RNA-seq 2024 Ute from raw counts
gene_x 0 like s 989 view s
Tags: processing, pipeline, RNA-seq
1, input files (use R 4.3.3 (/home/jhuang/miniconda3/bin/R))
merged_gene_counts_40samples.txt
merged_gene_counts_WaGa_virus_rounded.txt
merged_gene_counts_MKL-1_virus_rounded.txt
#!! Problem: merge the two files and check if the read number in Dox samples are really lower than that of DMSO samples --> Not really!
2, background knowledge
#Figure 4. MCPyV sT alters the expression of cell surface proteins in WaGa cells. WaGa shRNA scr and shRNA sT cell lines were induced with 1 m g/ml Dox or DMSO 3 d before the experiment. #Surface proteins upregulated during WaGa sT KD, indicating downregulation by sT in WaGa cells, are depicted in a.
#Surface markers downregulated during sT KD, indicating an upregulation by sT in WaGa cells, are displayed in b. The threshold of sT-dependent differential regulation was set to 0.1 log 2diff.
(c) The independent, confirmatory FACS experiments for the surface markers ADAM10, CD44, CD47, and CD95 in WaGa cells described in a and b Dox/DMSO addition.
(d) CD47 surface marker expression in HEK293, WaGa, or nHDF cells overexpressing sT. All experiments were performed in triplicates,
grown as described (Czech-Sioli et al., 2020a).
For knockdown experiments, cells were induced with 1 mg/ml Dox or DMSO.
Plasmids, shRNAs, and lentiviral transduction
- MCPyV sT cDNA was cloned into lentiviral expression plasmid LeGo-iC2 through EcoRI and NotI restriction sites.
- Small interfering RNA transfection
- Analysis was performed using www.webgestalt.org.
- Complete data of transcriptome analysis can be found in Supplementary Tables S1 and S2.
d, day;
Dox, doxycycline;
FDR, false discovery rate;
GO, gene ontology;
GSEA, gene set enrichment analysis;
KD, knockdown;
RNA-Seq, RNA sequencing;
scr, scrambled;
#For knockdown experiments, cells were induced with 1 mg/ml Dox or DMSO.
#shRNA scr, control cell line expressing a scrambled short hairpin RNA;
#shRNA sT, small T antigen‒specific short hairpin RNA;
#sT, small T antigen.
#Figure 1. (b) Immunoblot analysis of total cell lysates from WaGa cells transduced with shRNA scr, shRNA sT, or shRNA sT/LT 3 and 5 d after Dox induction.
#sT and LTtrunc protein expression was detected using the sT/LT-recognizing antibody 2T2 (first panel) and the LT-specific antibody Cm2B4 antibody (third panel); antibody-recognizing actin was used as a loading control.
!Only "shRNA sT with Dox induction" can low the expression of sT; the DMSO control or shRNA scrambled both cannot low the expression.
#Only "shRNA sT with Dox induction" specifically targets and reduces the expression of small T antigen (sT). The DMSO control and the scrambled shRNA do not have this effect because they do not specifically target the sT gene. The Dox induction is necessary to activate the shRNA sT, leading to the knockdown of sT expression.
#In the figure, "shRNA scr" not knocked down, "shRNA sT and DMSO" not knocked down!
#Yes, typically, in systems where doxycycline (Dox) is used to control shRNA expression, all shRNAs under the control of a Dox-inducible promoter would require doxycycline to be induced. Doxycycline is often used in inducible systems like the Tet-On or Tet-Off systems to regulate gene expression, including the expression of short hairpin RNAs (shRNAs). In these systems, without doxycycline, there should be minimal to no expression of the shRNA. When doxycycline is added, it induces the expression of the shRNA, leading to the targeted knockdown of gene expression.
#shRNA (小干扰RNA)敲减的原理是通过介导RNA干扰(RNA interference, RNAi)过程来降低特定基因的表达。shRNA是一种双链RNA分子,其中一条链与目标基因的mRNA序列互补。当shRNA进入细胞后,它被DICER酶切割成小分子干扰RNA (siRNA)。然后siRNA被纳入到RNA诱导沉默复合体(RISC)中。在RISC中,siRNA的一条链被降解,而另一条与目标mRNA互补的链则被用来寻找相对应的mRNA分子。
当siRNA找到与其互补的目标mRNA后,RISC会切割这个mRNA,从而阻止它被翻译成蛋白质。这种降解过程减少了目标基因在细胞内的表达量,从而实现了基因表达的敲减。通过选择特定的目标基因进行敲减,研究人员可以研究这些基因的功能,或者在疾病治疗中靶向这些基因。
#shRNA scrambled(杂乱或无特定靶向的小干扰RNA)是一种设计用于不特定地靶向任何基因的短链RNA。它通常用作对照,以确保观察到的效果是特定于靶向的shRNA(如针对特定基因的shRNA)的结果,而不是由于RNA干扰技术本身或细胞对处理的非特异性反应。shRNA scrambled不会靶向或降低任何特定基因的表达,因此在实验中,它用于与特定靶向的shRNA处理的细胞进行比较,以证明任何观察到的变化是由于特定基因表达的降低。
#DMSO 是二甲基亚砜(Dimethyl sulfoxide)的缩写,这是一种有机溶剂,常用于生物学实验,可以增加细胞膜的渗透性,帮助药物或化合物进入细胞。DMSO 也常作为对照物质使用,在实验中用于对比特定处理的效果。
#Doxycycline(Dox)是一种抗生素,属于四环素类,通常用于治疗各种感染症,如呼吸道感染、尿路感染、眼睛感染等。在分子生物学研究中,Doxycycline 常用于诱导表达系统,如在doxycycline-inducible shRNA表达系统中,Doxycycline 的添加可以诱导特定基因的沉默或表达。
#Difference between design=~shRNA+treatment+shRNA:treatment and design=~shRNA+treatment
#在这两个设计公式中,design=~shRNA+treatment 表示一个没有交互作用项的模型,其中 shRNA 和 treatment 作为独立的因素被考虑。这意味着每个因素的效果是单独评估的,不考虑这两个因素之间可能的相互作用。
#而 design=~shRNA+treatment+shRNA:treatment 这个设计公式包含了一个交互作用项(shRNA:treatment),这表示除了考虑 shRNA 和 treatment 作为独立因素的效果之外,还要考虑它们之间的相互作用。换句话说,该模型会评估 treatment(处理方式)如何依赖于不同 shRNA 的情况下有所不同。
#总之,第一个设计只考虑了独立效果,而第二个设计还考虑了这两个因素的相互作用。在进行实验设计和统计分析时,选择哪一个取决于你的研究假设和数据的特点。
3, common processing for the data MKL+1 + WaGa
#The pipeline finished successfully, but the following samples were skipped,
# - 0505_MKL-1_wt_EV,EV.RNA
#### -------
setwd("/home/jhuang/DATA/Data_Ute/Data_RNA-Seq_MKL-1_WaGa/results_2024_2/featureCounts")
# ---- when BiocManager::install("ggtree") doesn't work, uisng devtools install it (Successful!) ----
#update.packages(ask = FALSE, checkBuilt = TRUE)
#.libPaths()
#install.packages(c("rlang", "cli"))
#rm(list = ls())
#options(repos = BiocManager::repositories())
#if (!requireNamespace("devtools", quietly = TRUE))
# install.packages("devtools")
#devtools::install_github("YuLab-SMU/ggtree")
#install.packages("gplots")
#if (!requireNamespace("BiocManager", quietly = TRUE))
# install.packages("BiocManager")
#BiocManager::install(c("clusterProfiler", "ReactomePA", "DESeq2", "AnnotationDbi", "GenomeInfoDb", "Biostrings"), force=TRUE)
# R(4.3) works well of the saga server!
library("AnnotationDbi")
library("clusterProfiler")
library("ReactomePA")
#library("org.Mm.eg.db")
library(DESeq2)
library(gplots)
[1] gene_name
[2] X042_MKL.1_wt_EV
[3] MKL.1_RNA_147
[4] X042_MKL.1_sT_DMSO #EIGENTLICH _EV
[5] MKL.1_RNA
[6] X0505_MKL.1_scr_DMSO_EV
[7] X0505_MKL.1_sT_DMSO_EV
[8] MKL.1_EV.RNA_87
[9] MKL.1_EV.RNA_27
[10] X042_MKL.1_scr_Dox_EV
[11] X042_MKL.1_scr_DMSO_EV
[12] MKL.1_EV.RNA_118
[13] X0505_MKL.1_scr_Dox_EV
[14] MKL.1_EV.RNA
[15] X042_MKL.1_sT_Dox #EIGENTLICH _EV
[16] X0505_MKL.1_sT_Dox_EV
[17] MKL.1_RNA_118
[18] MKL.1_EV.RNA_2
[19] Geneid.1
[20] gene_name.1
[21] X1605_WaGa_sT_DMSO_EV
[22] WaGa_EV.RNA_118
[23] WaGa_EV.RNA
[24] X2706_WaGa_scr_Dox_EV
[25] X2706_WaGa_sT_DMSO_EV
[26] X1107_WaGa_wt_EV
[27] X1107_WaGa_sT_Dox_EV
[28] WaGa_RNA
[29] X1605_WaGa_scr_DMSO_EV
[30] X2706_WaGa_scr_DMSO_EV
[31] WaGa_EV.RNA_226
[32] X2706_WaGa_sT_Dox_EV
[33] X1605_WaGa_scr_Dox_EV
[34] X1605_WaGa_wt_EV
[35] X1605_WaGa_sT_Dox_EV
[36] X1107_WaGa_scr_DMSO_EV
[37] WaGa_EV.RNA_2
[38] WaGa_EV.RNA_147
[39] WaGa_RNA_118
[40] X1107_WaGa_scr_Dox_EV
[41] X1107_WaGa_sT_DMSO_EV
[42] WaGa_RNA_147
[43] X2706_WaGa_wt_EV
#Geneid gene_name 1605_WaGa_sT_DMSO_EV WaGa_EV-RNA_118 WaGa_EV-RNA 2706_WaGa_scr_Dox_EV 2706_WaGa_sT_DMSO_EV 1107_WaGa_wt_EV 1107_WaGa_sT_Dox_EVWaGa_RNA 1605_WaGa_scr_DMSO_EV 2706_WaGa_scr_DMSO_EV WaGa_EV-RNA_226 2706_WaGa_sT_Dox_EV 1605_WaGa_scr_Dox_EV 1605_WaGa_wt_EV 1605_WaGa_sT_Dox_EV1107_WaGa_scr_DMSO_EV WaGa_EV-RNA_2 WaGa_EV-RNA_147 WaGa_RNA_118 1107_WaGa_scr_Dox_EV 1107_WaGa_sT_DMSO_EV WaGa_RNA_147 2706_WaGa_wt_EV
d.raw_human<- read.delim2("merged_gene_counts_40samples.txt",sep="\t", header=TRUE, row.names=1)
colnames(d.raw_human)<- c("gene_name","X042_MKL.1_wt_EV","MKL.1_RNA_147","X042_MKL.1_sT_DMSO","MKL.1_RNA","X0505_MKL.1_scr_DMSO_EV","X0505_MKL.1_sT_DMSO_EV","MKL.1_EV.RNA_87","MKL.1_EV.RNA_27","X042_MKL.1_scr_Dox_EV","X042_MKL.1_scr_DMSO_EV","MKL.1_EV.RNA_118","X0505_MKL.1_scr_Dox_EV","MKL.1_EV.RNA","X042_MKL.1_sT_Dox","X0505_MKL.1_sT_Dox_EV","MKL.1_RNA_118","MKL.1_EV.RNA_2", "Geneid.1","gene_name.1","X1605_WaGa_sT_DMSO_EV","WaGa_EV.RNA_118","WaGa_EV.RNA","X2706_WaGa_scr_Dox_EV","X2706_WaGa_sT_DMSO_EV","X1107_WaGa_wt_EV","X1107_WaGa_sT_Dox_EV","WaGa_RNA","X1605_WaGa_scr_DMSO_EV","X2706_WaGa_scr_DMSO_EV","WaGa_EV.RNA_226","X2706_WaGa_sT_Dox_EV","X1605_WaGa_scr_Dox_EV","X1605_WaGa_wt_EV","X1605_WaGa_sT_Dox_EV","X1107_WaGa_scr_DMSO_EV","WaGa_EV.RNA_2","WaGa_EV.RNA_147","WaGa_RNA_118","X1107_WaGa_scr_Dox_EV","X1107_WaGa_sT_DMSO_EV","WaGa_RNA_147","X2706_WaGa_wt_EV")
col_order <- c("gene_name", "MKL.1_RNA","MKL.1_RNA_118","MKL.1_RNA_147","MKL.1_EV.RNA","MKL.1_EV.RNA_2","MKL.1_EV.RNA_118","MKL.1_EV.RNA_87","MKL.1_EV.RNA_27","X042_MKL.1_wt_EV","X042_MKL.1_sT_DMSO","X0505_MKL.1_sT_DMSO_EV","X042_MKL.1_scr_DMSO_EV","X0505_MKL.1_scr_DMSO_EV","X042_MKL.1_sT_Dox","X0505_MKL.1_sT_Dox_EV","X042_MKL.1_scr_Dox_EV","X0505_MKL.1_scr_Dox_EV", "Geneid.1","gene_name.1", "WaGa_RNA","WaGa_RNA_118","WaGa_RNA_147", "WaGa_EV.RNA","WaGa_EV.RNA_2","WaGa_EV.RNA_118","WaGa_EV.RNA_147","WaGa_EV.RNA_226","X1107_WaGa_wt_EV","X1605_WaGa_wt_EV","X2706_WaGa_wt_EV", "X1107_WaGa_sT_DMSO_EV","X1605_WaGa_sT_DMSO_EV","X2706_WaGa_sT_DMSO_EV", "X1107_WaGa_scr_DMSO_EV","X1605_WaGa_scr_DMSO_EV","X2706_WaGa_scr_DMSO_EV", "X1107_WaGa_sT_Dox_EV","X1605_WaGa_sT_Dox_EV","X2706_WaGa_sT_Dox_EV", "X1107_WaGa_scr_Dox_EV","X1605_WaGa_scr_Dox_EV","X2706_WaGa_scr_Dox_EV")
reordered.raw_human <- d.raw_human[,col_order]
d.raw_virus <- read.delim2("merged_gene_counts_virus_rounded.txt",sep="\t", header=TRUE, row.names=1)
reordered.raw_virus <- d.raw_virus[,col_order]
identical(colnames(reordered.raw_human), colnames(reordered.raw_virus))
reordered.raw <- rbind(reordered.raw_human, reordered.raw_virus)
#rename
#colnames(reordered.raw) <- c("gene_name", "MKL-1 RNA","MKL-1 RNA 118","MKL-1 RNA 147", "MKL-1 EV","MKL-1 EV 2","MKL-1 EV 118","MKL-1 EV 87","MKL-1 EV 27","MKL-1 EV 042", "MKL-1 EV sT DMSO 042","MKL-1 EV sT DMSO 0505","MKL-1 EV scr DMSO 042","MKL-1 EV scr DMSO 0505","MKL-1 EV sT Dox 042","MKL-1 EV sT Dox 0505","MKL-1 EV scr Dox 042","MKL-1 EV scr Dox 0505", "Geneid.1","gene_name.1", "WaGa RNA","WaGa RNA 118","WaGa RNA 147", "WaGa EV","WaGa EV 2","WaGa EV 118","WaGa EV 147","WaGa EV 226","WaGa EV 1107","WaGa EV 1605","WaGa EV 2706", "WaGa EV sT DMSO 1107","WaGa EV sT DMSO 1605","WaGa EV sT DMSO 2706", "WaGa EV scr DMSO 1107","WaGa EV scr DMSO 1605","WaGa EV scr DMSO 2706", "WaGa EV sT Dox 1107","WaGa EV sT Dox 1605","WaGa EV sT Dox 2706", "WaGa EV scr Dox 1107","WaGa EV scr Dox 1605","WaGa EV scr Dox 2706")
colnames(reordered.raw) <- c("gene_name", "MKL-1 parental cell RNA","MKL-1 parental cell RNA 118","MKL-1 parental cell RNA 147", "MKL-1 wt EV RNA","MKL-1 wt EV RNA 2","MKL-1 wt EV RNA 118","MKL-1 wt EV RNA 87","MKL-1 wt EV RNA 27","MKL-1 wt EV RNA 042", "MKL-1 sT DMSO EV RNA 042","MKL-1 sT DMSO EV RNA 0505", "MKL-1 scr DMSO EV RNA 042","MKL-1 scr DMSO EV RNA 0505", "MKL-1 sT Dox EV RNA 042","MKL-1 sT Dox EV RNA 0505", "MKL-1 scr Dox EV RNA 042","MKL-1 scr Dox EV RNA 0505", "Geneid.1","gene_name.1", "WaGa parental cell RNA","WaGa parental cell RNA 118","WaGa parental cell RNA 147", "WaGa wt EV RNA","WaGa wt EV RNA 2","WaGa wt EV RNA 118","WaGa wt EV RNA 147","WaGa wt EV RNA 226","WaGa wt EV RNA 1107","WaGa wt EV RNA 1605","WaGa wt EV RNA 2706", "WaGa sT DMSO EV RNA 1107","WaGa sT DMSO EV RNA 1605","WaGa sT DMSO EV RNA 2706", "WaGa scr DMSO EV RNA 1107","WaGa scr DMSO EV RNA 1605","WaGa scr DMSO EV RNA 2706", "WaGa sT Dox EV RNA 1107","WaGa sT Dox EV RNA 1605","WaGa sT Dox EV RNA 2706", "WaGa scr Dox EV RNA 1107","WaGa scr Dox EV RNA 1605","WaGa scr Dox EV RNA 2706")
reordered.raw$gene_name <- NULL
reordered.raw$Geneid.1 <- NULL
reordered.raw$gene_name.1 <- NULL
write.csv(reordered.raw, file="counts.txt")
#IMPORTANT that we should filter the data with the counts in the STEP!
d <- reordered.raw[rowSums(reordered.raw>3)>2,]
condition_for_pca = as.factor(c("RNA","RNA","RNA","EV","EV","EV","EV","EV","EV","sT.DMSO","sT.DMSO","scr.DMSO","scr.DMSO","sT.Dox","sT.Dox","scr.Dox","scr.Dox", "RNA","RNA","RNA","EV","EV","EV","EV","EV","EV","EV","EV","sT.DMSO","sT.DMSO","sT.DMSO","scr.DMSO","scr.DMSO","scr.DMSO","sT.Dox","sT.Dox","sT.Dox","scr.Dox","scr.Dox","scr.Dox"))
condition = as.factor(c("MKL1.RNA","MKL1.RNA","MKL1.RNA","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.sT.DMSO","MKL1.sT.DMSO","MKL1.scr.DMSO","MKL1.scr.DMSO","MKL1.sT.Dox","MKL1.sT.Dox","MKL1.scr.Dox","MKL1.scr.Dox", "WaGa.RNA","WaGa.RNA","WaGa.RNA","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.sT.DMSO","WaGa.sT.DMSO","WaGa.sT.DMSO","WaGa.scr.DMSO","WaGa.scr.DMSO","WaGa.scr.DMSO","WaGa.sT.Dox","WaGa.sT.Dox","WaGa.sT.Dox","WaGa.scr.Dox","WaGa.scr.Dox","WaGa.scr.Dox"))
#not sure if they rep1 in the first time is the same to the second time.
donor = as.factor(c("1","118","147", "1","2","118","87","27","042", "042","0505","042","0505","042","0505","042","0505", "1","118","147", "1","2","118","147","226","1107","1605","2706", "1107","1605","2706","1107","1605","2706","1107","1605","2706","1107","1605","2706"))
batch = as.factor(c("2021.08","2021.09","2021.09","2021.08","2021.08","2021.09","2021.09","2021.09","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08", "2021.08","2021.09","2021.09","2021.08","2021.08","2021.09","2021.09","2021.09","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11"))
cell.line = as.factor(c("MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1", "WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa"))
ids = as.factor(c("MKL1.RNA","MKL1.RNA.118","MKL1.RNA.147", "MKL1.EV","MKL1.EV.2","MKL1.EV.118","MKL1.EV.87","MKL1.EV.27","MKL1.EV.042", "MKL1.EV.sT.DMSO.042","MKL1.EV.sT.DMSO.0505", "MKL1.EV.scr.DMSO.042","MKL1.EV.scr.DMSO.0505", "MKL1.EV.sT.Dox.042","MKL1.EV.sT.Dox.0505", "MKL1.EV.scr.Dox.042","MKL1.EV.scr.Dox.0505", "WaGa.RNA","WaGa.RNA.118","WaGa.RNA.147", "WaGa.EV","WaGa.EV.2","WaGa.EV.118","WaGa.EV.147","WaGa.EV.226","WaGa.EV.1107","WaGa.EV.1605","WaGa.EV.2706", "WaGa.EV.sT.DMSO.1107","WaGa.EV.sT.DMSO.1605","WaGa.EV.sT.DMSO.2706", "WaGa.EV.scr.DMSO.1107","WaGa.EV.scr.DMSO.1605","WaGa.EV.scr.DMSO.2706", "WaGa.EV.sT.Dox.1107","WaGa.EV.sT.Dox.1605","WaGa.EV.sT.Dox.2706", "WaGa.EV.scr.Dox.1107","WaGa.EV.scr.Dox.1605","WaGa.EV.scr.Dox.2706"))
#DEL ids = as.factor(c("MKL.1_RNA","MKL.1_RNA_118","MKL.1_RNA_147","MKL.1_EV.RNA","MKL.1_EV.RNA_2","MKL.1_EV.RNA_118","MKL.1_EV.RNA_87","MKL.1_EV.RNA_27","042_MKL.1_wt_EV","042_MKL.1_sT_DMSO","0505_MKL.1_sT_DMSO_EV","042_MKL.1_scr_DMSO_EV","0505_MKL.1_scr_DMSO_EV","042_MKL.1_sT_Dox","0505_MKL.1_sT_Dox_EV","042_MKL.1_scr_Dox_EV","0505_MKL.1_scr_Dox_EV"))
#IMPORTANT: using d instead of reordered.raw.
#cData = data.frame(row.names=colnames(d), condition=condition, batch=batch, ids=ids)
#dds<-DESeqDataSetFromMatrix(countData=d, colData=cData, design=~batch+condition)
#cData = data.frame(row.names=colnames(d), condition=condition, ids=ids)
#dds<-DESeqDataSetFromMatrix(countData=d, colData=cData, design=~condition)
cData = data.frame(row.names=colnames(d), condition=condition, donor=donor, batch=batch, cell.line=cell.line, ids=ids)
#dds<-DESeqDataSetFromMatrix(countData=d, colData=cData, design=~batch+condition_for_pca)
dds<-DESeqDataSetFromMatrix(countData=d, colData=cData, design=~batch+condition)
#rld <- rlogTransformation(dds)
rld <- vst(dds)
#--> [OPTION] goto DEG_Heatmap_Drawing!
4, preparing the data for PCA_MKL1 and PCA_WaGa drawing
d_WaGa <- d[, !grepl("parental|MKL-1", names(d))]
condition = as.factor(c("WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.EV","WaGa.sT.DMSO","WaGa.sT.DMSO","WaGa.sT.DMSO","WaGa.scr.DMSO","WaGa.scr.DMSO","WaGa.scr.DMSO","WaGa.sT.Dox","WaGa.sT.Dox","WaGa.sT.Dox","WaGa.scr.Dox","WaGa.scr.Dox","WaGa.scr.Dox"))
donor = as.factor(c("1","2","118","147","226","1107","1605","2706", "1107","1605","2706","1107","1605","2706","1107","1605","2706","1107","1605","2706"))
batch = as.factor(c("2021.08","2021.08","2021.09","2021.09","2021.09","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11","2022.11"))
cell.line = as.factor(c("WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa"))
ids = as.factor(c("WaGa.EV","WaGa.EV.2","WaGa.EV.118","WaGa.EV.147","WaGa.EV.226","WaGa.EV.1107","WaGa.EV.1605","WaGa.EV.2706", "WaGa.EV.sT.DMSO.1107","WaGa.EV.sT.DMSO.1605","WaGa.EV.sT.DMSO.2706", "WaGa.EV.scr.DMSO.1107","WaGa.EV.scr.DMSO.1605","WaGa.EV.scr.DMSO.2706", "WaGa.EV.sT.Dox.1107","WaGa.EV.sT.Dox.1605","WaGa.EV.sT.Dox.2706", "WaGa.EV.scr.Dox.1107","WaGa.EV.scr.Dox.1605","WaGa.EV.scr.Dox.2706"))
cData = data.frame(row.names=colnames(d_WaGa), condition=condition, donor=donor, batch=batch, cell.line=cell.line, ids=ids)
dds_WaGa<-DESeqDataSetFromMatrix(countData=d_WaGa, colData=cData, design=~batch+condition)
rld_WaGa <- vst(dds_WaGa)
d_MKL1 <- d[, !grepl("parental|WaGa", names(d))]
condition = as.factor(c("MKL1.EV","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.EV","MKL1.sT.DMSO","MKL1.sT.DMSO","MKL1.scr.DMSO","MKL1.scr.DMSO","MKL1.sT.Dox","MKL1.sT.Dox","MKL1.scr.Dox","MKL1.scr.Dox"))
donor = as.factor(c("1","2","118","87","27","042", "042","0505","042","0505","042","0505","042","0505"))
batch = as.factor(c("2021.08","2021.08","2021.09","2021.09","2021.09","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08","2022.08"))
cell.line = as.factor(c("MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1"))
ids = as.factor(c("MKL1.EV","MKL1.EV.2","MKL1.EV.118","MKL1.EV.87","MKL1.EV.27","MKL1.EV.042", "MKL1.EV.sT.DMSO.042","MKL1.EV.sT.DMSO.0505", "MKL1.EV.scr.DMSO.042","MKL1.EV.scr.DMSO.0505", "MKL1.EV.sT.Dox.042","MKL1.EV.sT.Dox.0505", "MKL1.EV.scr.Dox.042","MKL1.EV.scr.Dox.0505"))
cData = data.frame(row.names=colnames(d_MKL1), condition=condition, donor=donor, batch=batch, cell.line=cell.line, ids=ids)
dds_MKL1<-DESeqDataSetFromMatrix(countData=d_MKL1, colData=cData, design=~batch+condition)
rld_MKL1 <- vst(dds_MKL1)
# -- before pca --
png("pca_before_batch_correction2.png", 1200, 800)
#plotPCA(rld, intgroup=c("condition"))
plotPCA(rld, intgroup = c("condition", "batch"))
#plotPCA(rld, intgroup = c("condition", "ids"))
#plotPCA(rld, "batch")
dev.off()
#73% (PC1), 11% (PC2) 8% (PC3)
5, drawing PCA MKL1+WaGa
# -- construct a data structure (merged_df) as above with data and pc --
library(ggplot2)
data <- plotPCA(rld, intgroup=c("condition", "batch", "cell.line"), returnData=TRUE)
write.csv(data, file="plotPCA_data.csv")
#calculate all PCs including PC3 with the following codes
library(genefilter)
ntop <- 500
rv <- rowVars(assay(rld))
select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))]
mat <- t( assay(rld)[select, ] )
pc <- prcomp(mat)
summary(pc)
#Cumulative Proportion 0.7651 0.07746 0.05608 0.01405 0.00984 0.00857 0.00689 (with rld <- rlogTransformation(dds))
#Proportion of Variance 0.7286 0.1060 0.07657 0.01445 0.01042 0.00705 0.00578 (with rld <- vst(dds))
pc$x[,1:3]
#df_pc <- data.frame(pc$x[,1:3])
df_pc <- data.frame(pc$x)
#> head(data)
# PC1 PC2 group condition batch
#MKL-1 RNA -71.108137 -2.054660 MKL1.RNA:2021.08 MKL1.RNA 2021.08
#MKL-1 RNA 118 -62.978513 -6.906138 MKL1.RNA:2021.09 MKL1.RNA 2021.09
#MKL-1 RNA 147 -77.698768 -3.355581 MKL1.RNA:2021.09 MKL1.RNA 2021.09
#MKL-1 EV -49.482607 -25.469602 MKL1.EV:2021.08 MKL1.EV 2021.08
#MKL-1 EV 2 -19.805802 -23.850122 MKL1.EV:2021.08 MKL1.EV 2021.08
#MKL-1 EV 118 2.264943 -22.114427 MKL1.EV:2021.09 MKL1.EV 2021.09
#> head(df_pc)
# PC1 PC2 PC3 PC4 PC5 PC6
#MKL-1 RNA -71.108137 -2.054660 16.617315 -1.7281595 6.275895 -2.3597341
#MKL-1 RNA 118 -62.978513 -6.906138 9.973149 -2.4119718 7.778461 -2.3797116
#MKL-1 RNA 147 -77.698768 -3.355581 17.062958 -2.0865184 6.100746 -0.6303835
#MKL-1 EV -49.482607 -25.469602 -2.410902 2.2679166 5.300198 10.5329350
#MKL-1 EV 2 -19.805802 -23.850122 -1.704857 -2.3895896 -1.742060 -3.7750239
#MKL-1 EV 118 2.264943 -22.114427 -4.991651 -0.3336808 -2.973397 -4.5959970
#Note it is a little different since we added the virus-gene-expression in the table!
identical(rownames(data), rownames(df_pc)) #-->TRUE
data$PC1 <- NULL
data$PC2 <- NULL
merged_df <- merge(data, df_pc, by = "row.names")
#merged_df <- merged_df[, -1]
row.names(merged_df) <- merged_df$Row.names
merged_df$Row.names <- NULL # remove the "name" column
merged_df$name <- NULL
merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","PC11","PC12","PC13","PC14","PC15","PC16","PC17","PC18","PC19","PC20","PC21","PC22","PC23","PC24","PC25","PC26","PC27","PC28","PC29","PC30","PC31","PC32","PC33","PC34","PC35","PC36","PC37","PC38","PC39","PC40","group","condition","batch","cell.line")]
write.csv(merged_df, file="merged_df_40PCs.csv")
#using the python script to draw the 3D PCA-plot.
# need to install plotly with pip ~/.local/bin/pip3 in .local/bin/python3.10/site-packages
#python3 ~/Scripts/PCA_3D_drawing.py
python3 ~/Scripts/PCA_3D_drawing.py merged_df_40PCs.csv
6, drawing PCA WaGa
library(ggplot2)
data <- plotPCA(rld_WaGa, intgroup=c("condition", "batch", "cell.line"), returnData=TRUE)
write.csv(data, file="plotPCA_data_WaGa.csv")
#calculate all PCs including PC3 with the following codes
library(genefilter)
ntop <- 500
rv <- rowVars(assay(rld_WaGa))
select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))]
mat <- t( assay(rld_WaGa)[select, ] )
pc <- prcomp(mat)
summary(pc)
#Proportion of Variance 0.6342 0.06491 0.06044 --> 0.63, 0.06, 0.06
pc$x[,1:3]
#df_pc <- data.frame(pc$x[,1:3])
df_pc <- data.frame(pc$x)
#Note it is a little different since we added the virus-gene-expression in the table!
identical(rownames(data), rownames(df_pc)) #-->TRUE
data$PC1 <- NULL
data$PC2 <- NULL
merged_df <- merge(data, df_pc, by = "row.names")
#merged_df <- merged_df[, -1]
row.names(merged_df) <- merged_df$Row.names
merged_df$Row.names <- NULL # remove the "name" column
merged_df$name <- NULL
merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","PC11","PC12","PC13","PC14","PC15","PC16","PC17","PC18","PC19","PC20","group","condition","batch","cell.line")]
write.csv(merged_df, file="merged_df_WaGa.csv")
python3 ~/Scripts/PCA_3D_drawing_WaGa.py
7, drawing PCA MKL1
library(ggplot2)
data <- plotPCA(rld_MKL1, intgroup=c("condition", "batch", "cell.line"), returnData=TRUE)
write.csv(data, file="plotPCA_data_MKL1.csv")
#calculate all PCs including PC3 with the following codes
library(genefilter)
ntop <- 500
rv <- rowVars(assay(rld_MKL1))
select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))]
mat <- t( assay(rld_MKL1)[select, ] )
pc <- prcomp(mat)
summary(pc)
#Proportion of Variance 0.7549 0.05717 0.04503 --> 0.75, 0.06, 0.05
pc$x[,1:3]
#df_pc <- data.frame(pc$x[,1:3])
df_pc <- data.frame(pc$x)
#Note it is a little different since we added the virus-gene-expression in the table!
identical(rownames(data), rownames(df_pc)) #-->TRUE
data$PC1 <- NULL
data$PC2 <- NULL
merged_df <- merge(data, df_pc, by = "row.names")
#merged_df <- merged_df[, -1]
row.names(merged_df) <- merged_df$Row.names
merged_df$Row.names <- NULL # remove the "name" column
merged_df$name <- NULL
merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","PC11","PC12","PC13","PC14","group","condition","batch","cell.line")]
write.csv(merged_df, file="merged_df_MKL1.csv")
python3 ~/Scripts/PCA_3D_drawing_MKL1.py
# -- before heatmap --
## generate the pairwise comparison between samples
library(gplots)
library("RColorBrewer")
png("heatmap_before_donor_correction.png", 1200, 800)
distsRL <- dist(t(assay(rld)))
mat <- as.matrix(distsRL)
#paste( rld$dex, rld$cell, sep="-" )
#rownames(mat) <- colnames(mat) <- with(colData(dds),paste(condition,batch, sep=":"))
#rownames(mat) <- colnames(mat) <- with(colData(dds),paste(condition,ids, sep=":"))
hc <- hclust(distsRL)
hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100)
heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13))
dev.off()
# -- remove batch effect --
mat <- assay(rld)
mm <- model.matrix(~condition, colData(rld))
mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm)
assay(rld) <- mat
# -- after pca --
png("pca_after_batch_correction.png", 1200, 800)
svg("pca_after_batch_correction.svg")
plotPCA(rld, intgroup=c("condition"))
dev.off()
library(ggplot2)
data <- plotPCA(rld, intgroup=c("condition", "cell.line"), returnData=TRUE)
colnames(data) <- c("PC1","PC2","group2","Group","Cell.line","name")
percentVar <- round(100 * attr(data, "percentVar"))
#, shape=donor
#png("pca6.png", 1000, 1000)
svg("pca6.svg", 10, 8)
ggplot(data, aes(PC1, PC2, color=Group, shape=Cell.line)) +
geom_point(size=3) +
scale_color_manual(values = c("RNA" = "grey",
"EV"="cyan",
"scr.DMSO"="#b2df8a",
"sT.DMSO"="#33a02c",
"scr.Dox"="#fb9a99",
"sT.Dox"="#e31a1c")) +
xlab(paste0("PC1: ",percentVar[1],"% variance")) +
ylab(paste0("PC2: ",percentVar[2],"% variance")) +
coord_fixed()
dev.off()
# -- after heatmap --
## generate the pairwise comparison between samples
png("heatmap_after_donor_correction.png", 1200, 800)
distsRL <- dist(t(assay(rld)))
mat <- as.matrix(distsRL)
rownames(mat) <- colnames(mat) <- with(colData(dds),paste(condition,donor, sep=":"))
#rownames(mat) <- colnames(mat) <- with(colData(dds),paste(condition,ids, sep=":"))
hc <- hclust(distsRL)
hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100)
heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13))
dev.off()
#convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor
sizeFactors(dds)
#NULL
dds <- estimateSizeFactors(dds)
> sizeFactors(dds)
MKL-1 parental cell RNA MKL-1 parental cell RNA 118
2.5689118 1.6270659
MKL-1 parental cell RNA 147 MKL-1 wt EV RNA
1.9241464 1.5296877
MKL-1 wt EV RNA 2 MKL-1 wt EV RNA 118
1.2829153 0.6786713
MKL-1 wt EV RNA 87 MKL-1 wt EV RNA 27
0.6741590 0.4176156
MKL-1 wt EV RNA 042 MKL-1 sT DMSO EV RNA 042
1.3360156 0.9539241
MKL-1 sT DMSO EV RNA 0505 MKL-1 scr DMSO EV RNA 042
1.7357438 1.2957555
MKL-1 scr DMSO EV RNA 0505 MKL-1 sT Dox EV RNA 042
1.0908450 1.3657925
MKL-1 sT Dox EV RNA 0505 MKL-1 scr Dox EV RNA 042
1.1221456 1.4708191
MKL-1 scr Dox EV RNA 0505 WaGa parental cell RNA
1.1242096 2.1097851
WaGa parental cell RNA 118 WaGa parental cell RNA 147
1.6925780 1.6712182
WaGa wt EV RNA WaGa wt EV RNA 2
0.6021352 1.2486966
WaGa wt EV RNA 118 WaGa wt EV RNA 147
0.4518733 0.5695057
WaGa wt EV RNA 226 WaGa wt EV RNA 1107
0.5449802 1.4885064
WaGa wt EV RNA 1605 WaGa wt EV RNA 2706
1.2631181 0.7678146
WaGa sT DMSO EV RNA 1107 WaGa sT DMSO EV RNA 1605
0.7981256 0.9054709
WaGa sT DMSO EV RNA 2706 WaGa scr DMSO EV RNA 1107
1.0603277 0.7927572
WaGa scr DMSO EV RNA 1605 WaGa scr DMSO EV RNA 2706
0.8115785 1.0659662
WaGa sT Dox EV RNA 1107 WaGa sT Dox EV RNA 1605
0.8553457 0.9547341
WaGa sT Dox EV RNA 2706 WaGa scr Dox EV RNA 1107
0.9081678 0.7440897
WaGa scr Dox EV RNA 1605 WaGa scr Dox EV RNA 2706
0.8735490 1.0146989
raw_counts <- counts(dds)
normalized_counts <- counts(dds, normalized=TRUE)
write.table(raw_counts, file="raw_counts.txt", sep="\t", quote=F, col.names=NA)
write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA)
#bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_RNAAligned.sortedByCoord.out.markDups.bam -o WaGa_RNA.bw --binSize 10 --scaleFactor 0.4958732 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_RNA_118Aligned.sortedByCoord.out.markDups.bam -o WaGa_RNA_118.bw --binSize 10 --scaleFactor 0.6013898 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_RNA_147Aligned.sortedByCoord.out.markDups.bam -o WaGa_RNA_147.bw --binSize 10 --scaleFactor 0.6154516 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_RNAAligned.sortedByCoord.out.markDups.bam -o MKL1_RNA.bw --binSize 10 --scaleFactor 0.4078775 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_RNA_118Aligned.sortedByCoord.out.markDups.bam -o MKL1_RNA_118.bw --binSize 10 --scaleFactor 0.6525297 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_RNA_147Aligned.sortedByCoord.out.markDups.bam -o MKL1_RNA_147.bw --binSize 10 --scaleFactor 0.5331748 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_EV_RNAAligned.sortedByCoord.out.markDups.bam -o WaGa_EV_RNA.bw --binSize 10 --scaleFactor 1.733964 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_EV_RNA_2Aligned.sortedByCoord.out.markDups.bam -o WaGa_EV_RNA_2.bw --binSize 10 --scaleFactor 0.7761222 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_EV_RNA_118Aligned.sortedByCoord.out.markDups.bam -o WaGa_EV_RNA_118.bw --binSize 10 --scaleFactor 2.222971 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_EV_RNA_147Aligned.sortedByCoord.out.markDups.bam -o WaGa_EV_RNA_147.bw --binSize 10 --scaleFactor 1.679485 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/WaGa_EV_RNA_226Aligned.sortedByCoord.out.markDups.bam -o WaGa_EV_RNA_226.bw --binSize 10 --scaleFactor 1.901282 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_EV_RNAAligned.sortedByCoord.out.markDups.bam -o MKL1_EV_RNA.bw --binSize 10 --scaleFactor 0.6469583 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_EV_RNA_2Aligned.sortedByCoord.out.markDups.bam -o MKL1_EV_RNA_2.bw --binSize 10 --scaleFactor 0.6877478 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_EV_RNA_27Aligned.sortedByCoord.out.markDups.bam -o MKL1_EV_RNA_27.bw --binSize 10 --scaleFactor 2.462424 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_EV_RNA_87Aligned.sortedByCoord.out.markDups.bam -o MKL1_EV_RNA_87.bw --binSize 10 --scaleFactor 1.411154 --effectiveGenomeSize 2864785220
bamCoverage --bam ../markDuplicates/MKL1_EV_RNA_118Aligned.sortedByCoord.out.markDups.bam -o MKL1_EV_RNA_118.bw --binSize 10 --scaleFactor 1.544239 --effectiveGenomeSize 2864785220
setwd("degenes")
#---- * to untreated and wildtype ----
dds$condition <- relevel(dds$condition, "MKL1.RNA")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("MKL1.EV_vs_MKL1.RNA")
dds$condition <- relevel(dds$condition, "MKL1.EV")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("MKL1.sT.DMSO_vs_MKL1.EV", "MKL1.sT.Dox_vs_MKL1.EV", "MKL1.scr.DMSO_vs_MKL1.EV", "MKL1.scr.Dox_vs_MKL1.EV")
#For internal comparisons between sT.DMSO, sT.Dox, scr.DMSO and scr.Dox see the chapter "Do separate shRNA and treatment analysis"
dds$condition <- relevel(dds$condition, "WaGa.RNA")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("WaGa.EV_vs_WaGa.RNA")
dds$condition <- relevel(dds$condition, "WaGa.EV")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("WaGa.sT.DMSO_vs_WaGa.EV", "WaGa.sT.Dox_vs_WaGa.EV", "WaGa.scr.DMSO_vs_WaGa.EV", "WaGa.scr.Dox_vs_WaGa.EV")
#For internal comparisons between sT.DMSO, sT.Dox, scr.DMSO and scr.Dox see the chapter "Do separate shRNA and treatment analysis"
dds$condition <- relevel(dds$condition, "MKL1.RNA")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("WaGa.RNA_vs_MKL1.RNA")
dds$condition <- relevel(dds$condition, "MKL1.EV")
dds = DESeq(dds, betaPrior=FALSE)
resultsNames(dds)
clist <- c("WaGa.EV_vs_MKL1.EV")
##https://bioconductor.statistik.tu-dortmund.de/packages/3.7/data/annotation/
#BiocManager::install("EnsDb.Mmusculus.v79")
#library(EnsDb.Mmusculus.v79)
#edb <- EnsDb.Mmusculus.v79
#https://bioconductor.org/packages/release/bioc/vignettes/biomaRt/inst/doc/accessing_ensembl.html#selecting-an-ensembl-biomart-database-and-dataset
#https://bioconductor.org/packages/release/bioc/vignettes/biomaRt/inst/doc/accessing_ensembl.html#selecting-an-ensembl-biomart-database-and-dataset
library(biomaRt)
listEnsembl()
listMarts()
#ensembl <- useEnsembl(biomart = "genes", mirror="asia") # default is Mouse strains 104
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "mmusculus_gene_ensembl", mirror = "www")
#ensembl = useMart("ensembl_mart_44", dataset="hsapiens_gene_ensembl",archive=TRUE, mysql=TRUE)
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "mmusculus_gene_ensembl", version="104")
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="86")
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="GRCh37")
#--> total 69, 27 GRCh38.p7 and 39 GRCm38.p4
#DEBUG: use R version 4.3.3, the version 104 is not loadable, using version 112 instead!
#ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="104")
#Error in bmRequest(request = request, httr_config = httr_config, verbose = verbose) :
# Internal Server Error (HTTP 500).
#--> total 202 80 GRCh38.p13 107 GRCm39
#80 hsapiens_gene_ensembl Human genes (GRCh38.p13) GRCh38.p13
#107 mmusculus_gene_ensembl Mouse genes (GRCm39) GRCm39
ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="112")
#80 GRCh38.p14
#107 GRCm39
datasets <- listDatasets(ensembl)
> listEnsemblArchives()
name date url version
1 Ensembl GRCh37 Feb 2014 http://grch37.ensembl.org GRCh37 *
2 Ensembl 104 May 2021 http://may2021.archive.ensembl.org 104 *
3 Ensembl 103 Feb 2021 http://feb2021.archive.ensembl.org 103
4 Ensembl 102 Nov 2020 http://nov2020.archive.ensembl.org 102
attributes = listAttributes(ensembl)
attributes[1:25,]
#library("dplyr")
for (i in clist) {
#i<-clist[1]
contrast = paste("condition", i, sep="_")
res = results(dds, name=contrast)
res <- res[!is.na(res$log2FoldChange),]
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID"))
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID"))
# In the ENSEMBL-database, GENEID is ENSEMBL-ID.
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND"
#geness <- geness[!duplicated(geness$GENEID), ]
#using getBM replacing AnnotationDbi::select
#filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids.
geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = rownames(res),
mart = ensembl)
geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE)
#merge by column by common colunmn name, in the case "GENEID"
res$ENSEMBL = rownames(res)
identical(rownames(res), rownames(geness_uniq))
res_df <- as.data.frame(res)
geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL")
dim(geness_res)
rownames(geness_res) <- geness_res$ensembl_gene_id
geness_res$ensembl_gene_id <- NULL
write.csv(as.data.frame(geness_res[order(geness_res$pvalue),]), file = paste(i, "all.txt", sep="-"))
up <- subset(geness_res, padj<=0.05 & log2FoldChange>=2)
down <- subset(geness_res, padj<=0.05 & log2FoldChange<=-2)
write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-"))
write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-"))
}
8, do separate shRNA and treatment analysis
#In DESeq2, resultsNames(dds) gives you the names of the coefficients in your linear model that DESeq2 has fit to the count data. Each name corresponds to a comparison between levels of #your factors (or the interaction between them) that DESeq2 can test for differential expression. Here's what each of these results names likely means based on typical DESeq2 output:
# * "Intercept": This represents the base level of expression estimated by the model. If your factors are shRNA and treatment, and if the reference levels for these factors are scr and DMSO, respectively, then the "Intercept" represents the expression level for the reference group, which in this case is cells treated with scr and DMSO.
# * "shRNA_sT_vs_scr": This represents the comparison between the sT and scr levels of the shRNA factor. This means it represents the log2 fold change in expression from the scr condition to the sT condition, while keeping the treatment factor constant (at its reference level, likely DMSO).
# * "treatment_Dox_vs_DMSO": This represents the comparison between the Dox and DMSO levels of the treatment factor. This means it represents the log2 fold change in expression from the DMSO condition to the Dox condition, while keeping the shRNA factor constant (at its reference level, likely scr).
# * "shRNAsT.treatmentDox": This is the interaction term between the sT level of shRNA and the Dox level of treatment. This term tests whether the effect of the treatment (Dox vs. DMSO) is different in the sT condition compared to the scr condition. In other words, it tests whether the difference in expression due to the treatment (Dox) is different when the cells are treated with sT compared to when they are treated with scr.
#If you have an interaction term in your design, you should interpret main effects (shRNA_sT_vs_scr and treatment_Dox_vs_DMSO) cautiously because the main effects are evaluated at the reference level of the other factor, and their simple interpretation is complicated by the presence of interaction. Interaction term (shRNAsT.treatmentDox) can indicate if there's a specific combination of shRNA and treatment that has a differential expression different from what would be expected based on the individual effects of shRNA and treatment.
#当你运行 DESeq2 并使用 resultsNames(dds) 函数时,你将得到一系列的结果名称,这些名称代表了模型中的比较。在你给出的例子中:
# * "Intercept":截距,代表模型的基线水平。在生物统计学中,这通常指没有进行任何处理的条件。
# * "shRNA_sT_vs_scr":这代表 shRNA 的 sT 条件与 scr 条件的比较。如果你的实验设计中有两种不同的 shRNA 处理(sT 和 scr),这个比较会告诉你,与 scr 相比,sT 处理导致的基因表达量变化。
# * "treatment_Dox_vs_DMSO":这代表了处理条件 Dox 与 DMSO 的比较。这会显示在 DMSO(常作为对照组)和 Dox(可能是某种药物或处理)之间的差异。
# * "shRNAsT.treatmentDox":这是一个交互项,表示 shRNA 的 sT 处理和 Dox 处理的结合效应。这意味着它比较的是在 sT shRNA 影响下,Dox 对基因表达的影响与在 scr shRNA 影响下,Dox 对基因表达的影响之间的差异。
#简而言之,这些结果名称代表你的实验中不同条件或处理组合的比较,这有助于你了解不同实验条件下的基因表达如何变化。
d_MKL1 <- d[, !(grepl("parental cell|wt|WaGa", names(d)))]
cell.line = as.factor(c("MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1","MKL-1"))
shRNA = as.factor(c("sT","sT","scr","scr","sT","sT","scr","scr"))
treatment = as.factor(c("DMSO","DMSO","DMSO","DMSO","Dox","Dox","Dox","Dox"))
cData = data.frame(row.names=colnames(d_MKL1), shRNA=shRNA, treatment=treatment, cell.line=cell.line)
dds_MKL1<-DESeqDataSetFromMatrix(countData=d_MKL1, colData=cData, design=~shRNA+treatment+shRNA:treatment)
#png("pca_shRNA_treatment.png", 1200, 800)
#plotPCA(rld, intgroup = c("shRNA", "treatment"))
#dev.off()
#shRNA type (scr or sT) and the treatment (DMSO or Dox),
#design = ~ shRNA + treatment + shRNA:treatment
#DESeq2Res <- results(dds_MKL1, contrast = c("treatment","Dox","DMSO")) # scr_Dox vs scr_DMSO, effect of Dox in scr. "
#DESeq2Res = results(dds_MKL1, contrast = list( c("treatment_Dox_vs_DMSO","shRNAsT.treatmentDMSO") )) # sT_Dox vs sT_DMSO, effect of Dox in sT"
#DESeq2Res = results(dds_MKL1, name="shRNAsT.treatmentDMSO") # difference between sT and src, effect of Dox different in sT vs scr.
#dds_MKL1$treatment <- factor(dds_MKL1$treatment)
#dds_MKL1$treatment <- relevel(dds_MKL1$treatment, ref = "DMSO")
dds_MKL1 = DESeq(dds_MKL1, betaPrior=FALSE)
resultsNames(dds_MKL1)
contrasts <- c("shRNA_sT_vs_scr", "treatment_Dox_vs_DMSO", "shRNAsT.treatmentDox")
#library("dplyr")
for (contrast in contrasts) {
res = results(dds_MKL1, name=contrast)
res <- res[!is.na(res$log2FoldChange),]
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID"))
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID"))
# In the ENSEMBL-database, GENEID is ENSEMBL-ID.
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND"
#geness <- geness[!duplicated(geness$GENEID), ]
#using getBM replacing AnnotationDbi::select
#filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids.
geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = rownames(res),
mart = ensembl)
geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE)
#merge by column by common colunmn name, in the case "GENEID"
res$ENSEMBL = rownames(res)
identical(rownames(res), rownames(geness_uniq))
res_df <- as.data.frame(res)
geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL")
dim(geness_res)
rownames(geness_res) <- geness_res$ensembl_gene_id
geness_res$ensembl_gene_id <- NULL
write.csv(as.data.frame(geness_res[order(geness_res$pvalue),]), file = paste(contrast, "all.txt", sep="-"))
up <- subset(geness_res, padj<=0.05 & log2FoldChange>=2)
down <- subset(geness_res, padj<=0.05 & log2FoldChange<=-2)
write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(contrast, "up.txt", sep="-"))
write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(contrast, "down.txt", sep="-"))
}
mv shRNA_sT_vs_scr-all.txt MKL1_shRNA_sT_vs_scr-all.txt
mv shRNA_sT_vs_scr-up.txt MKL1_shRNA_sT_vs_scr-up.txt
mv shRNA_sT_vs_scr-down.txt MKL1_shRNA_sT_vs_scr-down.txt
mv treatment_Dox_vs_DMSO-all.txt MKL1_treatment_Dox_vs_DMSO-all.txt
mv treatment_Dox_vs_DMSO-up.txt MKL1_treatment_Dox_vs_DMSO-up.txt
mv treatment_Dox_vs_DMSO-down.txt MKL1_treatment_Dox_vs_DMSO-down.txt
mv shRNAsT.treatmentDox-all.txt MKL1_shRNAsT.treatmentDox-all.txt
mv shRNAsT.treatmentDox-up.txt MKL1_shRNAsT.treatmentDox-up.txt
mv shRNAsT.treatmentDox-down.txt MKL1_shRNAsT.treatmentDox-down.txt
d_WaGa <- d[, !(grepl("parental cell|wt|MKL", names(d)))]
cell.line = as.factor(c("WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa","WaGa"))
shRNA = as.factor(c("sT","sT","sT","scr","scr","scr","sT","sT","sT","scr","scr","scr"))
treatment = as.factor(c("DMSO","DMSO","DMSO","DMSO","DMSO","DMSO","Dox","Dox","Dox","Dox","Dox","Dox"))
cData = data.frame(row.names=colnames(d_WaGa), shRNA=shRNA, treatment=treatment, cell.line=cell.line)
dds_WaGa<-DESeqDataSetFromMatrix(countData=d_WaGa, colData=cData, design=~shRNA+treatment+shRNA:treatment)
dds_WaGa = DESeq(dds_WaGa, betaPrior=FALSE)
resultsNames(dds_WaGa)
contrasts <- c("shRNA_sT_vs_scr", "treatment_Dox_vs_DMSO", "shRNAsT.treatmentDox")
#library("dplyr")
for (contrast in contrasts) {
res = results(dds_WaGa, name=contrast)
res <- res[!is.na(res$log2FoldChange),]
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID"))
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID"))
# In the ENSEMBL-database, GENEID is ENSEMBL-ID.
#geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND"
#geness <- geness[!duplicated(geness$GENEID), ]
#using getBM replacing AnnotationDbi::select
#filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids.
geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = rownames(res),
mart = ensembl)
geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE)
#merge by column by common colunmn name, in the case "GENEID"
res$ENSEMBL = rownames(res)
identical(rownames(res), rownames(geness_uniq))
res_df <- as.data.frame(res)
geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL")
dim(geness_res)
rownames(geness_res) <- geness_res$ensembl_gene_id
geness_res$ensembl_gene_id <- NULL
write.csv(as.data.frame(geness_res[order(geness_res$pvalue),]), file = paste(contrast, "all.txt", sep="-"))
up <- subset(geness_res, padj<=0.05 & log2FoldChange>=2)
down <- subset(geness_res, padj<=0.05 & log2FoldChange<=-2)
write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(contrast, "up.txt", sep="-"))
write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(contrast, "down.txt", sep="-"))
}
mv shRNA_sT_vs_scr-all.txt WaGa_shRNA_sT_vs_scr-all.txt
mv shRNA_sT_vs_scr-up.txt WaGa_shRNA_sT_vs_scr-up.txt
mv shRNA_sT_vs_scr-down.txt WaGa_shRNA_sT_vs_scr-down.txt
mv treatment_Dox_vs_DMSO-all.txt WaGa_treatment_Dox_vs_DMSO-all.txt
mv treatment_Dox_vs_DMSO-up.txt WaGa_treatment_Dox_vs_DMSO-up.txt
mv treatment_Dox_vs_DMSO-down.txt WaGa_treatment_Dox_vs_DMSO-down.txt
mv shRNAsT.treatmentDox-all.txt WaGa_shRNAsT.treatmentDox-all.txt
mv shRNAsT.treatmentDox-up.txt WaGa_shRNAsT.treatmentDox-up.txt
mv shRNAsT.treatmentDox-down.txt WaGa_shRNAsT.treatmentDox-down.txt
~/Tools/csv2xls-0.4/csv_to_xls.py MKL1_shRNA_sT_vs_scr-up.txt MKL1_shRNA_sT_vs_scr-down.txt MKL1_shRNA_sT_vs_scr-all.txt -d$',' -o MKL1_shRNA_sT_vs_scr.xls
~/Tools/csv2xls-0.4/csv_to_xls.py MKL1_treatment_Dox_vs_DMSO-up.txt MKL1_treatment_Dox_vs_DMSO-down.txt MKL1_treatment_Dox_vs_DMSO-all.txt -d$',' -o MKL1_treatment_Dox_vs_DMSO.xls
~/Tools/csv2xls-0.4/csv_to_xls.py MKL1_shRNAsT.treatmentDox-up.txt MKL1_shRNAsT.treatmentDox-down.txt MKL1_shRNAsT.treatmentDox-all.txt -d$',' -o MKL1_shRNAsT.treatmentDox.xls
~/Tools/csv2xls-0.4/csv_to_xls.py WaGa_shRNA_sT_vs_scr-up.txt WaGa_shRNA_sT_vs_scr-down.txt WaGa_shRNA_sT_vs_scr-all.txt -d$',' -o WaGa_shRNA_sT_vs_scr.xls
~/Tools/csv2xls-0.4/csv_to_xls.py WaGa_treatment_Dox_vs_DMSO-up.txt WaGa_treatment_Dox_vs_DMSO-down.txt WaGa_treatment_Dox_vs_DMSO-all.txt -d$',' -o WaGa_treatment_Dox_vs_DMSO.xls
~/Tools/csv2xls-0.4/csv_to_xls.py WaGa_shRNAsT.treatmentDox-up.txt WaGa_shRNAsT.treatmentDox-down.txt WaGa_shRNAsT.treatmentDox-all.txt -d$',' -o WaGa_shRNAsT.treatmentDox.xls
9, volcano plots with automatically finding top_g
#A canonical visualization for interpreting differential gene expression results is the volcano plot.
library(ggrepel)
#"EV.RNA_vs_RNA" "scr.DMSO_vs_EV.RNA" "sT.DMSO_vs_EV.RNA" "scr.Dox_vs_EV.RNA" "sT.Dox_vs_EV.RNA" "sT.Dox_vs_scr.Dox" "sT.DMSO_vs_scr.DMSO"
geness_res <- read.csv(file = paste("WaGa.EV_vs_WaGa.RNA", "all.txt", sep="-"), row.names=1)
geness_res$Color <- "NS or log2FC < 2.0"
geness_res$Color[geness_res$pvalue < 0.05] <- "P < 0.05"
geness_res$Color[geness_res$padj < 0.05] <- "P-adj < 0.05"
geness_res$Color[geness_res$padj < 0.001] <- "P-adj < 0.001"
geness_res$Color[abs(geness_res$log2FoldChange) < 2.0] <- "NS or log2FC < 2.0"
geness_res$Color <- factor(geness_res$Color,
levels = c("NS or log2FC < 2.0", "P < 0.05",
"P-adj < 0.05", "P-adj < 0.001"))
geness_res$invert_P <- (-log10(geness_res$pvalue)) * sign(geness_res$log2FoldChange)
top_g <- c()
top_g <- c(top_g, geness_res[, 'external_gene_name'][order(geness_res[, 'invert_P'], decreasing = TRUE)[1:200]], geness_res[, 'external_gene_name'][order(geness_res[, 'invert_P'], decreasing = FALSE)[1:200]])
top_g <- unique(top_g)
geness_res <- geness_res[, -1*ncol(geness_res)] #remove invert_P from matrix
png("WaGa.EV_vs_WaGa.RNA.png",width=1400, height=1000)
ggplot(geness_res,
aes(x = log2FoldChange, y = -log10(pvalue),
color = Color, label = external_gene_name)) +
geom_vline(xintercept = c(2.0, -2.0), lty = "dashed") +
geom_hline(yintercept = -log10(0.05), lty = "dashed") +
geom_point() +
labs(x = "log2(FC)",
y = "Significance, -log10(P)",
color = "Significance") +
scale_color_manual(values = c(`P-adj < 0.001` = "dodgerblue",
`P-adj < 0.05` = "lightblue",
`P < 0.05` = "orange2",
`NS or log2FC < 2.0` = "gray"),
guide = guide_legend(override.aes = list(size = 4))) +
scale_y_continuous(expand = expansion(mult = c(0,0.05))) +
geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & pvalue < 0.05 & (abs(geness_res$log2FoldChange) >= 2.0)),
size = 4, point.padding = 0.15, color = "black",
min.segment.length = .1, box.padding = .2, lwd = 2) +
theme_bw(base_size = 16) +
theme(legend.position = "bottom")
dev.off()
#sed -i -e 's/Color/Category/g' *_Category.csv
#x <- data.frame(k1 = c(NA,3,4,5,2), k2 = c(1,NA,4,5,2), data = 6:10)
#merge(x, y, by = "k2")
for cmp in "EV.RNA_vs_RNA" "scr.DMSO_vs_EV.RNA" "sT.DMSO_vs_EV.RNA" "scr.Dox_vs_EV.RNA" "sT.Dox_vs_EV.RNA" "sT.Dox_vs_scr.Dox" "sT.DMSO_vs_scr.DMSO"; do
echo "~/Tools/csv2xls-0.4/csv_to_xls.py ${cmp}-all.txt ${cmp}-up.txt ${cmp}-down.txt -d$',' -o ${cmp}.xls"
done
~/Tools/csv2xls-0.4/csv_to_xls.py EV.RNA_vs_RNA-all.txt EV.RNA_vs_RNA-up.txt EV.RNA_vs_RNA-down.txt -d$',' -o EV.RNA_vs_RNA.xls
~/Tools/csv2xls-0.4/csv_to_xls.py scr.DMSO_vs_EV.RNA-all.txt scr.DMSO_vs_EV.RNA-up.txt scr.DMSO_vs_EV.RNA-down.txt -d$',' -o scr.DMSO_vs_EV.RNA.xls
~/Tools/csv2xls-0.4/csv_to_xls.py sT.DMSO_vs_EV.RNA-all.txt sT.DMSO_vs_EV.RNA-up.txt sT.DMSO_vs_EV.RNA-down.txt -d$',' -o sT.DMSO_vs_EV.RNA.xls
~/Tools/csv2xls-0.4/csv_to_xls.py scr.Dox_vs_EV.RNA-all.txt scr.Dox_vs_EV.RNA-up.txt scr.Dox_vs_EV.RNA-down.txt -d$',' -o scr.Dox_vs_EV.RNA.xls
~/Tools/csv2xls-0.4/csv_to_xls.py sT.Dox_vs_EV.RNA-all.txt sT.Dox_vs_EV.RNA-up.txt sT.Dox_vs_EV.RNA-down.txt -d$',' -o sT.Dox_vs_EV.RNA.xls
~/Tools/csv2xls-0.4/csv_to_xls.py sT.Dox_vs_scr.Dox-all.txt sT.Dox_vs_scr.Dox-up.txt sT.Dox_vs_scr.Dox-down.txt -d$',' -o sT.Dox_vs_scr.Dox.xls
~/Tools/csv2xls-0.4/csv_to_xls.py sT.DMSO_vs_scr.DMSO-all.txt sT.DMSO_vs_scr.DMSO-up.txt sT.DMSO_vs_scr.DMSO-down.txt -d$',' -o sT.DMSO_vs_scr.DMSO.xls
#WaGa.EV_vs_WaGa.RNA-all.txt
~/Tools/csv2xls-0.4/csv_to_xls.py WaGa.EV_vs_WaGa.RNA-up.txt WaGa.EV_vs_WaGa.RNA-down.txt -d$',' -o WaGa.EV_vs_WaGa.RNA.xls
10, clustering the genes and draw heatmap
install.packages("gplots")
library("gplots")
#Option3: as paper described, A heatmap showing expression values of all DEGs which are significant between any pair conditions.
all_genes <- c(rownames(mock_sT_d8_vs_mock_sT_d3_sig),rownames(sT_d3_vs_mock_sT_d3_sig),rownames(sT_d8_vs_mock_sT_d8_sig),rownames(sT_d8_vs_sT_d3_sig)) #873
all_genes <- unique(all_genes) #663
#all_genes2 <- c(rownames(WAC_vs_mock_sig),rownames(WAP_vs_mock_sig),rownames(WAC_vs_WAP_sig)) #3917
#all_genes2 <- unique(all_genes2) #2608
#intersected_genes <- intersect(all_genes, all_genes2) # 2608
#RNASeq.NoCellLine <- read.csv(file ="gene_expression_keeping_condition.txt", row.names=1)
RNASeq.NoCellLine_ <- RNASeq.NoCellLine[all_genes,]
write.csv(as.data.frame(RNASeq.NoCellLine_), file ="gene_expression_keeping_condition.txt")
RNASeq.NoCellLine_ <- cbind(RNASeq.NoCellLine_, mock_sT_d3 = rowMeans(RNASeq.NoCellLine_[, 1:2]))
RNASeq.NoCellLine_ <- cbind(RNASeq.NoCellLine_, mock_sT_d8 = rowMeans(RNASeq.NoCellLine_[, 3:4]))
RNASeq.NoCellLine_ <- cbind(RNASeq.NoCellLine_, sT_d3 = rowMeans(RNASeq.NoCellLine_[, 5:6]))
RNASeq.NoCellLine_ <- cbind(RNASeq.NoCellLine_, sT_d8 = rowMeans(RNASeq.NoCellLine_[, 7:8]))
RNASeq.NoCellLine_ <- RNASeq.NoCellLine_[,c(-1:-8)] #663x4
#RNASeq.NoCellLine__ <- read.csv(file ="gene_expression_keeping_condition.txt", row.names=1)
write.csv(as.data.frame(RNASeq.NoCellLine_), file ="gene_expression_merging_condition.txt")
cut -d',' -f1-1 ./WaGa.EV_vs_WaGa.RNA-up.txt > WaGa.EV_vs_WaGa.RNA-up.id
cut -d',' -f1-1 ./WaGa.sT.DMSO_vs_WaGa.EV-up.txt > WaGa.sT.DMSO_vs_WaGa.EV-up.id
cut -d',' -f1-1 ./WaGa.sT.Dox_vs_WaGa.EV-up.txt > WaGa.sT.Dox_vs_WaGa.EV-up.id
cut -d',' -f1-1 ./WaGa.scr.DMSO_vs_WaGa.EV-up.txt > WaGa.scr.DMSO_vs_WaGa.EV-up.id
cut -d',' -f1-1 ./WaGa.scr.Dox_vs_WaGa.EV-up.txt > WaGa.scr.Dox_vs_WaGa.EV-up.id
cut -d',' -f1-1 ./WaGa_shRNA_sT_vs_scr-up.txt > WaGa_shRNA_sT_vs_scr-up.id
cut -d',' -f1-1 ./WaGa_treatment_Dox_vs_DMSO-up.txt > WaGa_treatment_Dox_vs_DMSO-up.id
cut -d',' -f1-1 ./WaGa_shRNAsT.treatmentDox-up.txt > WaGa_shRNAsT.treatmentDox-up.id
cut -d',' -f1-1 ./WaGa.EV_vs_WaGa.RNA-down.txt > WaGa.EV_vs_WaGa.RNA-down.id
cut -d',' -f1-1 ./WaGa.sT.DMSO_vs_WaGa.EV-down.txt > WaGa.sT.DMSO_vs_WaGa.EV-down.id
cut -d',' -f1-1 ./WaGa.sT.Dox_vs_WaGa.EV-down.txt > WaGa.sT.Dox_vs_WaGa.EV-down.id
cut -d',' -f1-1 ./WaGa.scr.DMSO_vs_WaGa.EV-down.txt > WaGa.scr.DMSO_vs_WaGa.EV-down.id
cut -d',' -f1-1 ./WaGa.scr.Dox_vs_WaGa.EV-down.txt > WaGa.scr.Dox_vs_WaGa.EV-down.id
cut -d',' -f1-1 ./WaGa_shRNA_sT_vs_scr-down.txt > WaGa_shRNA_sT_vs_scr-down.id
cut -d',' -f1-1 ./WaGa_treatment_Dox_vs_DMSO-down.txt > WaGa_treatment_Dox_vs_DMSO-down.id
cut -d',' -f1-1 ./WaGa_shRNAsT.treatmentDox-down.txt > WaGa_shRNAsT.treatmentDox-down.id
cut -d',' -f1-1 ./MKL1.EV_vs_MKL1.RNA-up.txt > MKL1.EV_vs_MKL1.RNA-up.id
cut -d',' -f1-1 ./MKL1.sT.DMSO_vs_MKL1.EV-up.txt > MKL1.sT.DMSO_vs_MKL1.EV-up.id
cut -d',' -f1-1 ./MKL1.sT.Dox_vs_MKL1.EV-up.txt > MKL1.sT.Dox_vs_MKL1.EV-up.id
cut -d',' -f1-1 ./MKL1.scr.DMSO_vs_MKL1.EV-up.txt > MKL1.scr.DMSO_vs_MKL1.EV-up.id
cut -d',' -f1-1 ./MKL1.scr.Dox_vs_MKL1.EV-up.txt > MKL1.scr.Dox_vs_MKL1.EV-up.id
cut -d',' -f1-1 ./MKL1_shRNA_sT_vs_scr-up.txt > MKL1_shRNA_sT_vs_scr-up.id
cut -d',' -f1-1 ./MKL1_treatment_Dox_vs_DMSO-up.txt > MKL1_treatment_Dox_vs_DMSO-up.id
cut -d',' -f1-1 ./MKL1_shRNAsT.treatmentDox-up.txt > MKL1_shRNAsT.treatmentDox-up.id
cut -d',' -f1-1 ./MKL1.EV_vs_MKL1.RNA-down.txt > MKL1.EV_vs_MKL1.RNA-down.id
cut -d',' -f1-1 ./MKL1.sT.DMSO_vs_MKL1.EV-down.txt > MKL1.sT.DMSO_vs_MKL1.EV-down.id
cut -d',' -f1-1 ./MKL1.sT.Dox_vs_MKL1.EV-down.txt > MKL1.sT.Dox_vs_MKL1.EV-down.id
cut -d',' -f1-1 ./MKL1.scr.DMSO_vs_MKL1.EV-down.txt > MKL1.scr.DMSO_vs_MKL1.EV-down.id
cut -d',' -f1-1 ./MKL1.scr.Dox_vs_MKL1.EV-down.txt > MKL1.scr.Dox_vs_MKL1.EV-down.id
cut -d',' -f1-1 ./MKL1_shRNA_sT_vs_scr-down.txt > MKL1_shRNA_sT_vs_scr-down.id
cut -d',' -f1-1 ./MKL1_treatment_Dox_vs_DMSO-down.txt > MKL1_treatment_Dox_vs_DMSO-down.id
cut -d',' -f1-1 ./MKL1_shRNAsT.treatmentDox-down.txt > MKL1_shRNAsT.treatmentDox-down.id
cut -d',' -f1-1 WaGa.RNA_vs_MKL1.RNA-up.txt > WaGa.RNA_vs_MKL1.RNA-up.id
cut -d',' -f1-1 WaGa.RNA_vs_MKL1.RNA-down.txt > WaGa.RNA_vs_MKL1.RNA-down.id
cut -d',' -f1-1 WaGa.EV_vs_MKL1.EV-up.txt > WaGa.EV_vs_MKL1.EV-up.id
cut -d',' -f1-1 WaGa.EV_vs_MKL1.EV-down.txt > WaGa.EV_vs_MKL1.EV-down.id
cat MKL1.EV_vs_MKL1.RNA-down.id MKL1.EV_vs_MKL1.RNA-up.id MKL1.scr.DMSO_vs_MKL1.EV-down.id MKL1.scr.DMSO_vs_MKL1.EV-up.id MKL1.scr.Dox_vs_MKL1.EV-down.id MKL1.scr.Dox_vs_MKL1.EV-up.id MKL1_shRNAsT.treatmentDox-down.id MKL1_shRNAsT.treatmentDox-up.id MKL1_shRNA_sT_vs_scr-down.id MKL1_shRNA_sT_vs_scr-up.id MKL1.sT.DMSO_vs_MKL1.EV-down.id MKL1.sT.DMSO_vs_MKL1.EV-up.id MKL1.sT.Dox_vs_MKL1.EV-down.id MKL1.sT.Dox_vs_MKL1.EV-up.id MKL1_treatment_Dox_vs_DMSO-down.id MKL1_treatment_Dox_vs_DMSO-up.id | sort -u > MKL1.ids
cat WaGa.EV_vs_WaGa.RNA-down.id WaGa.EV_vs_WaGa.RNA-up.id WaGa.scr.DMSO_vs_WaGa.EV-down.id WaGa.scr.DMSO_vs_WaGa.EV-up.id WaGa.scr.Dox_vs_WaGa.EV-down.id WaGa.scr.Dox_vs_WaGa.EV-up.id WaGa_shRNAsT.treatmentDox-down.id WaGa_shRNAsT.treatmentDox-up.id WaGa_shRNA_sT_vs_scr-down.id WaGa_shRNA_sT_vs_scr-up.id WaGa.sT.DMSO_vs_WaGa.EV-down.id WaGa.sT.DMSO_vs_WaGa.EV-up.id WaGa.sT.Dox_vs_WaGa.EV-down.id WaGa.sT.Dox_vs_WaGa.EV-up.id WaGa_treatment_Dox_vs_DMSO-down.id WaGa_treatment_Dox_vs_DMSO-up.id | sort -u > WaGa.ids
#WaGa.RNA_vs_MKL1.RNA-up.id WaGa.RNA_vs_MKL1.RNA-down.id WaGa.EV_vs_MKL1.EV-up.id WaGa.EV_vs_MKL1.EV-down.id | sort -u > ids
# 28186 (new) vs 28193
cat MKL1.EV_vs_MKL1.RNA-down.id MKL1.EV_vs_MKL1.RNA-up.id MKL1.scr.DMSO_vs_MKL1.EV-down.id MKL1.scr.DMSO_vs_MKL1.EV-up.id MKL1.scr.Dox_vs_MKL1.EV-down.id MKL1.scr.Dox_vs_MKL1.EV-up.id MKL1_shRNAsT.treatmentDox-down.id MKL1_shRNAsT.treatmentDox-up.id MKL1_shRNA_sT_vs_scr-down.id MKL1_shRNA_sT_vs_scr-up.id MKL1.sT.DMSO_vs_MKL1.EV-down.id MKL1.sT.DMSO_vs_MKL1.EV-up.id MKL1.sT.Dox_vs_MKL1.EV-down.id MKL1.sT.Dox_vs_MKL1.EV-up.id MKL1_treatment_Dox_vs_DMSO-down.id MKL1_treatment_Dox_vs_DMSO-up.id WaGa.EV_vs_WaGa.RNA-down.id WaGa.EV_vs_WaGa.RNA-up.id WaGa.scr.DMSO_vs_WaGa.EV-down.id WaGa.scr.DMSO_vs_WaGa.EV-up.id WaGa.scr.Dox_vs_WaGa.EV-down.id WaGa.scr.Dox_vs_WaGa.EV-up.id WaGa_shRNAsT.treatmentDox-down.id WaGa_shRNAsT.treatmentDox-up.id WaGa_shRNA_sT_vs_scr-down.id WaGa_shRNA_sT_vs_scr-up.id WaGa.sT.DMSO_vs_WaGa.EV-down.id WaGa.sT.DMSO_vs_WaGa.EV-up.id WaGa.sT.Dox_vs_WaGa.EV-down.id WaGa.sT.Dox_vs_WaGa.EV-up.id WaGa_treatment_Dox_vs_DMSO-down.id WaGa_treatment_Dox_vs_DMSO-up.id WaGa.RNA_vs_MKL1.RNA-up.id WaGa.RNA_vs_MKL1.RNA-down.id WaGa.EV_vs_MKL1.EV-up.id WaGa.EV_vs_MKL1.EV-down.id | sort -u > ids
# ---- Draw DEGs_heatmap for parental_cell_RNA and wt_EV_RNA samples ----
#-- MKL-1.RNA and MKL-1.EV vs WaGa.RNA and WaGa.EV --
#add Gene_Id in the first line.
GOI <- read.csv("ids")$Gene_Id
RNASeq.NoCellLine <- assay(rld)
#clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman
datamat = RNASeq.NoCellLine[GOI, ]
hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete")
hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete")
mycl = cutree(hr, h=max(hr$height)/1.05)
mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN");
mycol = mycol[as.vector(mycl)]
sampleCols <- rep('GREY',ncol(datamat))
names(sampleCols) <- c("WaGa_RNA","WaGa_RNA_118","WaGa_RNA_147", "MKL1_RNA","MKL1_RNA_118","MKL1_RNA_147", "WaGa_EV_RNA","WaGa_EV_RNA_2","WaGa_EV_RNA_118","WaGa_EV_RNA_147","WaGa_EV_RNA_226", "MKL1_EV_RNA","MKL1_EV_RNA_2","MKL1_EV_RNA_27","MKL1_EV_RNA_87","MKL1_EV_RNA_118")
#sampleCols[substr(colnames(RNASeq.NoCellLine_),1,4)=='mock'] <- 'GREY'
sampleCols["WaGa_RNA"] <- 'DARKBLUE'
sampleCols["WaGa_RNA_118"] <- 'DARKBLUE'
sampleCols["WaGa_RNA_147"] <- 'DARKBLUE'
sampleCols["MKL1_RNA"] <- 'DARKRED'
sampleCols["MKL1_RNA_118"] <- 'DARKRED'
sampleCols["MKL1_RNA_147"] <- 'DARKRED'
sampleCols["WaGa_EV_RNA"] <- 'DARKORANGE'
sampleCols["WaGa_EV_RNA_2"] <- 'DARKORANGE'
sampleCols["WaGa_EV_RNA_118"] <- 'DARKORANGE'
sampleCols["WaGa_EV_RNA_147"] <- 'DARKORANGE'
sampleCols["WaGa_EV_RNA_226"] <- 'DARKORANGE'
sampleCols["MKL1_EV_RNA"] <- 'DARKGREEN'
sampleCols["MKL1_EV_RNA_2"] <- 'DARKGREEN'
sampleCols["MKL1_EV_RNA_27"] <- 'DARKGREEN'
sampleCols["MKL1_EV_RNA_87"] <- 'DARKGREEN'
sampleCols["MKL1_EV_RNA_118"] <- 'DARKGREEN'
png("DEGs_heatmap.png", width=1000, height=1200)
heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',
scale='row',trace='none',col=bluered(75),
RowSideColors = mycol, ColSideColors = sampleCols, labRow="", margins=c(22,10), cexRow=8, cexCol=2, srtCol=45, lwid=c(1,7), lhei = c(1, 8))
legend("top", title = "",legend=c("WaGa_RNA","MKL1_RNA","WaGa_EV_RNA","MKL1_EV_RNA"), fill=c("DARKBLUE","DARKRED","DARKORANGE","DARKGREEN"), cex=0.8, box.lty=0)
dev.off()
# ---- Draw DEGs_heatmap for the MKL-1 samples ----
#add Gene_Id in the first line.
setwd("degenes")
#BiocManager::install("biomaRt")
#install.packages("gplots")
#install.packages("writexl") # Install the writexl package
library(writexl) # Load the writexl package
library(gplots)
library(biomaRt)
library(dplyr)
GOI <- read.csv("MKL1.ids")$Gene_Id
RNASeq.NoCellLine <- assay(rld)
#clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman
datamat_ = RNASeq.NoCellLine[GOI, c("MKL-1 parental cell RNA", "MKL-1 parental cell RNA 118", "MKL-1 parental cell RNA 147", "MKL-1 wt EV RNA", "MKL-1 wt EV RNA 2", "MKL-1 wt EV RNA 118", "MKL-1 wt EV RNA 87", "MKL-1 wt EV RNA 27", "MKL-1 wt EV RNA 042", "MKL-1 sT DMSO EV RNA 042", "MKL-1 sT DMSO EV RNA 0505", "MKL-1 scr DMSO EV RNA 042", "MKL-1 scr DMSO EV RNA 0505", "MKL-1 sT Dox EV RNA 042", "MKL-1 sT Dox EV RNA 0505", "MKL-1 scr Dox EV RNA 042", "MKL-1 scr Dox EV RNA 0505")]
new_order <- c("MKL-1 parental cell RNA", "MKL-1 parental cell RNA 118", "MKL-1 parental cell RNA 147", "MKL-1 wt EV RNA", "MKL-1 wt EV RNA 2", "MKL-1 wt EV RNA 118", "MKL-1 wt EV RNA 87", "MKL-1 wt EV RNA 27", "MKL-1 wt EV RNA 042", "MKL-1 sT DMSO EV RNA 042", "MKL-1 sT DMSO EV RNA 0505", "MKL-1 sT Dox EV RNA 042", "MKL-1 sT Dox EV RNA 0505", "MKL-1 scr DMSO EV RNA 042", "MKL-1 scr DMSO EV RNA 0505", "MKL-1 scr Dox EV RNA 042", "MKL-1 scr Dox EV RNA 0505")
datamat <- datamat_[, new_order]
hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete")
hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete")
mycl = cutree(hr, h=max(hr$height)/1.05)
mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN");
mycol = mycol[as.vector(mycl)]
sampleCols <- rep('GREY',ncol(datamat))
names(sampleCols) <- c("MKL-1 parental cell RNA", "MKL-1 parental cell RNA 118", "MKL-1 parental cell RNA 147", "MKL-1 wt EV RNA", "MKL-1 wt EV RNA 2", "MKL-1 wt EV RNA 118", "MKL-1 wt EV RNA 87", "MKL-1 wt EV RNA 27", "MKL-1 wt EV RNA 042", "MKL-1 sT DMSO EV RNA 042", "MKL-1 sT DMSO EV RNA 0505", "MKL-1 sT Dox EV RNA 042", "MKL-1 sT Dox EV RNA 0505", "MKL-1 scr DMSO EV RNA 042", "MKL-1 scr DMSO EV RNA 0505", "MKL-1 scr Dox EV RNA 042", "MKL-1 scr Dox EV RNA 0505")
#sampleCols[substr(colnames(RNASeq.NoCellLine_),1,4)=='mock'] <- 'GREY'
sampleCols["MKL-1 parental cell RNA"] <- 'WHITE'
sampleCols["MKL-1 parental cell RNA 118"] <- 'WHITE'
sampleCols["MKL-1 parental cell RNA 147"] <- 'WHITE'
sampleCols["MKL-1 wt EV RNA"] <- 'GREY'
sampleCols["MKL-1 wt EV RNA 2"] <- 'GREY'
sampleCols["MKL-1 wt EV RNA 118"] <- 'GREY'
sampleCols["MKL-1 wt EV RNA 87"] <- 'GREY'
sampleCols["MKL-1 wt EV RNA 27"] <- 'GREY'
sampleCols["MKL-1 wt EV RNA 042"] <- 'GREY'
#"salmon", "lightcoral", or "mistyrose"
sampleCols["MKL-1 sT DMSO EV RNA 042"] <- 'mistyrose'
sampleCols["MKL-1 sT DMSO EV RNA 0505"] <- 'mistyrose'
sampleCols["MKL-1 sT Dox EV RNA 042"] <- 'RED'
sampleCols["MKL-1 sT Dox EV RNA 0505"] <- 'RED'
sampleCols["MKL-1 scr DMSO EV RNA 042"] <- 'LIGHTGREEN'
sampleCols["MKL-1 scr DMSO EV RNA 0505"] <- 'LIGHTGREEN'
sampleCols["MKL-1 scr Dox EV RNA 042"] <- 'GREEN'
sampleCols["MKL-1 scr Dox EV RNA 0505"] <- 'GREEN'
#legend("right", title = "",legend=c("MKL-1 parental cell RNA","MKL-1 wt EV RNA","MKL-1 sT DMSO EV RNA","MKL-1 sT Dox EV RNA","MKL-1 scr DMSO EV RNA","MKL-1 scr Dox EV RNA"), fill=c("WHITE", "GREY", "salmon","RED","LICHTGREEN","GREEN"), cex=0.8, box.lty=0)
png("DEGs_heatmap_MKL-1.png", width=1000, height=1200)
heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',
scale='row',trace='none',col=bluered(75),
RowSideColors = mycol, ColSideColors = sampleCols, labRow="", margins=c(22,10), cexRow=8, cexCol=2, srtCol=30, lwid=c(1,7), lhei = c(1, 8))
dev.off()
#rsync -a -P /home/jhuang/DATA/Data_Ute/Data_RNA-Seq_MKL-1+WaGa/results/featureCounts/degenes/DEGs_heatmap_MKL-1.png ./
# ---- cluster members ----
subset_1<-names(subset(mycl, mycl == '1'))
subset_1_ <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = subset_1,
mart = ensembl)
subset_1_uniq <- distinct(subset_1_, ensembl_gene_id, .keep_all= TRUE)
subset_1_expr <- datamat[subset_1,]
subset_1_expr <- as.data.frame(subset_1_expr)
subset_1_expr$ENSEMBL = rownames(subset_1_expr)
cluster1_YELLOW <- merge(subset_1_uniq, subset_1_expr, by.x="ensembl_gene_id", by.y="ENSEMBL")
#write.csv(cluster1_YELLOW,file='cluster1_YELLOW.txt')
subset_2<-names(subset(mycl, mycl == '2'))
subset_2_ <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = subset_2,
mart = ensembl)
subset_2_uniq <- distinct(subset_2_, ensembl_gene_id, .keep_all= TRUE)
subset_2_expr <- datamat[subset_2,]
subset_2_expr <- as.data.frame(subset_2_expr)
subset_2_expr$ENSEMBL = rownames(subset_2_expr)
cluster2_DARKBLUE <- merge(subset_2_uniq, subset_2_expr, by.x="ensembl_gene_id", by.y="ENSEMBL")
#write.csv(cluster2_DARKBLUE,file='cluster2_DARKBLUE.txt')
write_xlsx(list(
"Cluster 1 YELLOW" = cluster1_YELLOW,
"Cluster 2 DARKBLUE" = cluster2_DARKBLUE
), "DEGs_heatmap_data_MKL-1.xlsx")
#write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt')
#write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt')
#~/Tools/csv2xls-0.4/csv_to_xls.py \
#cluster1_YELLOW.txt \
#cluster2_DARKBLUE.txt \
#-d$',' -o gene_culsters.xls;
#TODO: save datamat as DEGs_heatmap_data.txt
#datamat = read.csv(file="DEGs_heatmap_data.txt", sep="\t", row.names=1)
#~/Tools/csv2xls-0.4/csv_to_xls.py \
#DEGs_heatmap_data.txt \
#-d',' -o DEGs_heatmap_data.xls;
#~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o genelist_clusters_MKL-1.xls
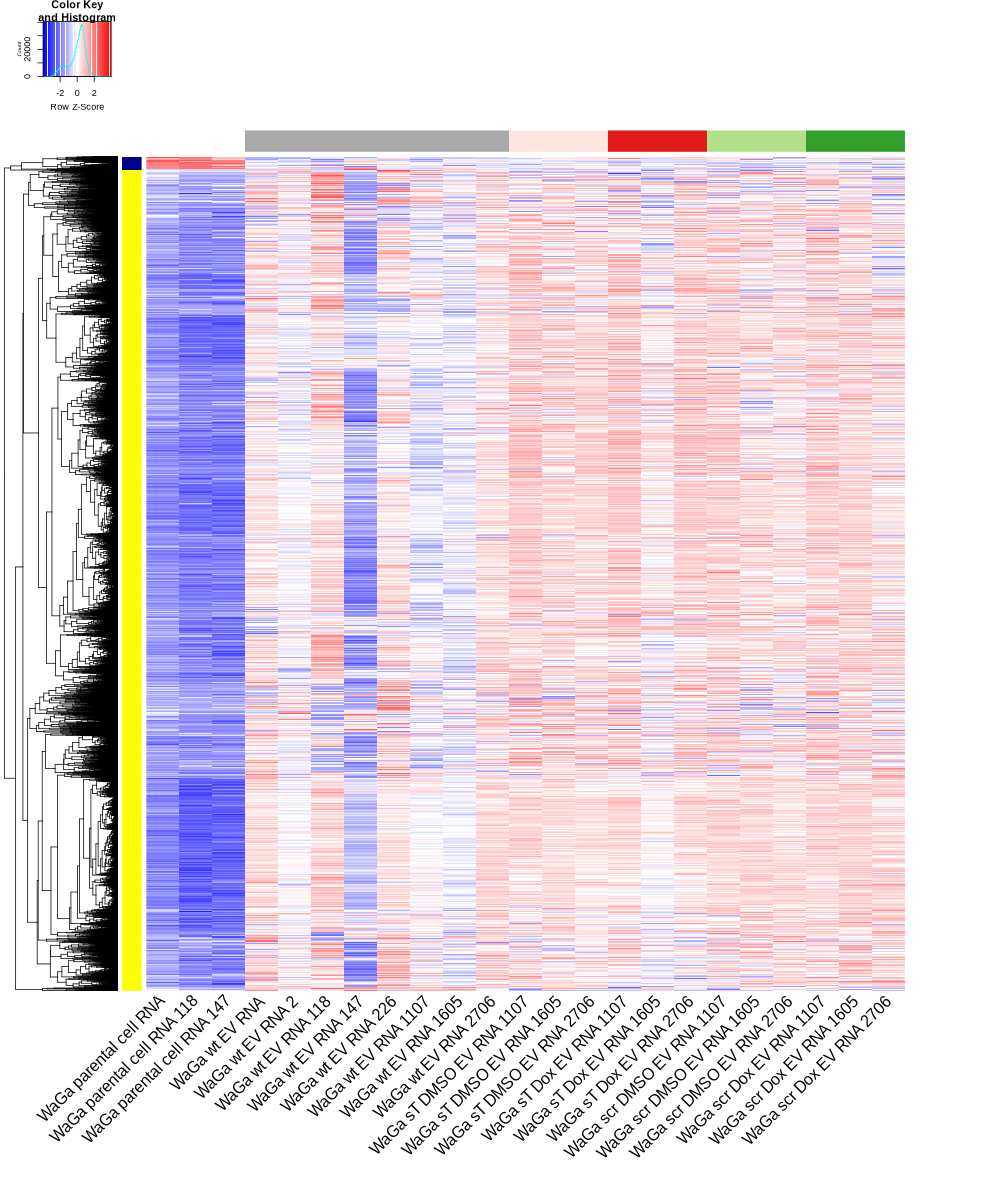
# ---- Draw DEGs_heatmap for the WaGa samples ----
#add Gene_Id in the first line.
GOI <- read.csv("WaGa.ids")$Gene_Id
RNASeq.NoCellLine <- assay(rld)
#clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman
datamat_ = RNASeq.NoCellLine[GOI, c("WaGa parental cell RNA", "WaGa parental cell RNA 118", "WaGa parental cell RNA 147", "WaGa wt EV RNA", "WaGa wt EV RNA 2", "WaGa wt EV RNA 118", "WaGa wt EV RNA 147", "WaGa wt EV RNA 226", "WaGa wt EV RNA 1107", "WaGa wt EV RNA 1605", "WaGa wt EV RNA 2706", "WaGa sT DMSO EV RNA 1107", "WaGa sT DMSO EV RNA 1605", "WaGa sT DMSO EV RNA 2706", "WaGa scr DMSO EV RNA 1107", "WaGa scr DMSO EV RNA 1605", "WaGa scr DMSO EV RNA 2706", "WaGa sT Dox EV RNA 1107", "WaGa sT Dox EV RNA 1605", "WaGa sT Dox EV RNA 2706", "WaGa scr Dox EV RNA 1107", "WaGa scr Dox EV RNA 1605", "WaGa scr Dox EV RNA 2706")]
new_order <- c("WaGa parental cell RNA", "WaGa parental cell RNA 118", "WaGa parental cell RNA 147", "WaGa wt EV RNA", "WaGa wt EV RNA 2", "WaGa wt EV RNA 118", "WaGa wt EV RNA 147", "WaGa wt EV RNA 226", "WaGa wt EV RNA 1107", "WaGa wt EV RNA 1605", "WaGa wt EV RNA 2706", "WaGa sT DMSO EV RNA 1107", "WaGa sT DMSO EV RNA 1605", "WaGa sT DMSO EV RNA 2706", "WaGa sT Dox EV RNA 1107", "WaGa sT Dox EV RNA 1605", "WaGa sT Dox EV RNA 2706", "WaGa scr DMSO EV RNA 1107", "WaGa scr DMSO EV RNA 1605", "WaGa scr DMSO EV RNA 2706", "WaGa scr Dox EV RNA 1107", "WaGa scr Dox EV RNA 1605", "WaGa scr Dox EV RNA 2706")
datamat <- datamat_[, new_order]
hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete")
hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete")
mycl = cutree(hr, h=max(hr$height)/1.05)
mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN");
mycol = mycol[as.vector(mycl)]
sampleCols <- rep('GREY',ncol(datamat))
names(sampleCols) <- c("WaGa parental cell RNA", "WaGa parental cell RNA 118", "WaGa parental cell RNA 147", "WaGa wt EV RNA", "WaGa wt EV RNA 2", "WaGa wt EV RNA 118", "WaGa wt EV RNA 147", "WaGa wt EV RNA 226", "WaGa wt EV RNA 1107", "WaGa wt EV RNA 1605", "WaGa wt EV RNA 2706", "WaGa sT DMSO EV RNA 1107", "WaGa sT DMSO EV RNA 1605", "WaGa sT DMSO EV RNA 2706", "WaGa sT Dox EV RNA 1107", "WaGa sT Dox EV RNA 1605", "WaGa sT Dox EV RNA 2706", "WaGa scr DMSO EV RNA 1107", "WaGa scr DMSO EV RNA 1605", "WaGa scr DMSO EV RNA 2706", "WaGa scr Dox EV RNA 1107", "WaGa scr Dox EV RNA 1605", "WaGa scr Dox EV RNA 2706")
#sampleCols[substr(colnames(RNASeq.NoCellLine_),1,4)=='mock'] <- 'GREY'
sampleCols["WaGa parental cell RNA"] <- 'WHITE'
sampleCols["WaGa parental cell RNA 118"] <- 'WHITE'
sampleCols["WaGa parental cell RNA 147"] <- 'WHITE'
sampleCols["WaGa wt EV RNA"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 2"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 118"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 147"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 226"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 1107"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 1605"] <- '#A9A9A9'
sampleCols["WaGa wt EV RNA 2706"] <- '#A9A9A9'
#fb9a99
sampleCols["WaGa sT DMSO EV RNA 1107"] <- 'mistyrose'
sampleCols["WaGa sT DMSO EV RNA 1605"] <- 'mistyrose'
sampleCols["WaGa sT DMSO EV RNA 2706"] <- 'mistyrose'
sampleCols["WaGa sT Dox EV RNA 1107"] <- '#e31a1c'
sampleCols["WaGa sT Dox EV RNA 1605"] <- '#e31a1c'
sampleCols["WaGa sT Dox EV RNA 2706"] <- '#e31a1c'
sampleCols["WaGa scr DMSO EV RNA 1107"] <- '#b2df8a'
sampleCols["WaGa scr DMSO EV RNA 1605"] <- '#b2df8a'
sampleCols["WaGa scr DMSO EV RNA 2706"] <- '#b2df8a'
sampleCols["WaGa scr Dox EV RNA 1107"] <- '#33a02c'
sampleCols["WaGa scr Dox EV RNA 1605"] <- '#33a02c'
sampleCols["WaGa scr Dox EV RNA 2706"] <- '#33a02c'
#legend("right", title = "",legend=c("WaGa parental cell RNA","WaGa wt EV RNA","WaGa sT DMSO EV RNA","WaGa sT Dox EV RNA","WaGa scr DMSO EV RNA","WaGa scr Dox EV RNA"), fill=c("WHITE", "GREY", "salmon","RED","LICHTGREEN","GREEN"), cex=0.8, box.lty=0)
png("DEGs_heatmap_WaGa.png", width=1000, height=1200)
heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',
scale='row',trace='none',col=bluered(75),
RowSideColors = mycol, ColSideColors = sampleCols, labRow="", margins=c(22,10), cexRow=8, cexCol=2, srtCol=45, lwid=c(1,7), lhei = c(1, 8))
dev.off()
# ---- cluster members ----
subset_1<-names(subset(mycl, mycl == '1'))
subset_1_ <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = subset_1,
mart = ensembl)
subset_1_uniq <- distinct(subset_1_, ensembl_gene_id, .keep_all= TRUE)
subset_1_expr <- datamat[subset_1,]
subset_1_expr <- as.data.frame(subset_1_expr)
subset_1_expr$ENSEMBL = rownames(subset_1_expr)
cluster1_YELLOW <- merge(subset_1_uniq, subset_1_expr, by.x="ensembl_gene_id", by.y="ENSEMBL")
#write.csv(cluster1_YELLOW,file='cluster1_YELLOW.txt')
subset_2<-names(subset(mycl, mycl == '2'))
subset_2_ <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'),
filters = 'ensembl_gene_id',
values = subset_2,
mart = ensembl)
subset_2_uniq <- distinct(subset_2_, ensembl_gene_id, .keep_all= TRUE)
subset_2_expr <- datamat[subset_2,]
subset_2_expr <- as.data.frame(subset_2_expr)
subset_2_expr$ENSEMBL = rownames(subset_2_expr)
cluster2_DARKBLUE <- merge(subset_2_uniq, subset_2_expr, by.x="ensembl_gene_id", by.y="ENSEMBL")
#write.csv(cluster2_DARKBLUE,file='cluster2_DARKBLUE.txt')
write_xlsx(list(
"Cluster 1 YELLOW" = cluster1_YELLOW,
"Cluster 2 DARKBLUE" = cluster2_DARKBLUE
), "DEGs_heatmap_data_WaGa.xlsx")
#write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt')
#write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt')
#write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt')
#write.csv(names(subset(mycl, mycl == '4')),file='cluster4_DARKMAGENTA.txt')
#write.csv(names(subset(mycl, mycl == '5')),file='cluster5_DARKCYAN.txt')
#~/Tools/csv2xls-0.4/csv_to_xls.py \
#cluster1_YELLOW.txt \
#cluster2_DARKBLUE.txt \
#-d$',' -o gene_culsters.xls;
#~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o genelist_clusters_WaGa.xls
# ---- Draw DEGs_heatmap for all samples ----
#add Gene_Id in the first line.
GOI <- read.csv("all.ids")$Gene_Id
RNASeq.NoCellLine <- assay(rld)
#clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman
datamat = RNASeq.NoCellLine[GOI, c("MKL-1 RNA", "MKL-1 RNA 118", "MKL-1 RNA 147", "MKL-1 EV", "MKL-1 EV 2", "MKL-1 EV 118", "MKL-1 EV 87", "MKL-1 EV 27", "MKL-1 EV 042", "MKL-1 EV sT DMSO 042", "MKL-1 EV sT DMSO 0505", "MKL-1 EV scr DMSO 042", "MKL-1 EV scr DMSO 0505", "MKL-1 EV sT Dox 042", "MKL-1 EV sT Dox 0505", "MKL-1 EV scr Dox 042", "MKL-1 EV scr Dox 0505", "WaGa RNA", "WaGa RNA 118", "WaGa RNA 147", "WaGa EV", "WaGa EV 2", "WaGa EV 118", "WaGa EV 147", "WaGa EV 226", "WaGa EV 1107", "WaGa EV 1605", "WaGa EV 2706", "WaGa EV sT DMSO 1107", "WaGa EV sT DMSO 1605", "WaGa EV sT DMSO 2706", "WaGa EV scr DMSO 1107", "WaGa EV scr DMSO 1605", "WaGa EV scr DMSO 2706", "WaGa EV sT Dox 1107", "WaGa EV sT Dox 1605", "WaGa EV sT Dox 2706", "WaGa EV scr Dox 1107", "WaGa EV scr Dox 1605", "WaGa EV scr Dox 2706")]
hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete")
hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete")
mycl = cutree(hr, h=max(hr$height)/1.05)
mycol = c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN");
mycol = mycol[as.vector(mycl)]
sampleCols <- rep('GREY',ncol(datamat))
names(sampleCols) <- c("MKL-1 RNA", "MKL-1 RNA 118", "MKL-1 RNA 147", "MKL-1 EV", "MKL-1 EV 2", "MKL-1 EV 118", "MKL-1 EV 87", "MKL-1 EV 27", "MKL-1 EV 042", "MKL-1 EV sT DMSO 042", "MKL-1 EV sT DMSO 0505", "MKL-1 EV scr DMSO 042", "MKL-1 EV scr DMSO 0505", "MKL-1 EV sT Dox 042", "MKL-1 EV sT Dox 0505", "MKL-1 EV scr Dox 042", "MKL-1 EV scr Dox 0505", "WaGa RNA", "WaGa RNA 118", "WaGa RNA 147", "WaGa EV", "WaGa EV 2", "WaGa EV 118", "WaGa EV 147", "WaGa EV 226", "WaGa EV 1107", "WaGa EV 1605", "WaGa EV 2706", "WaGa EV sT DMSO 1107", "WaGa EV sT DMSO 1605", "WaGa EV sT DMSO 2706", "WaGa EV scr DMSO 1107", "WaGa EV scr DMSO 1605", "WaGa EV scr DMSO 2706", "WaGa EV sT Dox 1107", "WaGa EV sT Dox 1605", "WaGa EV sT Dox 2706", "WaGa EV scr Dox 1107", "WaGa EV scr Dox 1605", "WaGa EV scr Dox 2706")
#sampleCols[substr(colnames(RNASeq.NoCellLine_),1,4)=='mock'] <- 'GREY'
sampleCols["MKL-1 RNA"] <- 'DARKBLUE'
sampleCols["MKL-1 RNA 118"] <- 'DARKBLUE'
sampleCols["MKL-1 RNA 147"] <- 'DARKBLUE'
sampleCols["MKL-1 EV"] <- 'DARKRED'
sampleCols["MKL-1 EV 2"] <- 'DARKRED'
sampleCols["MKL-1 EV 118"] <- 'DARKRED'
sampleCols["MKL-1 EV 87"] <- 'DARKRED'
sampleCols["MKL-1 EV 27"] <- 'DARKRED'
sampleCols["MKL-1 EV 042"] <- 'DARKRED'
sampleCols["MKL-1 EV sT DMSO 042"] <- 'DARKORANGE'
sampleCols["MKL-1 EV sT DMSO 0505"] <- 'DARKORANGE'
sampleCols["MKL-1 EV scr DMSO 042"] <- 'DARKORANGE'
sampleCols["MKL-1 EV scr DMSO 0505"] <- 'DARKORANGE'
sampleCols["MKL-1 EV sT Dox 042"] <- 'DARKGREEN'
sampleCols["MKL-1 EV sT Dox 0505"] <- 'DARKGREEN'
sampleCols["MKL-1 EV scr Dox 042"] <- 'DARKGREEN'
sampleCols["MKL-1 EV scr Dox 0505"] <- 'DARKGREEN'
sampleCols["WaGa RNA"] <- 'BLUE'
sampleCols["WaGa RNA 118"] <- 'BLUE'
sampleCols["WaGa RNA 147"] <- 'BLUE'
sampleCols["WaGa EV"] <- 'RED'
sampleCols["WaGa EV 2"] <- 'RED'
sampleCols["WaGa EV 118"] <- 'RED'
sampleCols["WaGa EV 147"] <- 'RED'
sampleCols["WaGa EV 226"] <- 'RED'
sampleCols["WaGa EV 1107"] <- 'RED'
sampleCols["WaGa EV 1605"] <- 'RED'
sampleCols["WaGa EV 2706"] <- 'RED'
sampleCols["WaGa EV sT DMSO 1107"] <- 'ORANGE'
sampleCols["WaGa EV sT DMSO 1605"] <- 'ORANGE'
sampleCols["WaGa EV sT DMSO 2706"] <- 'ORANGE'
sampleCols["WaGa EV scr DMSO 1107"] <- 'ORANGE'
sampleCols["WaGa EV scr DMSO 1605"] <- 'ORANGE'
sampleCols["WaGa EV scr DMSO 2706"] <- 'ORANGE'
sampleCols["WaGa EV sT Dox 1107"] <- 'GREEN'
sampleCols["WaGa EV sT Dox 1605"] <- 'GREEN'
sampleCols["WaGa EV sT Dox 2706"] <- 'GREEN'
sampleCols["WaGa EV scr Dox 1107"] <- 'GREEN'
sampleCols["WaGa EV scr Dox 1605"] <- 'GREEN'
sampleCols["WaGa EV scr Dox 2706"] <- 'GREEN'
#legend("right", title = "",legend=c("WaGa parental cell RNA","WaGa wt EV RNA","WaGa sT DMSO EV RNA","WaGa sT Dox EV RNA","WaGa scr DMSO EV RNA","WaGa scr Dox EV RNA"), fill=c("WHITE", "GREY", "salmon","RED","LICHTGREEN","GREEN"), cex=0.8, box.lty=0)
png("DEGs_heatmap_all.png", width=1000, height=1200)
heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',
scale='row',trace='none',col=bluered(75),
RowSideColors = mycol, ColSideColors = sampleCols, labRow="", margins=c(22,10), cexRow=8, cexCol=2, srtCol=45, lwid=c(1,7), lhei = c(1, 8))
dev.off()
# ---- cluster members ----
#c("YELLOW", "DARKBLUE", "DARKORANGE", "DARKMAGENTA", "DARKCYAN", "DARKRED", "MAROON", "DARKGREEN", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN","LIGHTGREEN", "BLUE", "ORANGE", "CYAN", "RED", "GREEN");
write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt')
write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt')
write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt')
write.csv(names(subset(mycl, mycl == '4')),file='cluster4_DARKMAGENTA.txt')
write.csv(names(subset(mycl, mycl == '5')),file='cluster5_DARKCYAN.txt')
#~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o genelist_clusters_all.xls
11, pathways
mkdir pathways
#--continue from BREAK POINT--
##
#source("https://bioconductor.org/biocLite.R")
#biocLite("AnnotationDbi")
library("clusterProfiler")
library("ReactomePA")
setwd("~/DATA/Data_Anastasia_RNASeq/results/featureCounts/pathways")
for sample in WaGa_RNA_vs_MKL1_RNA MKL1_EV_RNA_vs_MKL1_RNA WaGa_EV_RNA_vs_MKL1_EV_RNA WaGa_EV_RNA_vs_WaGa_RNA; do \
echo "${sample}_up <- read.csv('../degenes/${sample}-up.txt', row.names=1)"
echo "${sample}_up_KEGG <- enrichKEGG(${sample}_up\$entrezgene_id)"
echo "write.table(as.data.frame(${sample}_up_KEGG), file = '${sample}_up_KEGG.txt', sep = '\t', row.names = FALSE)"
echo "${sample}_down <- read.csv('../degenes/${sample}-down.txt', row.names=1)"
echo "${sample}_down_KEGG <- enrichKEGG(${sample}_down\$entrezgene_id)"
echo "write.table(as.data.frame(${sample}_down_KEGG), file = '${sample}_down_KEGG.txt', sep = '\t', row.names = FALSE)"
echo "${sample}_sig <- rbind(${sample}_up, ${sample}_down)"
echo "${sample}_sig_KEGG <- enrichKEGG(${sample}_sig\$entrezgene_id)"
echo "write.table(as.data.frame(${sample}_sig_KEGG), file = '${sample}_sig_KEGG.txt', sep = '\t', row.names = FALSE)"
done
WaGa_RNA_vs_MKL1_RNA_up <- read.csv('../degenes/WaGa_RNA_vs_MKL1_RNA-up.txt', row.names=1)
WaGa_RNA_vs_MKL1_RNA_up_KEGG <- enrichKEGG(WaGa_RNA_vs_MKL1_RNA_up$entrezgene_id)
write.table(as.data.frame(WaGa_RNA_vs_MKL1_RNA_up_KEGG), file = 'WaGa_RNA_vs_MKL1_RNA_up_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_RNA_vs_MKL1_RNA_down <- read.csv('../degenes/WaGa_RNA_vs_MKL1_RNA-down.txt', row.names=1)
WaGa_RNA_vs_MKL1_RNA_down_KEGG <- enrichKEGG(WaGa_RNA_vs_MKL1_RNA_down$entrezgene_id)
write.table(as.data.frame(WaGa_RNA_vs_MKL1_RNA_down_KEGG), file = 'WaGa_RNA_vs_MKL1_RNA_down_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_RNA_vs_MKL1_RNA_sig <- rbind(WaGa_RNA_vs_MKL1_RNA_up, WaGa_RNA_vs_MKL1_RNA_down)
WaGa_RNA_vs_MKL1_RNA_sig_KEGG <- enrichKEGG(WaGa_RNA_vs_MKL1_RNA_sig$entrezgene_id)
write.table(as.data.frame(WaGa_RNA_vs_MKL1_RNA_sig_KEGG), file = 'WaGa_RNA_vs_MKL1_RNA_sig_KEGG.txt', sep = '\t', row.names = FALSE)
MKL1_EV_RNA_vs_MKL1_RNA_up <- read.csv('../degenes/MKL1_EV_RNA_vs_MKL1_RNA-up.txt', row.names=1)
MKL1_EV_RNA_vs_MKL1_RNA_up_KEGG <- enrichKEGG(MKL1_EV_RNA_vs_MKL1_RNA_up$entrezgene_id)
write.table(as.data.frame(MKL1_EV_RNA_vs_MKL1_RNA_up_KEGG), file = 'MKL1_EV_RNA_vs_MKL1_RNA_up_KEGG.txt', sep = '\t', row.names = FALSE)
MKL1_EV_RNA_vs_MKL1_RNA_down <- read.csv('../degenes/MKL1_EV_RNA_vs_MKL1_RNA-down.txt', row.names=1)
MKL1_EV_RNA_vs_MKL1_RNA_down_KEGG <- enrichKEGG(MKL1_EV_RNA_vs_MKL1_RNA_down$entrezgene_id)
write.table(as.data.frame(MKL1_EV_RNA_vs_MKL1_RNA_down_KEGG), file = 'MKL1_EV_RNA_vs_MKL1_RNA_down_KEGG.txt', sep = '\t', row.names = FALSE)
MKL1_EV_RNA_vs_MKL1_RNA_sig <- rbind(MKL1_EV_RNA_vs_MKL1_RNA_up, MKL1_EV_RNA_vs_MKL1_RNA_down)
MKL1_EV_RNA_vs_MKL1_RNA_sig_KEGG <- enrichKEGG(MKL1_EV_RNA_vs_MKL1_RNA_sig$entrezgene_id)
write.table(as.data.frame(MKL1_EV_RNA_vs_MKL1_RNA_sig_KEGG), file = 'MKL1_EV_RNA_vs_MKL1_RNA_sig_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_EV_RNA_vs_MKL1_EV_RNA_up <- read.csv('../degenes/WaGa_EV_RNA_vs_MKL1_EV_RNA-up.txt', row.names=1)
WaGa_EV_RNA_vs_MKL1_EV_RNA_up_KEGG <- enrichKEGG(WaGa_EV_RNA_vs_MKL1_EV_RNA_up$entrezgene_id)
write.table(as.data.frame(WaGa_EV_RNA_vs_MKL1_EV_RNA_up_KEGG), file = 'WaGa_EV_RNA_vs_MKL1_EV_RNA_up_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_EV_RNA_vs_MKL1_EV_RNA_down <- read.csv('../degenes/WaGa_EV_RNA_vs_MKL1_EV_RNA-down.txt', row.names=1)
WaGa_EV_RNA_vs_MKL1_EV_RNA_down_KEGG <- enrichKEGG(WaGa_EV_RNA_vs_MKL1_EV_RNA_down$entrezgene_id)
write.table(as.data.frame(WaGa_EV_RNA_vs_MKL1_EV_RNA_down_KEGG), file = 'WaGa_EV_RNA_vs_MKL1_EV_RNA_down_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_EV_RNA_vs_MKL1_EV_RNA_sig <- rbind(WaGa_EV_RNA_vs_MKL1_EV_RNA_up, WaGa_EV_RNA_vs_MKL1_EV_RNA_down)
WaGa_EV_RNA_vs_MKL1_EV_RNA_sig_KEGG <- enrichKEGG(WaGa_EV_RNA_vs_MKL1_EV_RNA_sig$entrezgene_id)
write.table(as.data.frame(WaGa_EV_RNA_vs_MKL1_EV_RNA_sig_KEGG), file = 'WaGa_EV_RNA_vs_MKL1_EV_RNA_sig_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_EV_RNA_vs_WaGa_RNA_up <- read.csv('../degenes/WaGa_EV_RNA_vs_WaGa_RNA-up.txt', row.names=1)
WaGa_EV_RNA_vs_WaGa_RNA_up_KEGG <- enrichKEGG(WaGa_EV_RNA_vs_WaGa_RNA_up$entrezgene_id)
write.table(as.data.frame(WaGa_EV_RNA_vs_WaGa_RNA_up_KEGG), file = 'WaGa_EV_RNA_vs_WaGa_RNA_up_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_EV_RNA_vs_WaGa_RNA_down <- read.csv('../degenes/WaGa_EV_RNA_vs_WaGa_RNA-down.txt', row.names=1)
WaGa_EV_RNA_vs_WaGa_RNA_down_KEGG <- enrichKEGG(WaGa_EV_RNA_vs_WaGa_RNA_down$entrezgene_id)
write.table(as.data.frame(WaGa_EV_RNA_vs_WaGa_RNA_down_KEGG), file = 'WaGa_EV_RNA_vs_WaGa_RNA_down_KEGG.txt', sep = '\t', row.names = FALSE)
WaGa_EV_RNA_vs_WaGa_RNA_sig <- rbind(WaGa_EV_RNA_vs_WaGa_RNA_up, WaGa_EV_RNA_vs_WaGa_RNA_down)
WaGa_EV_RNA_vs_WaGa_RNA_sig_KEGG <- enrichKEGG(WaGa_EV_RNA_vs_WaGa_RNA_sig$entrezgene_id)
write.table(as.data.frame(WaGa_EV_RNA_vs_WaGa_RNA_sig_KEGG), file = 'WaGa_EV_RNA_vs_WaGa_RNA_sig_KEGG.txt', sep = '\t', row.names = FALSE)
png("pathways_KEGG.png",width=1260, height=1000)
merged_list <- merge_result(list('WaGa_RNA vs MKL1_RNA'=WaGa_RNA_vs_MKL1_RNA_sig_KEGG, 'MKL1_EV_RNA vs MKL1_RNA'=MKL1_EV_RNA_vs_MKL1_RNA_sig_KEGG, 'WaGa_EV_RNA vs MKL1_EV_RNA'=WaGa_EV_RNA_vs_MKL1_EV_RNA_sig_KEGG, 'WaGa_EV_RNA vs WaGa_RNA'=WaGa_EV_RNA_vs_WaGa_RNA_sig_KEGG))
dotplot(merged_list, showCategory=1000) #, font.size=6, srt = 35
dev.off()
# under CONSOLE
cd pathways_KEGG
~/Tools/csv2xls-0.4/csv_to_xls.py WaGa_RNA_vs_MKL1_RNA_sig_KEGG.txt MKL1_EV_RNA_vs_MKL1_RNA_sig_KEGG.txt WaGa_EV_RNA_vs_MKL1_EV_RNA_sig_KEGG.txt WaGa_EV_RNA_vs_WaGa_RNA_sig_KEGG.txt -d$'\t' -o pathways_KEGG.xls
12, GOs
setwd("~/DATA/Data_Anastasia_RNASeq/results/featureCounts/GOs")
for sample in IKK1_vs_Cre IKK2_vs_Cre Nemo_vs_Cre NIK_vs_Cre; do \
echo "${sample}_up <- read.csv('../degenes/${sample}-up.txt', row.names=1)"
echo "${sample}_up_GO_BP <- enrichGO(${sample}_up\$entrezgene_id, 'org.Mm.eg.db', ont='BP')"
echo "${sample}_up_GO_MF <- enrichGO(${sample}_up\$entrezgene_id, 'org.Mm.eg.db', ont='MF')"
echo "${sample}_up_GO_CC <- enrichGO(${sample}_up\$entrezgene_id, 'org.Mm.eg.db', ont='CC')"
#echo "${sample}_up_GO_ALL <- enrichGO(${sample}_up\$entrezgene_id, 'org.Mm.eg.db', ont='ALL')"
echo "write.table(as.data.frame(${sample}_up_GO_BP), file = '${sample}_up_GO_BP.txt', sep = '\t', row.names = FALSE)"
echo "write.table(as.data.frame(${sample}_up_GO_MF), file = '${sample}_up_GO_MF.txt', sep = '\t', row.names = FALSE)"
echo "write.table(as.data.frame(${sample}_up_GO_CC), file = '${sample}_up_GO_CC.txt', sep = '\t', row.names = FALSE)"
#echo "write.table(as.data.frame(${sample}_up_GO_ALL), file = '${sample}_up_GO_ALL.txt', sep = '\t', row.names = FALSE)"
echo "${sample}_down <- read.csv('../degenes/${sample}-down.txt', row.names=1)"
echo "${sample}_down_GO_BP <- enrichGO(${sample}_down\$entrezgene_id, 'org.Mm.eg.db', ont='BP')"
echo "${sample}_down_GO_MF <- enrichGO(${sample}_down\$entrezgene_id, 'org.Mm.eg.db', ont='MF')"
echo "${sample}_down_GO_CC <- enrichGO(${sample}_down\$entrezgene_id, 'org.Mm.eg.db', ont='CC')"
#echo "${sample}_down_GO_ALL <- enrichGO(${sample}_down\$entrezgene_id, 'org.Mm.eg.db', ont='ALL')"
echo "write.table(as.data.frame(${sample}_down_GO_BP), file = '${sample}_down_GO_BP.txt', sep = '\t', row.names = FALSE)"
echo "write.table(as.data.frame(${sample}_down_GO_MF), file = '${sample}_down_GO_MF.txt', sep = '\t', row.names = FALSE)"
echo "write.table(as.data.frame(${sample}_down_GO_CC), file = '${sample}_down_GO_CC.txt', sep = '\t', row.names = FALSE)"
#echo "write.table(as.data.frame(${sample}_down_GO_ALL), file = '${sample}_down_GO_ALL.txt', sep = '\t', row.names = FALSE)"
echo "${sample}_sig <- rbind(${sample}_up, ${sample}_down)"
echo "${sample}_sig_GO_BP <- enrichGO(${sample}_sig\$entrezgene_id, 'org.Mm.eg.db', ont='BP')"
echo "${sample}_sig_GO_MF <- enrichGO(${sample}_sig\$entrezgene_id, 'org.Mm.eg.db', ont='MF')"
echo "${sample}_sig_GO_CC <- enrichGO(${sample}_sig\$entrezgene_id, 'org.Mm.eg.db', ont='CC')"
#echo "${sample}_sig_GO_ALL <- enrichGO(${sample}_sig\$entrezgene_id, 'org.Mm.eg.db', ont='ALL')"
echo "write.table(as.data.frame(${sample}_sig_GO_BP), file = '${sample}_sig_GO_BP.txt', sep = '\t', row.names = FALSE)"
echo "write.table(as.data.frame(${sample}_sig_GO_MF), file = '${sample}_sig_GO_MF.txt', sep = '\t', row.names = FALSE)"
echo "write.table(as.data.frame(${sample}_sig_GO_CC), file = '${sample}_sig_GO_CC.txt', sep = '\t', row.names = FALSE)"
#echo "write.table(as.data.frame(${sample}_sig_GO_ALL), file = '${sample}_sig_GO_ALL.txt', sep = '\t', row.names = FALSE)"
done
png("GOs_BP.png",width=3000, height=24000)
merged_list <- merge_result(list('IKK1_vs_Cre_up'=IKK1_vs_Cre_up_GO_BP, 'IKK1_vs_Cre_down'=IKK1_vs_Cre_down_GO_BP, 'IKK1_vs_Cre_sig'=IKK1_vs_Cre_sig_GO_BP, 'IKK2_vs_Cre_up'=IKK2_vs_Cre_up_GO_BP, 'IKK2_vs_Cre_down'=IKK2_vs_Cre_down_GO_BP, 'IKK2_vs_Cre_sig'=IKK2_vs_Cre_sig_GO_BP, 'Nemo_vs_Cre_up'= Nemo_vs_Cre_up_GO_BP, 'Nemo_vs_Cre_down'=Nemo_vs_Cre_down_GO_BP, 'Nemo_vs_Cre_sig'=Nemo_vs_Cre_sig_GO_BP, 'NIK_vs_Cre_up'=NIK_vs_Cre_up_GO_BP, 'NIK_vs_Cre_down'=NIK_vs_Cre_down_GO_BP, 'NIK_vs_Cre_sig'=NIK_vs_Cre_sig_GO_BP))
dotplot(merged_list, showCategory=10000)
dev.off()
# under CONSOLE
#
cd GOs
~/Tools/csv2xls-0.4/csv_to_xls.py IKK1_vs_Cre_up_GO_BP.txt IKK1_vs_Cre_down_GO_BP.txt IKK1_vs_Cre_sig_GO_BP.txt IKK2_vs_Cre_up_GO_BP.txt IKK2_vs_Cre_down_GO_BP.txt IKK2_vs_Cre_sig_GO_BP.txt Nemo_vs_Cre_up_GO_BP.txt Nemo_vs_Cre_down_GO_BP.txt Nemo_vs_Cre_sig_GO_BP.txt NIK_vs_Cre_up_GO_BP.txt NIK_vs_Cre_down_GO_BP.txt NIK_vs_Cre_sig_GO_BP.txt -d$'\t' -o GOs_BP.xls
~/Tools/csv2xls-0.4/csv_to_xls.py IKK1_vs_Cre_up_GO_MF.txt IKK1_vs_Cre_down_GO_MF.txt IKK1_vs_Cre_sig_GO_MF.txt IKK2_vs_Cre_up_GO_MF.txt IKK2_vs_Cre_down_GO_MF.txt IKK2_vs_Cre_sig_GO_MF.txt Nemo_vs_Cre_up_GO_MF.txt Nemo_vs_Cre_down_GO_MF.txt Nemo_vs_Cre_sig_GO_MF.txt NIK_vs_Cre_up_GO_MF.txt NIK_vs_Cre_down_GO_MF.txt NIK_vs_Cre_sig_GO_MF.txt -d$'\t' -o GOs_MF.xls
~/Tools/csv2xls-0.4/csv_to_xls.py IKK1_vs_Cre_up_GO_CC.txt IKK1_vs_Cre_down_GO_CC.txt IKK1_vs_Cre_sig_GO_CC.txt IKK2_vs_Cre_up_GO_CC.txt IKK2_vs_Cre_down_GO_CC.txt IKK2_vs_Cre_sig_GO_CC.txt Nemo_vs_Cre_up_GO_CC.txt Nemo_vs_Cre_down_GO_CC.txt Nemo_vs_Cre_sig_GO_CC.txt NIK_vs_Cre_up_GO_CC.txt NIK_vs_Cre_down_GO_CC.txt NIK_vs_Cre_sig_GO_CC.txt -d$'\t' -o GOs_CC.xls
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Kraken2 Installation and Usage Guide
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
- 基因组与表型特性研究:一株药物敏感的鲍曼不动杆菌显示出增强的毒力相关特性和应激耐受性
- A. baumannii HKAB-1 Point-by-point response to Reviewers
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
RNA-seq data analysis of Yersinia on GRCh38