDRNA_12_K05.fna contig00030:2451-12616 > HDRNA_12_K05_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K05/HDRNA_12_K05.fna contig00016:11-1901 >> HDRNA_12_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K05_sub.fna
#HDRNA_12_K06
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 51961..53451
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00014 57293..58642
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00014 58643..60292
#IS1272:3:AM292304 99.95 1844/1843 contig00014 62134..63977
#dmecR1:1:AB033763 100.00 987/987 contig00014 63966..64952
#mecA:12:AB505628 100.00 2010/2010 contig00014 65049..67058
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K06/HDRNA_12_K06.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K06/HDRNA_12_K06.fna contig00014:51761-67258 > HDRNA_12_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K06_sub.fna
#HDRNA_12_K07
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 40711..42201
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00014 46153..47502
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00014 47503..49152
#IS1272:3:AM292304 99.95 1844/1843 contig00014 50994..52837
#dmecR1:1:AB033763 100.00 987/987 contig00014 52826..53812
#mecA:12:AB505628 100.00 2010/2010 contig00014 53909..55918
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K07/HDRNA_12_K07.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K07/HDRNA_12_K07.fna contig00014:40511-56118 > HDRNA_12_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K07_sub.fna
#HDRNA_12_K08
#dmecR1:1:AB033763 100.00 987/987 contig00027 10441..11427
#mecA:12:AB505628 100.00 2010/2010 contig00027 11524..13533
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3768..5117
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5118..6767
#IS1272:3:AM292304 99.95 1844/1843 contig00027 8609..10452
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00005 137459..138949
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K08/HDRNA_12_K08.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K08/HDRNA_12_K08.fna contig00027:3568-13733 > HDRNA_12_K08_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K08/HDRNA_12_K08.fna contig00005:137259-139149 >> HDRNA_12_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K08_sub.fna
#HDRNA_12_K09
#dmecR1:1:AB033763 100.00 987/987 contig00028 10368..11354
#mecA:12:AB505628 100.00 2010/2010 contig00028 11451..13460
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 3695..5044
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 5045..6694
#IS1272:3:AM292304 99.95 1844/1843 contig00028 8536..10379
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 52207..53697
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K09/HDRNA_12_K09.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K09/HDRNA_12_K09.fna contig00028:3495-13660 > HDRNA_12_K09_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K09/HDRNA_12_K09.fna contig00014:52007-53897 >> HDRNA_12_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K09_sub.fna
#HDRNA_12_K10
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 11067..12416
#mecA:12:AB505628 100.00 2010/2010 contig00028 2651..4660
#dmecR1:1:AB033763 100.00 987/987 contig00028 4757..5743
#IS1272:3:AM292304 99.95 1844/1843 contig00028 5732..7575
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 9417..11066
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 211..1701
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K10/HDRNA_12_K10.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K10/HDRNA_12_K10.fna contig00028:2451-12616 > HDRNA_12_K10_sub.fna
samtools faidx ../snippy_CP133684/prokka/HDRNA_12_K10/HDRNA_12_K10.fna contig00014:11-1901 >> HDRNA_12_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_12_K10_sub.fna
#CP133688
#mecA:12:AB505628 100.00 2010/2010 CP133688 37716..39725
#dmecR1:1:AB033763 100.00 987/987 CP133688 39822..40808
#IS1272:3:AM292304 100.00 1843/1843 CP133688 40797..42639
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133688 44481..46130
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133688 46131..47480
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133688 51377..52867
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133688.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133688.gb_converted.gbff 37516 53067 CP133688_sub.gbff
#HDRNA_16_K02
#dmecR1:1:AB033763 100.00 987/987 contig00012 10440..11426
#mecA:12:AB505628 100.00 2010/2010 contig00012 11523..13532
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00012 3768..5117
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00012 5118..6767
#IS1272:3:AM292304 100.00 1843/1843 contig00012 8609..10451
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 71060..72550
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K02/HDRNA_16_K02.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K02/HDRNA_16_K02.fna contig00012:3568-13732 > HDRNA_16_K02_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K02/HDRNA_16_K02.fna contig00013:70860-72750 >> HDRNA_16_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K02_sub.fna
#HDRNA_16_K03
#dmecR1:1:AB033763 100.00 987/987 contig00014 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00014 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00014 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00014 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00014 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 56677..58167
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K03/HDRNA_16_K03.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K03/HDRNA_16_K03.fna contig00014:3550-13714 > HDRNA_16_K03_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K03/HDRNA_16_K03.fna contig00015:56477-58367 >> HDRNA_16_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K03_sub.fna
#HDRNA_16_K04
#dmecR1:1:AB033763 100.00 987/987 contig00017 10367..11353
#mecA:12:AB505628 100.00 2010/2010 contig00017 11450..13459
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00017 3695..5044
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00017 5045..6694
#IS1272:3:AM292304 100.00 1843/1843 contig00017 8536..10378
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00012 56677..58167
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K04/HDRNA_16_K04.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K04/HDRNA_16_K04.fna contig00017:3495-13659 > HDRNA_16_K04_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K04/HDRNA_16_K04.fna contig00012:56477-58367 >> HDRNA_16_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K04_sub.fna
#HDRNA_16_K05
#mecA:12:AB505628 100.00 2010/2010 contig00013 64162..66171
#dmecR1:1:AB033763 100.00 987/987 contig00013 66268..67254
#IS1272:3:AM292304 100.00 1843/1843 contig00013 67243..69085
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00013 70927..72576
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00013 72577..73926
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00014 67300..68790
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K05/HDRNA_16_K05.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K05/HDRNA_16_K05.fna contig00013:63962-74126 > HDRNA_16_K05_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K05/HDRNA_16_K05.fna contig00014:67100-68990 >> HDRNA_16_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K05_sub.fna
#HDRNA_16_K06
#dmecR1:1:AB033763 100.00 987/987 contig00004 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00004 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00004 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00004 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00004 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 67555..69045
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K06/HDRNA_16_K06.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K06/HDRNA_16_K06.fna contig00004:3550-13714 > HDRNA_16_K06_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K06/HDRNA_16_K06.fna contig00013:67355-69245 >> HDRNA_16_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K06_sub.fna
#HDRNA_16_K07
#mecA:12:AB505628 100.00 2010/2010 contig00003 156854..158863
#dmecR1:1:AB033763 100.00 987/987 contig00003 158960..159946
#IS1272:3:AM292304 100.00 1843/1843 contig00003 159935..161777
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00003 163619..165268
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00003 165269..166618
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00003 170515..172005
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K07/HDRNA_16_K07.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K07/HDRNA_16_K07.fna contig00003:156654-172205 > HDRNA_16_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K07_sub.fna
#HDRNA_16_K08
#dmecR1:1:AB033763 100.00 987/987 contig00011 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00011 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00011 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00011 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00011 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 67300..68790
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K08/HDRNA_16_K08.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K08/HDRNA_16_K08.fna contig00011:3550-13714 > HDRNA_16_K08_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K08/HDRNA_16_K08.fna contig00013:67100-68990 >> HDRNA_16_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K08_sub.fna
#HDRNA_16_K09
#dmecR1:1:AB033763 100.00 987/987 contig00017 10497..11483
#mecA:12:AB505628 100.00 2010/2010 contig00017 11580..13589
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00017 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00017 5175..6824
#IS1272:3:AM292304 100.00 1843/1843 contig00017 8666..10508
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00012 52313..53803
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K09/HDRNA_16_K09.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K09/HDRNA_16_K09.fna contig00017:3625-13789 > HDRNA_16_K09_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K09/HDRNA_16_K09.fna contig00012:52113-54003 >> HDRNA_16_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K09_sub.fna
#HDRNA_16_K10
#mecA:12:AB505628 100.00 2010/2010 contig00005 156908..158917
#dmecR1:1:AB033763 100.00 987/987 contig00005 159014..160000
#IS1272:3:AM292304 100.00 1843/1843 contig00005 159989..161831
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00005 163673..165322
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00005 165323..166672
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 211..1701
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K10/HDRNA_16_K10.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K10/HDRNA_16_K10.fna contig00005:156708-166872 > HDRNA_16_K10_sub.fna
samtools faidx ../snippy_CP133688/prokka/HDRNA_16_K10/HDRNA_16_K10.fna contig00013:11-1901 >> HDRNA_16_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_16_K10_sub.fna
#CP133693
#mecA:12:AB505628 100.00 2010/2010 CP133693 37128..39137
#dmecR1:1:AB033763 100.00 987/987 CP133693 39234..40220
#IS1272:3:AM292304 100.00 1843/1843 CP133693 40209..42051
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133693 43893..45542
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133693 45543..46892
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133693 51009..52499
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133693.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133693.gb_converted.gbff 36928 52699 CP133693_sub.gbff
#HDRNA_17_K02
#dmecR1:1:AB033763 100.00 987/987 contig00028 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00028 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00028 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00031 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K02/HDRNA_17_K02.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K02/HDRNA_17_K02.fna contig00028:3550-13714 > HDRNA_17_K02_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K02/HDRNA_17_K02.fna contig00031:4666-6556 >> HDRNA_17_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K02_sub.fna
#HDRNA_17_K03
#dmecR1:1:AB033763 100.00 987/987 contig00025 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00025 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00025 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00025 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00025 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00028 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K03/HDRNA_17_K03.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K03/HDRNA_17_K03.fna contig00025:3550-13714 > HDRNA_17_K03_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K03/HDRNA_17_K03.fna contig00028:4666-6556 >> HDRNA_17_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K03_sub.fna
#HDRNA_17_K04
#dmecR1:1:AB033763 100.00 987/987 contig00027 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00027 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00027 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00030 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K04/HDRNA_17_K04.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K04/HDRNA_17_K04.fna contig00027:3550-13714 > HDRNA_17_K04_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K04/HDRNA_17_K04.fna contig00030:4666-6556 >> HDRNA_17_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K04_sub.fna
#HDRNA_17_K05
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00024 11321..12670
#mecA:12:AB505628 100.00 2010/2010 contig00024 2906..4915
#dmecR1:1:AB033763 100.00 987/987 contig00024 5012..5998
#IS1272:3:AM292304 100.00 1843/1843 contig00024 5987..7829
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00024 9671..11320
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00024 16567..18057
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K05/HDRNA_17_K05.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K05/HDRNA_17_K05.fna contig00024:2706-18257 > HDRNA_17_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K05_sub.fna
#HDRNA_17_K06
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 11301..12650
#mecA:12:AB505628 100.00 2010/2010 contig00027 2886..4895
#dmecR1:1:AB033763 100.00 987/987 contig00027 4992..5978
#IS1272:3:AM292304 100.00 1843/1843 contig00027 5967..7809
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 9651..11300
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00029 211..1701
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K06/HDRNA_17_K06.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K06/HDRNA_17_K06.fna contig00027:2686-12850 > HDRNA_17_K06_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K06/HDRNA_17_K06.fna contig00029:11-1901 >> HDRNA_17_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K06_sub.fna
#HDRNA_17_K07
#dmecR1:1:AB033763 100.00 987/987 contig00026 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00026 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00026 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00026 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00026 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00030 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K07/HDRNA_17_K07.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K07/HDRNA_17_K07.fna contig00026:3550-13714 > HDRNA_17_K07_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K07/HDRNA_17_K07.fna contig00030:4666-6556 >> HDRNA_17_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K07_sub.fna
#HDRNA_17_K08
#dmecR1:1:AB033763 100.00 987/987 contig00027 10422..11408
#mecA:12:AB505628 100.00 2010/2010 contig00027 11505..13514
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5100..6749
#IS1272:3:AM292304 100.00 1843/1843 contig00027 8591..10433
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00031 4866..6356
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K08/HDRNA_17_K08.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K08/HDRNA_17_K08.fna contig00027:3550-13714 > HDRNA_17_K08_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K08/HDRNA_17_K08.fna contig00031:4666-6556 >> HDRNA_17_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K08_sub.fna
#HDRNA_17_K09
#dmecR1:1:AB033763 100.00 987/987 contig00025 10497..11483
#mecA:12:AB505628 100.00 2010/2010 contig00025 11580..13589
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00025 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00025 5175..6824
#IS1272:3:AM292304 100.00 1843/1843 contig00025 8666..10508
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00028 4886..6376
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K09/HDRNA_17_K09.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K09/HDRNA_17_K09.fna contig00025:3625-13789 > HDRNA_17_K09_sub.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K09/HDRNA_17_K09.fna contig00028:4686-6576 >> HDRNA_17_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K09_sub.fna
#HDRNA_17_K10
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00024 10273..11622
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00024 11623..13272
#IS1272:3:AM292304 100.00 1843/1843 contig00024 15114..16956
#dmecR1:1:AB033763 100.00 987/987 contig00024 16945..17931
#mecA:12:AB505628 100.00 2010/2010 contig00024 18028..20037
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00024 4886..6376
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K10/HDRNA_17_K10.fna
samtools faidx ../snippy_CP133693/prokka/HDRNA_17_K10/HDRNA_17_K10.fna contig00024:4686-20237 > HDRNA_17_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_17_K10_sub.fna
#CP133696
#ccrB2:1:N315:D86934 98.96 1629/1629 CP133696 42277..43905
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 CP133696 43927..45276
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133696.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133696.gb_converted.gbff 42077 45476 CP133696_sub.gbff
#HDRNA_19_K02
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K02/HDRNA_19_K02.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K02/HDRNA_19_K02.fna contig00030:2360-5759 > HDRNA_19_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K02_sub.fna
#HDRNA_19_K03
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K03/HDRNA_19_K03.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K03/HDRNA_19_K03.fna contig00031:2360-5759 > HDRNA_19_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K03_sub.fna
#HDRNA_19_K04
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K04/HDRNA_19_K04.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K04/HDRNA_19_K04.fna contig00031:2360-5759 > HDRNA_19_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K04_sub.fna
#HDRNA_19_K05
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K05/HDRNA_19_K05.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K05/HDRNA_19_K05.fna contig00031:2360-5759 > HDRNA_19_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K05_sub.fna
#HDRNA_19_K06
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2580..3929
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3951..5579
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K06/HDRNA_19_K06.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K06/HDRNA_19_K06.fna contig00031:2380-5779 > HDRNA_19_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K06_sub.fna
#HDRNA_19_K07
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00031 2580..3929
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00031 3951..5579
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K07/HDRNA_19_K07.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K07/HDRNA_19_K07.fna contig00031:2380-5779 > HDRNA_19_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K07_sub.fna
#HDRNA_19_K08
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2577..3926
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3948..5576
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K08/HDRNA_19_K08.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K08/HDRNA_19_K08.fna contig00030:2377-5776 > HDRNA_19_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K08_sub.fna
#HDRNA_19_K09
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K09/HDRNA_19_K09.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K09/HDRNA_19_K09.fna contig00030:2360-5759 > HDRNA_19_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K09_sub.fna
#HDRNA_19_K10
#ccrA2:13:JCSC6668:AB425823 98.67 1350/1350 contig00030 2560..3909
#ccrB2:1:N315:D86934 98.96 1629/1629 contig00030 3931..5559
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K10/HDRNA_19_K10.fna
samtools faidx ../snippy_CP133696/prokka/HDRNA_19_K10/HDRNA_19_K10.fna contig00030:2360-5759 > HDRNA_19_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_19_K10_sub.fna
#CP133700
#mecA:12:AB505628 100.00 2010/2010 CP133700 37713..39722
#dmecR1:1:AB033763 100.00 987/987 CP133700 39819..40805
#IS1272:3:AM292304 99.95 1844/1843 CP133700 40794..42637
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 CP133700 44479..46128
#ccrA2:7:81108:AB096217 100.00 1350/1350 CP133700 46129..47478
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 CP133700 51595..53085
bakta --db /mnt/nvme0n1p1/REFs/bakta_db ~/Tools/bacto/db/CP133700.gb_converted.fna
python3 ~/Scripts/extract_subregion.py CP133700.gb_converted.gbff 37513 53285 CP133700_sub.gbff
#HDRNA_20_K02
#subtyppe-Vc(5C2&5):10:AB505629 99.90 1935/1935 contig00009 2050..3984
#ccrA4:2:BK20781:FJ670542 100.00 1362/1362 contig00009 6472..7833
#ccrB4:2:BK20781:FJ670542 100.00 1629/1629 contig00009 7830..9458
##IS1272:2:AB033763 91.06 1577/1585 contig00030 7..1583
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K02/HDRNA_20_K02.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K02/HDRNA_20_K02.fna contig00009:1850-9658 > HDRNA_20_K02_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K02_sub.fna
#HDRNA_20_K03
#dmecR1:1:AB033763 100.00 987/987 contig00028 10498..11484
#mecA:12:AB505628 100.00 2010/2010 contig00028 11581..13590
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00028 3825..5174
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00028 5175..6824
#IS1272:3:AM292304 99.95 1844/1843 contig00028 8666..10509
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 51961..53451
#subtyppe-Vc(5C2&5):10:AB505629 99.79 1936/1935 contig00004 4168..6103
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 8591..9952
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 9949..11583
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna contig00028:3625-13790 > HDRNA_20_K03_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna contig00015:51761-53651 >> HDRNA_20_K03_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K03/HDRNA_20_K03.fna contig00004:3968-11783 >> HDRNA_20_K03_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K03_sub.fna
#HDRNA_20_K04
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00013 51961..53451
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00013 57348..58697
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00013 58698..60347
#IS1272:3:AM292304 99.95 1844/1843 contig00013 62189..64032
#dmecR1:1:AB033763 100.00 987/987 contig00013 64021..65007
#mecA:12:AB505628 100.00 2010/2010 contig00013 65104..67113
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K04/HDRNA_20_K04.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K04/HDRNA_20_K04.fna contig00013:51761-67313 > HDRNA_20_K04_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K04_sub.fna
#HDRNA_20_K05
#dmecR1:1:AB033763 100.00 987/987 contig00032 10423..11409
#mecA:12:AB505628 100.00 2010/2010 contig00032 11506..13515
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00032 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00032 5100..6749
#IS1272:3:AM292304 99.95 1844/1843 contig00032 8591..10434
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 51407..52897
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 3319..5253
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 7741..9102
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 9099..10733
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna contig00032:3550-13715 > HDRNA_20_K05_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna contig00016:51207-53097 >> HDRNA_20_K05_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K05/HDRNA_20_K05.fna contig00004:3119-10933 >> HDRNA_20_K05_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K05_sub.fna
#HDRNA_20_K06
#dmecR1:1:AB033763 100.00 987/987 contig00027 10441..11427
#mecA:12:AB505628 100.00 2010/2010 contig00027 11524..13533
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00027 3768..5117
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00027 5118..6767
#IS1272:3:AM292304 99.95 1844/1843 contig00027 8609..10452
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00006 159812..161302
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 14074..16008
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 18496..19857
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 19854..21488
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna contig00027:3568-13733 > HDRNA_20_K06_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna contig00006:159612-161502 >> HDRNA_20_K06_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K06/HDRNA_20_K06.fna contig00004:13874-21688 >> HDRNA_20_K06_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K06_sub.fna
#HDRNA_20_K07
##IS1272:2:AB033763 91.01 1524/1585 contig00032 1..1524
#subtyppe-Vc(5C2&5):10:AB505629 99.90 1935/1935 contig00021 2048..3982
#ccrA4:2:BK20781:FJ670542 100.00 1362/1362 contig00021 6470..7831
#ccrB4:2:BK20781:FJ670542 100.00 1629/1629 contig00021 7828..9456
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K07/HDRNA_20_K07.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K07/HDRNA_20_K07.fna contig00021:1848-9656 > HDRNA_20_K07_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K07_sub.fna
#HDRNA_20_K08
#dmecR1:1:AB033763 100.00 987/987 contig00034 10423..11409
#mecA:12:AB505628 100.00 2010/2010 contig00034 11506..13515
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00034 3750..5099
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00034 5100..6749
#IS1272:3:AM292304 99.95 1844/1843 contig00034 8591..10434
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00016 51407..52897
#ccrB4:2:BK20781:FJ670542 91.68 1635/1629 contig00004 142407..144041
#ccrA4:2:BK20781:FJ670542 90.53 1362/1362 contig00004 144038..145399
#subtyppe-Vc(5C2&5):10:AB505629 99.84 1935/1935 contig00004 147887..149821
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna contig00034:3550-13715 > HDRNA_20_K08_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna contig00016:51207-53097 >> HDRNA_20_K08_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K08/HDRNA_20_K08.fna contig00004:142207-150021 >> HDRNA_20_K08_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K08_sub.fna
cp HDRNA_20_K08_sub.gbff gbff_HDRNA_20/HDRNA_20_K08.gbff
#HDRNA_20_K09
#ccrA2:7:81108:AB096217 100.00 1350/1350 contig00029 11177..12526
#mecA:12:AB505628 100.00 2010/2010 contig00029 2761..4770
#dmecR1:1:AB033763 100.00 987/987 contig00029 4867..5853
#IS1272:3:AM292304 99.95 1844/1843 contig00029 5842..7685
#ccrB2:9:JCSC4469:AB097677 99.94 1650/1650 contig00029 9527..11176
#subtype-IVa(2B):1:CA05:AB063172 100.00 1491/1491 contig00015 211..1701
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K09/HDRNA_20_K09.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K09/HDRNA_20_K09.fna contig00029:2561-12726 > HDRNA_20_K09_sub.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K09/HDRNA_20_K09.fna contig00015:11-1901 >> HDRNA_20_K09_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K09_sub.fna
#TODO_3: HOW to draw an empty unit?
#HDRNA_20_K10
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K10/HDRNA_20_K10.fna
samtools faidx ../snippy_CP133700/prokka/HDRNA_20_K10/HDRNA_20_K10.fna contig00001:1-401 > HDRNA_20_K10_sub.fna
bakta --db /mnt/nvme0n1p1/REFs/bakta_db HDRNA_20_K10_sub.fna
#write in the result-mail: In total, we have 4 different subtype
# - subtype-IVa(2B):1:CA05:AB063172
- HDRNA_01_K01 subtype-IVa(2B)
- HDRNA_03_K01 subtype-IVa(2B)
- HDRNA_12_K01 subtype-IVa(2B)
- HDRNA_16_K01 subtype-IVa(2B)
- HDRNA_17_K01 subtype-IVa(2B)
# - subtype-IVc(2B):3:81108:AB096217
- HDRNA_06_K01
# - subtype-IVd(2B):4:JCSC4469:AB097677
- HDRNA_08_K01
# - subtyppe-Vc(5C2&5):10:AB505629 + subtype-IVa(2B):1:CA05:AB063172
- HDRNA_07_K01
- HDRNA_20_K01
mv *_sub.gbff gbff_all
cd gbff_all
mv CP133676_sub.gbff HDRNA_01_K01.gbff
mv CP133677_sub.gbff HDRNA_03_K01.gbff
mv CP133678_sub.gbff HDRNA_06_K01.gbff
mv CP133680_sub.gbff HDRNA_07_K01.gbff
mv CP133682_sub.gbff HDRNA_08_K01.gbff
mv CP133684_sub.gbff HDRNA_12_K01.gbff
mv CP133688_sub.gbff HDRNA_16_K01.gbff
mv CP133693_sub.gbff HDRNA_17_K01.gbff
mv CP133696_sub.gbff HDRNA_19_K01.gbff
mv CP133700_sub.gbff HDRNA_20_K01.gbff
for f in *_sub.gbff; do mv "$f" "${f/_sub.gbff/.gbff}"; done
#mkdir ../subtype-IVa_2B ../subtype-IVc_2B ../subtype-IVd_2B ../subtype-Vc_5C2and5_subtype-IVa_2B
mkdir gbff_HDRNA_01 gbff_HDRNA_03 gbff_HDRNA_06 gbff_HDRNA_07 gbff_HDRNA_08 gbff_HDRNA_12 gbff_HDRNA_16 gbff_HDRNA_17 gbff_HDRNA_19 gbff_HDRNA_20
# copy or move the corresponding gbff to its directory
rm *.json
clinker *_sub.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_01
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_03
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_06
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_07
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cd gbff_HDRNA_08
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
...
cd gbff_HDRNA_20
rm *.json
clinker *.gbff -p plot_HDRNA.html --dont_set_origin -s session_HDRNA.json -o alignments_HDRNA.csv -dl "," -dc 4
cp ./gbff_HDRNA_01/clinker.png HDRNA_01_clinker.png
cp ./gbff_HDRNA_03/clinker.png HDRNA_03_clinker.png
cp ./gbff_HDRNA_06/clinker.png HDRNA_06_clinker.png
cp ./gbff_HDRNA_07/clinker.png HDRNA_07_clinker.png
cp ./gbff_HDRNA_08/clinker.png HDRNA_08_clinker.png
cp ./gbff_HDRNA_12/clinker.png HDRNA_12_clinker.png
cp ./gbff_HDRNA_16/clinker.png HDRNA_16_clinker.png
cp ./gbff_HDRNA_17/clinker.png HDRNA_17_clinker.png
cp ./gbff_HDRNA_19/clinker.png HDRNA_19_clinker.png
cp ./gbff_HDRNA_20/clinker.png HDRNA_20_clinker.png
#TODO_4: rearrange the Reihenfolge of Isolate in the alignments zu den Reihenfolge in the phylogenetic tree! --> B-region.
# generate a C-region for ACME (similar as 1-s2.0-S1368764622001108-gr1.jpg)
# !!!!!! or generate a new plot for ACME region with the phylogenetic tree !!!!!!
# Evolution of RND efflux pumps in the development of a successful pathogen
# #change the SNP-tables by controlling the REF and isolate1 identical, change the REF--> isolate1, delete the isolate1 column!
# 先自己calculate ACME!
-
code of extract_gb_from_gbk.py
import argparse from Bio import SeqIO def extract_contig(genbank_file, contig_id, output_file): with open(genbank_file, "r") as input_handle: # Iterate over each record (contig) in the GenBank file for record in SeqIO.parse(input_handle, "genbank"): if record.id == contig_id: # Write the specific contig to a new file with open(output_file, "w") as output_handle: SeqIO.write(record, output_handle, "genbank") print(f"Contig {contig_id} has been extracted to {output_file}") return print(f"Contig {contig_id} not found in {genbank_file}") def main(): parser = argparse.ArgumentParser(description="Extract a specific contig from a GenBank file.") parser.add_argument("genbank_file", help="The input GenBank file.") parser.add_argument("contig_id", help="The ID of the contig to extract.") parser.add_argument("output_file", help="The output file to save the extracted contig.") args = parser.parse_args() extract_contig(args.genbank_file, args.contig_id, args.output_file) if __name__ == "__main__": main() #python3 extract_gb_from_gbk.py snippy_CP133676/prokka/HDRNA_01_K02/HDRNA_01_K02.gbk contig00005 HDRNA_01_K02_contig00005.gb -
code of extract_subregion.py
#!/usr/bin/env python3 import sys from Bio import SeqIO def extract_subregion(genbank_path, start, end, output_file): for record in SeqIO.parse(genbank_path, "genbank"): subregion = record[start:end] SeqIO.write(subregion, output_file, "genbank") if __name__ == "__main__": if len(sys.argv) < 5: print("Usage: python extract_subregion.py <GenBank file> <start> <end> <output file>") sys.exit(1) genbank_file = sys.argv[1] start = int(sys.argv[2]) end = int(sys.argv[3]) output_file = sys.argv[4] extract_subregion(genbank_file, start, end, output_file) -
(optional) merge images
from Bio import Phylo import matplotlib.pyplot as plt from PIL import Image, ImageDraw def generate_tree_image(newick, tree_image_path): # Parse the Newick format tree tree = Phylo.read(newick, "newick") # Create a matplotlib figure fig = plt.figure(figsize=(10, 10)) ax = fig.add_subplot(1, 1, 1) # Draw the tree Phylo.draw(tree, do_show=False, axes=ax) # Save the figure plt.savefig(tree_image_path) plt.close() def merge_images(tree_image_path, sccmec_image_path, output_image_path): # Load the images tree_image = Image.open(tree_image_path) sccmec_image = Image.open(sccmec_image_path) # Resize images to have the same height tree_width, tree_height = tree_image.size sccmec_width, sccmec_height = sccmec_image.size new_height = max(tree_height, sccmec_height) new_tree_width = int(tree_width * (new_height / tree_height)) new_sccmec_width = int(sccmec_width * (new_height / sccmec_height)) tree_image = tree_image.resize((new_tree_width, new_height), Image.ANTIALIAS) sccmec_image = sccmec_image.resize((new_sccmec_width, new_height), Image.ANTIALIAS) # Create a new image with a white background total_width = new_tree_width + new_sccmec_width combined_image = Image.new('RGB', (total_width, new_height), (255, 255, 255)) # Paste the images into the combined image combined_image.paste(tree_image, (0, 0)) combined_image.paste(sccmec_image, (new_tree_width, 0)) # Optionally, draw connecting lines (example with a single line for demonstration) draw = ImageDraw.Draw(combined_image) draw.line((new_tree_width, 100, new_tree_width + new_sccmec_width, 100), fill="black", width=2) # Save the combined image combined_image.save(output_image_path) if __name__ == "__main__": # Example Newick format tree (replace with your own tree) newick = "((A:0.1,B:0.2,(C:0.3,D:0.4):0.5):0.6,E:0.7);" tree_image_path = 'tree_image.png' sccmec_image_path = 'sccmec_image.png' # Replace with your actual SCCmec region image path output_image_path = 'combined_image.png' # Generate the tree image generate_tree_image(newick, tree_image_path) # Merge images merge_images(tree_image_path, sccmec_image_path, output_image_path) print(f"Combined image saved to {output_image_path}") #pip3 install biopython matplotlib pillow #python3 generate_and_merge_images.py
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
- MCV病毒中的LT与sT蛋白功能
- Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)