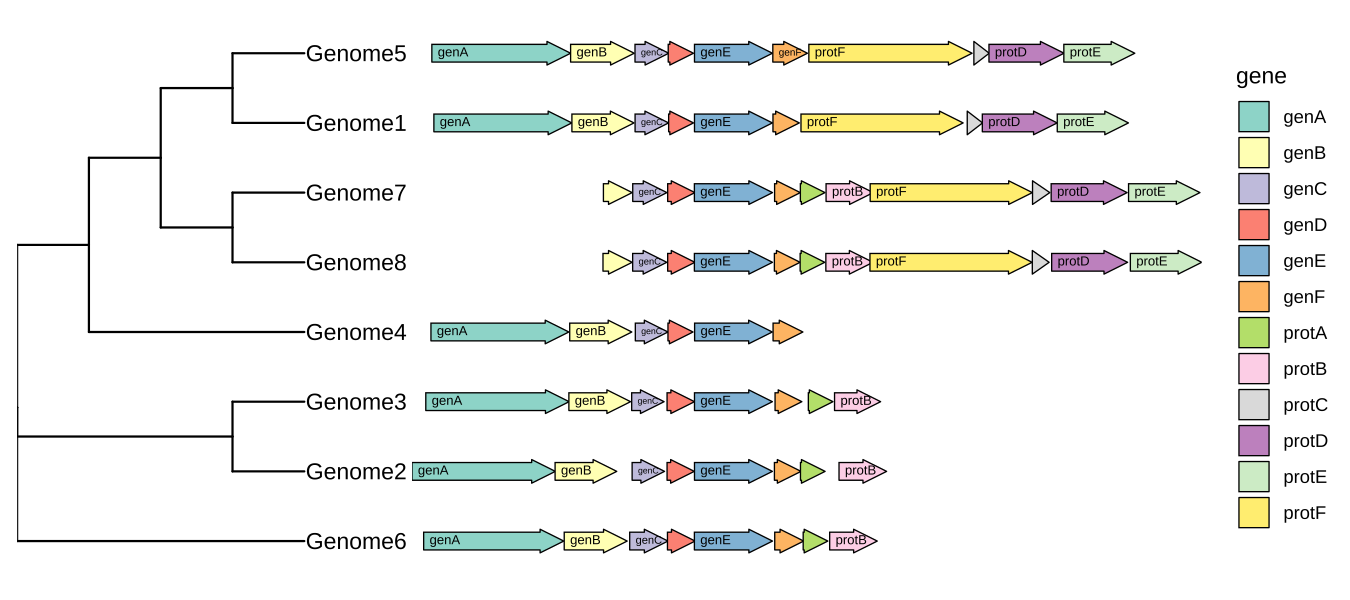
Plot comparative genomics using R-ggtree
gene_x 0 like s 663 view s
Tags: pipeline
https://guangchuangyu.github.io/2016/10/facet_plot-a-general-solution-to-associate-data-with-phylogenetic-tree/
https://yulab-smu.top/treedata-book/chapter13.html
# Load necessary libraries
library(dplyr)
library(ggplot2)
#library(gggenes)
library(ggtree)
library(ape)
# Reconstruct example_genes data
example_genes <- data.frame(
molecule = c(rep("Genome1", 10), rep("Genome2", 8), rep("Genome3", 8), rep("Genome4", 6),
rep("Genome5", 10), rep("Genome6", 8), rep("Genome7", 11), rep("Genome8", 9)),
gene = c("genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF",
"genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD", "protE", "protF",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD"),
start = c(15389, 17301, 18176, 18641, 18999, 20086, 22777, 22986, 24024, 20474,
8345, 10327, 11394, 11878, 12258, 13365, 13726, 14260,
-67849, -65867, -64997, -64507, -64127, -63011, -62550, -62187,
-47353, -45431, -44522, -44070, -43701, -42614,
405113, 407035, 407927, 408387, 408751, 409836, 412621, 412830, 413867, 410335,
65751, 67698, 68605, 69128, 69501, 70614, 71008, 71375,
-9390, -8984, -8500, -8130, -7019, -6662, -6306, -3446, -3188, -2116, -5695,
2, 413, 898, 1268, 2376, 2733, 3089, 5949, 6217),
end = c(17299, 18161, 18640, 18985, 20078, 20451, 22989, 24023, 25010, 22720,
10330, 11181, 11843, 12255, 13337, 13733, 14067, 14919,
-65864, -65013, -64548, -64127, -63048, -62640, -62209, -61549,
-45443, -44571, -44070, -43723, -42625, -42201,
407035, 407916, 408394, 408737, 409830, 410315, 412833, 413870, 414850, 412596,
67691, 68570, 69135, 69511, 70583, 71015, 71349, 72034,
-8992, -8511, -8123, -7048, -6663, -6321, -5653, -3207, -2136, -1127, -3449,
406, 886, 1275, 2350, 2732, 3074, 3742, 6182, 7269),
strand = c("reverse", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse",
"reverse", "reverse", "reverse", "forward", "reverse", "reverse", "reverse", "reverse",
"reverse", "reverse", "forward", "reverse", "forward", "forward",
"forward", "forward", "forward", "reverse", "forward", "forward", "forward", "forward", "forward", "reverse",
"forward", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward",
"reverse", "forward", "reverse", "reverse", "forward", "reverse", "forward", "reverse", "reverse", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse", "reverse"),
orientation = c(1, 0, 1, 0, 1, 1, 1, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1,
0, 1, 0, 0, 1, 1, 1, 0,
1, 1, 1, 0, 0, 0,
0, 0, 0, 0, 1, 0, 1, 0, 0, 1,
0, 1, 0, 0, 1, 1, 0, 0,
1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0,
1, 1, 1, 0, 0, 0, 0, 1, 1)
)
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
#for (i in 1:n) {
# for (j in 1:i) {
# jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
# length(union(genes[[i]], genes[[j]]))
# d[j, i] <- d[i, j] <- 1 - jaccard_sim
# }
#}
#tree <- ape::bionj(d)
# Define the tree structure in Newick format
newick <- "((Genome1:0.1, Genome2:0.2):0.3, (Genome3:0.4, Genome4:0.5):0.6, Genome5:0.7, Genome6:0.8, Genome7:0.9, Genome8:1.0);"
tree <- read.tree(text = newick)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))
#TODO: plot the image combining tree and SNP!!!!
# Load necessary libraries
library(dplyr)
library(ggplot2)
#library(gggenes)
library(ggtree)
library(ape)
# Reconstruct example_genes data
example_genes <- data.frame(
molecule = c(rep("Genome1", 10), rep("Genome2", 8), rep("Genome3", 8), rep("Genome4", 6),
rep("Genome5", 10), rep("Genome6", 8), rep("Genome7", 11), rep("Genome8", 9)),
gene = c("genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genA", "genB", "genC", "genD", "genE", "genF",
"genA", "genB", "genC", "genD", "genE", "genF", "protC", "protD", "protE", "protF",
"genA", "genB", "genC", "genD", "genE", "genF", "protA", "protB",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD", "protE", "protF",
"genB", "genC", "genD", "genE", "genF", "protA", "protB", "protC", "protD"),
start = c(15389, 17301, 18176, 18641, 18999, 20086, 22777, 22986, 24024, 20474,
8345, 10327, 11394, 11878, 12258, 13365, 13726, 14260,
-67849, -65867, -64997, -64507, -64127, -63011, -62550, -62187,
-47353, -45431, -44522, -44070, -43701, -42614,
405113, 407035, 407927, 408387, 408751, 409836, 412621, 412830, 413867, 410335,
65751, 67698, 68605, 69128, 69501, 70614, 71008, 71375,
-9390, -8984, -8500, -8130, -7019, -6662, -6306, -3446, -3188, -2116, -5695,
2, 413, 898, 1268, 2376, 2733, 3089, 5949, 6217),
end = c(17299, 18161, 18640, 18985, 20078, 20451, 22989, 24023, 25010, 22720,
10330, 11181, 11843, 12255, 13337, 13733, 14067, 14919,
-65864, -65013, -64548, -64127, -63048, -62640, -62209, -61549,
-45443, -44571, -44070, -43723, -42625, -42201,
407035, 407916, 408394, 408737, 409830, 410315, 412833, 413870, 414850, 412596,
67691, 68570, 69135, 69511, 70583, 71015, 71349, 72034,
-8992, -8511, -8123, -7048, -6663, -6321, -5653, -3207, -2136, -1127, -3449,
406, 886, 1275, 2350, 2732, 3074, 3742, 6182, 7269),
strand = c("reverse", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse",
"reverse", "reverse", "reverse", "forward", "reverse", "reverse", "reverse", "reverse",
"reverse", "reverse", "forward", "reverse", "forward", "forward",
"forward", "forward", "forward", "reverse", "forward", "forward", "forward", "forward", "forward", "reverse",
"forward", "forward", "reverse", "forward", "reverse", "forward", "forward", "forward",
"reverse", "forward", "reverse", "reverse", "forward", "reverse", "forward", "reverse", "reverse", "forward", "forward",
"forward", "forward", "forward", "forward", "forward", "reverse", "forward", "reverse", "reverse"),
orientation = c(1, 0, 1, 0, 1, 1, 1, 0, 0, 0,
0, 0, 1, 0, 1, 1, 1, 1,
0, 1, 0, 0, 1, 1, 1, 0,
1, 1, 1, 0, 0, 0,
0, 0, 0, 0, 1, 0, 1, 0, 0, 1,
0, 1, 0, 0, 1, 1, 0, 0,
1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0,
1, 1, 1, 0, 0, 0, 0, 1, 1)
)
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
#for (i in 1:n) {
# for (j in 1:i) {
# jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
# length(union(genes[[i]], genes[[j]]))
# d[j, i] <- d[i, j] <- 1 - jaccard_sim
# }
#}
#tree <- ape::bionj(d)
# Define the tree structure in Newick format
newick <- "((Genome1:0.1, Genome2:0.2):0.3, (Genome3:0.4, Genome4:0.5):0.6, Genome5:0.7, Genome6:0.8, Genome7:0.9, Genome8:1.0);"
tree <- read.tree(text = newick)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))
#----
# Install and load necessary packages
install.packages(c("Biostrings", "ape", "ggplot2", "gggenes", "ggtree", "dplyr", "patchwork"))
library(Biostrings)
library(ape)
library(ggplot2)
library(gggenes)
library(ggtree)
library(dplyr)
library(patchwork)
# Function to extract SCCmec/ACME regions
extract_regions <- function(genome, regions_of_interest) {
# Example function to simulate extraction of regions based on provided coordinates
extracted_regions <- genome[genome$start %in% regions_of_interest$start & genome$end %in% regions_of_interest$end,]
return(extracted_regions)
}
# Hypothetical function to read genome data
read_genome <- function(filepath) {
# Read the genome data from file
# Here you would implement code to read your genomic data
# Example placeholder for genomic data
genome <- data.frame(
molecule = rep("Genome1", 10),
gene = paste0("gene", 1:10),
start = seq(100, 1000, by = 100),
end = seq(200, 1100, by = 100),
strand = rep(c("forward", "reverse"), 5),
orientation = sample(0:1, 10, replace = TRUE)
)
return(genome)
}
# Example file paths (replace with your actual file paths)
file_paths <- c("path/to/genome1.fasta", "path/to/genome2.fasta", ..., "path/to/genome10.fasta")
# Example regions of interest (replace with actual coordinates)
regions_of_interest <- data.frame(
start = c(100, 300, 500),
end = c(200, 400, 600)
)
# Extract SCCmec/ACME regions for each genome
all_extracted_regions <- lapply(file_paths, function(file) {
genome <- read_genome(file)
extracted_regions <- extract_regions(genome, regions_of_interest)
return(extracted_regions)
})
# Combine all extracted regions into a single data frame
combined_regions <- do.call(rbind, all_extracted_regions)
# Add a 'genome' column to identify the source genome
combined_regions$genome <- rep(paste0("Genome", 1:length(file_paths)), each = nrow(regions_of_interest))
# Display the combined data frame
print(combined_regions)
#install.packages("gggenes")
get_genes <- function(data, genome) {
filter(data, molecule == genome) %>% pull(gene)
}
g <- unique(example_genes[,1])
n <- length(g)
d <- matrix(nrow = n, ncol = n)
rownames(d) <- colnames(d) <- g
genes <- lapply(g, get_genes, data = example_genes)
for (i in 1:n) {
for (j in 1:i) {
jaccard_sim <- length(intersect(genes[[i]], genes[[j]])) /
length(union(genes[[i]], genes[[j]]))
d[j, i] <- d[i, j] <- 1 - jaccard_sim
}
}
tree <- ape::bionj(d)
p <- ggtree(tree, branch.length='none') +
geom_tiplab() + xlim_tree(5.5) +
geom_facet(mapping = aes(xmin = start, xmax = end, fill = gene),
data = example_genes, geom = geom_motif, panel = 'Alignment',
on = 'genE', label = 'gene', align = 'left') +
scale_fill_brewer(palette = "Set3") +
scale_x_continuous(expand=c(0,0)) +
theme(strip.text=element_blank(),
panel.spacing=unit(0, 'cm'))
facet_widths(p, widths=c(1,2))
#adapt the following code to use a tree file and a vcf-file to draw the figure.
# Install required packages if they are not already installed
if (!requireNamespace("ggtree", quietly = TRUE)) {
BiocManager::install("ggtree")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("vcfR", quietly = TRUE)) {
install.packages("vcfR")
}
# Load libraries
library(tidyverse)
library(vcfR)
library(ggtree)
#library(TDbook)
library(cowplot)
# Define input and output file paths CP133676
#cp raxml-ng/snippy.core.aln.raxml.bestTree raxml-ng/snippy.core.aln.raxml.bestTree_
tree_file <- "raxml-ng/snippy.core.aln.raxml.bestTree_"
vcf_file <- "variants/snippy.core.vcf"
output_image <- "output_image.png"
# Function to load VCF file and extract SNP data
load_vcf <- function(vcf_file) {
vcf <- read.vcfR(vcf_file, verbose = FALSE)
gt <- extract.gt(vcf)
snp_data <- as.data.frame(t(gt))
colnames(snp_data) <- sub("X", "", colnames(snp_data))
rownames(snp_data) <- NULL
snp_data <- snp_data %>%
rownames_to_column("pos") %>%
gather(key = "name", value = "allele", -pos) %>%
mutate(pos = as.numeric(pos), allele = as.character(allele))
return(snp_data)
}
# Function to plot the tree and SNP data
plot_variants <- function(tree_file, vcf_file, output_image) {
# Load the tree from a Newick file
tree <- read.tree(tree_file)
# Load the VCF file and extract SNP data
snp_data <- load_vcf(vcf_file)
# Debug: Print the first few rows of SNP data
print("SNP data:")
print(head(snp_data))
# Visualize the tree
tree_plot <- ggtree(tree)
# Debug: Print the tree plot object
print(tree_plot)
# Filter SNP data to include only variable positions
gapChar <- "?"
snp_filtered <- snp_data %>%
group_by(pos) %>%
filter(any(allele != first(allele) & allele != gapChar & first(allele) != gapChar))
# Debug: Print the first few rows of filtered SNP data
print("Filtered SNP data:")
print(head(snp_filtered))
if (nrow(snp_filtered) == 0) {
warning("No variable SNP positions found.")
} else {
# Plot SNPs
snp_plot <- ggplot(snp_filtered, aes(x = pos, y = name, color = allele)) +
geom_point(shape = '|', size = 5) +
scale_color_manual(values = c("0" = "blue", "1" = "red")) +
theme_minimal() +
theme(axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank())
# Combine the plots
combined_plot <- plot_grid(tree_plot, snp_plot, ncol = 1, align = "v", rel_heights = c(2, 1))
# Save the plot
ggsave(output_image, plot = combined_plot, height = 10, width = 8)
}
}
# Plot the tree and variants
plot_variants(tree_file, vcf_file, output_image)
#------
## load `tree_nwk`, `df_info`, `df_alleles`, and `df_bar_data` from 'TDbook'
tree <- tree_nwk
snps <- df_alleles
snps_strainCols <- snps[1,]
snps<-snps[-1,] # drop strain names
colnames(snps) <- snps_strainCols
gapChar <- "?"
snp <- t(snps)
lsnp <- apply(snp, 1, function(x) {
x != snp[1,] & x != gapChar & snp[1,] != gapChar
})
lsnp <- as.data.frame(lsnp)
lsnp$pos <- as.numeric(rownames(lsnp))
lsnp <- tidyr::gather(lsnp, name, value, -pos)
snp_data <- lsnp[lsnp$value, c("name", "pos")]
## visualize the tree
p <- ggtree(tree)
## attach the sampling information data set
## and add symbols colored by location
p <- p %<+% df_info + geom_tippoint(aes(color=location))
## visualize SNP and Trait data using dot and bar charts,
## and align them based on tree structure
p + geom_facet(panel = "SNP", data = snp_data, geom = geom_point,
mapping=aes(x = pos, color = location), shape = '|') +
geom_facet(panel = "Trait", data = df_bar_data, geom = geom_col,
aes(x = dummy_bar_value, color = location,
fill = location), orientation = 'y', width = .6) +
theme_tree2(legend.position=c(.05, .85))
#------
# Install required packages if they are not already installed
if (!requireNamespace("ggtree", quietly = TRUE)) {
BiocManager::install("ggtree")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("vcfR", quietly = TRUE)) {
install.packages("vcfR")
}
if (!requireNamespace("cowplot", quietly = TRUE)) {
install.packages("cowplot")
}
# Load libraries
library(ggtree)
library(tidyverse)
library(vcfR)
library(cowplot)
# Define input and output file paths
#cp raxml-ng/snippy.core.aln.raxml.bestTree raxml-ng/snippy.core.aln.raxml.bestTree_
tree_file <- "raxml-ng/snippy.core.aln.raxml.bestTree_"
vcf_file <- "variants/snippy.core.vcf"
output_image <- "output_image.png"
# Function to load VCF file and extract SNP data
load_vcf <- function(vcf_file) {
vcf <- read.vcfR(vcf_file, verbose = FALSE)
gt <- extract.gt(vcf)
snp_data <- as.data.frame(t(gt))
colnames(snp_data) <- sub("X", "", colnames(snp_data))
rownames(snp_data) <- NULL
snp_data <- snp_data %>%
rownames_to_column("pos") %>%
gather(key = "name", value = "allele", -pos) %>%
mutate(pos = as.numeric(pos), allele = as.character(allele))
return(snp_data)
}
# Function to plot the tree and SNP data
plot_variants <- function(tree_file, vcf_file, output_image) {
# Load the tree from a Newick file
tree <- read.tree(tree_file)
# Load the VCF file and extract SNP data
snp_data <- load_vcf(vcf_file)
# Debug: Print the first few rows of SNP data
print("SNP data:")
print(head(snp_data))
# Sampling information, ensuring the length of locations matches the number of tree tips
df_info <- data.frame(
name = c("HDRNA_01_K01", "HDRNA_01_K02", "HDRNA_01_K03", "HDRNA_01_K04", "HDRNA_01_K05", "HDRNA_01_K06", "HDRNA_01_K07", "HDRNA_01_K08", "HDRNA_01_K09", "HDRNA_01_K10"),
location = c("Location1", "Location2", "Location3", "Location4", "Location5", "Location6", "Location7", "Location8", "Location9", "Location10")
)
# Debug: Print the df_info data frame
print("Sampling information:")
print(df_info)
# Visualize the tree
p <- ggtree(tree)
# Attach the sampling information data set and add symbols colored by location
p <- p %<+% df_info + geom_tippoint(aes(color = location))
# Debug: Print the tree plot object
print(p)
# Filter SNP data to include only variable positions
gapChar <- "?"
snp_filtered <- snp_data %>%
group_by(pos) %>%
filter(any(allele != first(allele) & allele != gapChar & first(allele) != gapChar))
# Debug: Print the first few rows of filtered SNP data
print("Filtered SNP data:")
print(head(snp_filtered))
if (nrow(snp_filtered) == 0) {
warning("No variable SNP positions found.")
} else {
# Plot tree
tree_plot <- ggtree(tree) + geom_tippoint(aes(color = location))
# Plot SNPs
snp_plot <- ggplot(snp_filtered, aes(x = pos, y = name, color = allele)) +
geom_point(shape = '|', size = 5) +
scale_color_manual(values = c("0" = "blue", "1" = "red")) +
theme_minimal() +
theme(axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank())
# Combine the plots
combined_plot <- plot_grid(tree_plot, snp_plot, ncol = 1, align = "v", rel_heights = c(2, 1))
# Save the plot
ggsave(output_image, plot = combined_plot, height = 10, width = 8)
}
}
# Plot the tree and variants
plot_variants(tree_file, vcf_file, output_image)
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Stihl Elektro-Heckenschneider
- Processing Data_Tam_RNAseq_2024_MHB_vs_Urine_ATCC19606
- Workflow using PICRUSt2 for Data_Karoline_16S_2025
- Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Processing Data_Tam_RNAseq_2024_MHB_vs_Urine_ATCC19606
Workflow using PICRUSt2 for Data_Karoline_16S_2025
Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences