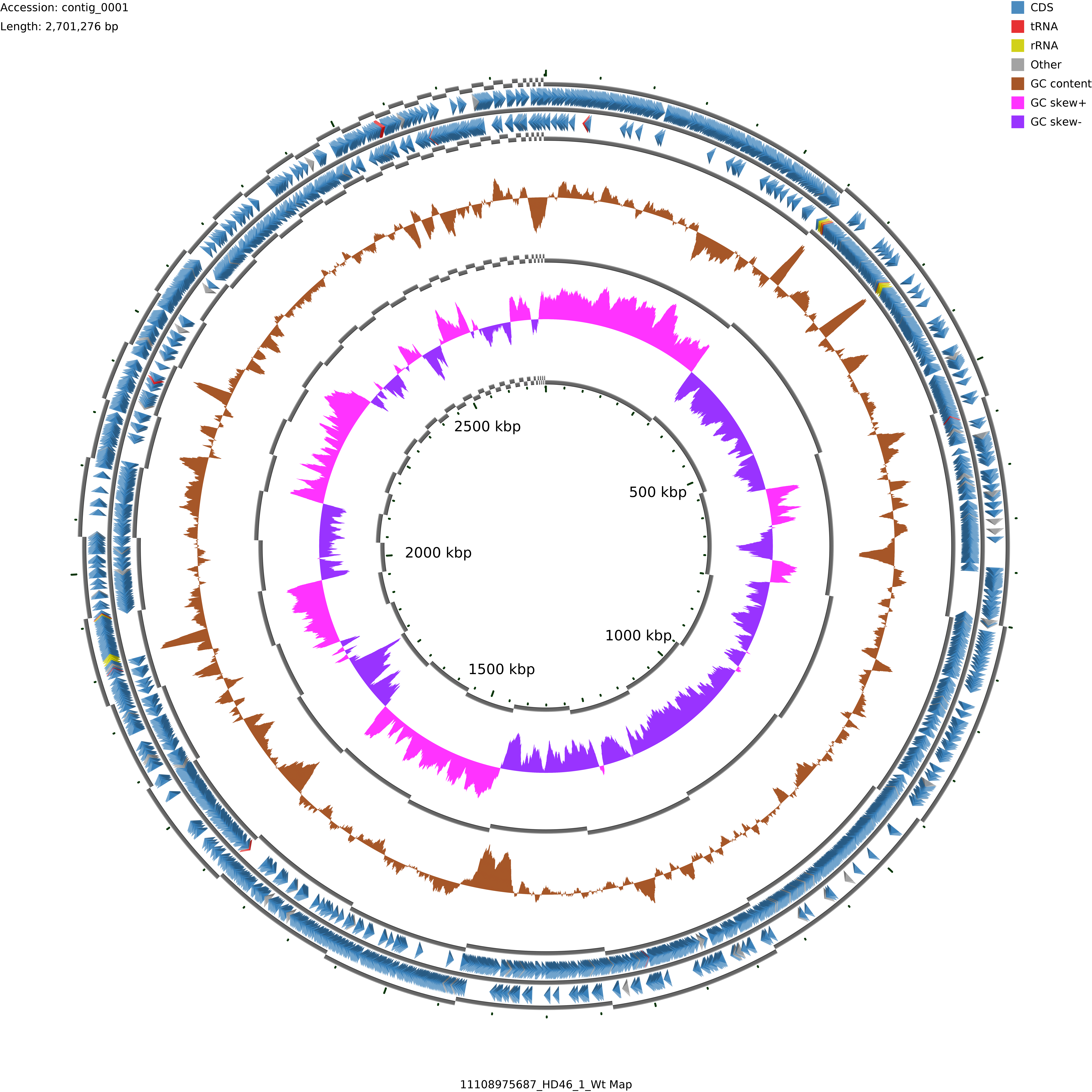
Bacterial Genome Assembly - Analysis Method
gene_x 0 like s 724 view s
Tags: pipeline
- Raw nanopore sequencing reads are assessed for quality and filtering.
- Filtlong v0.2.1 [1] is used to remove short and low-quality reads from the raw nanopore sequencing data.
- The high-quality nanopore sequencing reads are then used for de novo assembly of the bacterial genome.
- The assembly is performed using Flye v2.9.3 [2] with parameters optimized for bacterial genomes.
- The resulting contigs are further polished using Medaka v1.8 [3] to improve base accuracy.
- The assembled genomes are annotated using Bakta v.1.8.2 [4].
- The annotation includes the prediction of coding sequences, tRNAs, rRNAs, and other genomic features based on published databases such as RefSeq and UniProt.
- The quality of the assembled genomes is assessed using various quality assessment tools (QUAST v5.2 [5], CheckM2 v1.0.1 [6], Mash v2.3 [7]).
- Genome completeness, contiguity, and accuracy are evaluated to ensure the reliability of the assemblies.
- A final QC step is included to ensure the purity of samples, which uses minimap2 v2.24 [8] to map the sequence-cleaned reads onto the assembly and finally employs Clair3 v1.0.4 [9] to call variations (SNPs and INDELS) within the assembled genome.
- Any variants detected along with the location in the bacterial assembly are reported.
References
- Filtlong: Ryan R Wick, https://github.com/rrwick/Filtlong
- Flye: Kolmogorov, Mikhail, Jeffrey Yuan, Yu Lin, und Pavel A. Pevzner. "Assembly of Long, Error-Prone Reads Using Repeat Graphs". Nature Biotechnology 37, Nr. 5 (Mai 2019): 540–46. https://doi.org/10.1038/s41587-019-0072-8.
- Medaka: ONT Research, https://github.com/nanoporetech/medaka
- Bakta: Schwengers, Oliver, Lukas Jelonek, Marius Alfred Dieckmann, Sebastian Beyvers, Jochen Blom, und Alexander Goesmann. "Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification". Microbial Genomics 7, Nr. 11 (5. November 2021): 000685. https://doi.org/10.1099/mgen.0.000685.
- Quast: Gurevich, Alexey, Vladislav Saveliev, Nikolay Vyahhi, und Glenn Tesler. "QUAST: Quality Assessment Tool for Genome Assemblies". Bioinformatics (Oxford, England) 29, Nr. 8 (15. April 2013): 1072–75. https://doi.org/10.1093/bioinformatics/btt086.
- CheckM2: Chklovski, Alex, Donovan H. Parks, Ben J. Woodcroft, und Gene W. Tyson. "CheckM2: A Rapid, Scalable and Accurate Tool for Assessing Microbial Genome Quality Using Machine Learning". Nature Methods 20, Nr. 8 (August 2023): 1203–12. https://doi.org/10.1038/s41592-023-01940-w.
- Mash: Ondov, Brian D., Todd J. Treangen, Páll Melsted, Adam B. Mallonee, Nicholas H. Bergman, Sergey Koren, und Adam M. Phillippy. "Mash: Fast Genome and Metagenome Distance Estimation Using MinHash". Genome Biology 17, Nr. 1 (20. Juni 2016): 132. https://doi.org/10.1186/s13059-016-0997-x.
- Minimap2: Li, Heng. "Minimap2: Pairwise Alignment for Nucleotide Sequences". Bioinformatics (Oxford, England) 34, Nr. 18 (15. September 2018): 3094–3100. https://doi.org/10.1093/bioinformatics/bty191.
- Clair3: Zheng, Zhenxian, Shumin Li, Junhao Su, Amy Wing-Sze Leung, Tak-Wah Lam, und Ruibang Luo. "Symphonizing Pileup and Full-Alignment for Deep Learning-Based Long-Read Variant Calling". Nature Computational Science 2, Nr. 12 (Dezember 2022): 797–803. https://doi.org/10.1038/s43588-022-00387-x.
To compare long Nanopore sequencing data with a reference genome, you can follow these steps:
* Align the Nanopore reads to the reference genome.
* Convert and sort the alignment file.
* Call variants.
* Analyze the variants to understand the differences.
Here's a detailed step-by-step guide:
Step 1: Align Nanopore Reads to the Reference Genome
Use a long-read aligner such as minimap2 to align your Nanopore FASTQ reads to the reference genome.
Install minimap2 (if not already installed):
sudo apt-get install minimap2
Align the reads:
minimap2 -ax map-ont reference.fasta nanopore_reads.fastq > aligned.sam
Step 2: Convert SAM to BAM and Sort
Convert the SAM file to BAM format and sort it using samtools.
Install samtools (if not already installed):
sudo apt-get install samtools
Convert SAM to BAM:
samtools view -S -b aligned.sam > aligned.bam
Sort the BAM file:
samtools sort aligned.bam -o aligned_sorted.bam
Index the sorted BAM file:
samtools index aligned_sorted.bam
Step 3: Call Variants
Call variants using bcftools.
Install bcftools (if not already installed):
sudo apt-get install bcftools
Generate a VCF file:
bcftools mpileup -f reference.fasta aligned_sorted.bam | bcftools call -mv -Oz -o variants.vcf.gz
Index the VCF file:
tabix -p vcf variants.vcf.gz
Step 4: Analyze Variants
Analyze the variants to understand the differences between your sequencing data and the reference genome.
View the VCF file:
bcftools view variants.vcf.gz
Filter the variants (if needed):
bcftools filter -i 'QUAL>20' variants.vcf.gz -Oz -o filtered_variants.vcf.gz
tabix -p vcf filtered_variants.vcf.gz
Compare with another VCF file (if you have another VCF file for comparison):
bcftools isec -p output_prefix -n=2 -c all variants.vcf.gz another_variants.vcf.gz
Step 5: Visualize and Interpret
Use visualization tools like IGV (Integrative Genomics Viewer) to visualize the alignments and variants.
Download and install IGV from IGV's official website.
Load your reference genome in IGV.
Load your sorted BAM file (aligned_sorted.bam) and the VCF file (variants.vcf.gz) in IGV.
By following these steps, you can align your long Nanopore reads to the reference genome, call and analyze variants, and visualize the results to identify the differences between your sequencing data and the reference. This process will help you determine if the sequencing data matches the reference genome and identify any variations.
To compare your reassembled contigs with a reference genome to determine if they are from the same sample, you can use various bioinformatics tools and approaches. Here's a step-by-step guide:
Step 1: Align Contigs to the Reference Genome
First, align your reassembled contigs to the reference genome. Tools like BWA, Bowtie2, or minimap2 are commonly used for this purpose. Using BWA:
Index the reference genome:
bwa index CP052959-CP052961.fasta
Align the contigs to the reference genome:
bwa mem CP052959-CP052961.fasta 11108975687_HD46_1_Wt.assembly.fasta > aligned.sam
Step 2: Convert SAM to BAM and Sort
Convert the resulting SAM file to BAM format and sort it using samtools.
Convert SAM to BAM:
samtools view -S -b aligned.sam > aligned.bam
Sort the BAM file:
samtools sort aligned.bam -o aligned_sorted.bam
Index the sorted BAM file:
samtools index aligned_sorted.bam
Step 3: Variant Calling
Call variants using tools like bcftools or GATK.
Using bcftools:
Generate a VCF file:
bcftools mpileup -f CP052959-CP052961.fasta aligned_sorted.bam | bcftools call -mv -Oz -o variants.vcf.gz
Index the VCF file:
tabix -p vcf variants.vcf.gz
Step 4: Analyze Variants
Compare the variants in your reassembled contigs with the reference genome. You can use tools like bcftools to filter and compare these variants.
View and filter the VCF file:
bcftools view variants.vcf.gz
Compare VCF files (if you have another VCF file for a different sample for comparison):
bcftools isec -p output_prefix -n=2 -c all variants1.vcf.gz variants2.vcf.gz
Step 5: Visualize and Interpret
Use visualization tools like IGV (Integrative Genomics Viewer) to visualize the alignments and variants. This can help you manually inspect regions of interest and ensure that your contigs align well with the reference genome.
Load BAM and VCF files in IGV:
Open IGV and load your reference genome.
Load the sorted BAM file (aligned_sorted.bam).
Load the VCF file (variants.vcf.gz).
By following these steps, you can align your reassembled contigs to the reference genome, call and analyze variants, and visualize the results to determine if they are from the same sample. If your contigs align well and have similar variants as the reference genome, it is likely they are from the same sample.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- MicrobiotaProcess_PCA_Group3-4.R processing Data_Karoline_16S_2025
- Phyloseq.Rmd processing Data_Karoline_16S_2025
- Processing RNAseq_2025_WT_vs_ΔIJ_on_ATCC19606
- Stihl Elektro-Heckenschneider
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing RNAseq_2025_WT_vs_ΔIJ_on_ATCC19606
Processing Data_Tam_RNAseq_2024_MHB_vs_Urine_ATCC19606
Workflow using PICRUSt2 for Data_Karoline_16S_2025
Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences