Long-Read Transposon Insertion-site Sequencing vs. short-read TIS methods
gene_x 0 like s 767 view s
Tags: sequencing
https://pubmed.ncbi.nlm.nih.gov/35241765/
- Transposon insertion site sequencing (TIS) is a powerful method for associating genotype to phenotype.
- However, all TIS methods described to date use short nucleotide sequence reads which cannot uniquely determine the locations of transposon insertions within repeating genomic sequences where the repeat units are longer than the sequence read length.
- To overcome this limitation, we have developed a TIS method using Oxford Nanopore sequencing technology that generates and uses long nucleotide sequence reads; we have called this method LoRTIS (Long-Read Transposon Insertion-site Sequencing).
- LoRTIS enabled the unique localisation of transposon insertion sites within long repetitive genetic elements of E. coli, such as the transposase genes of insertion sequences and copies of the ~ 5 kb ribosomal RNA operon.
- We demonstrate that LoRTIS is reproducible, gives comparable results to short-read TIS methods for essential genes, and better resolution around repeat elements.
- The Oxford Nanopore sequencing device that we used is cost-effective, small and easily portable.
- Thus, LoRTIS is an efficient means of uniquely identifying transposon insertion sites within long repetitive genetic elements and can be easily transported to, and used in, laboratories that lack access to expensive DNA sequencing facilities.
Introduction
- Transposon insertion site sequencing (TIS) is a robust tool used to identify the genetic loci associated with the phenotype of an organism1.
- The general approach for TIS methods has been developed over the last decade to understand the essential genes in many bacterial and eukaryotic species2,3. * TIS has helped the scientific community to understand genes involved in responses to a wide range of stress conditions and has enabled identification of many novel susceptibility and survival mechanisms1.
- The first step in TIS methods is the generation of a transposon mutant library in which the resulting pool of mutants contains many random transposon insertions in all genes (except essential genes) of the target organism at multiple positions4.
- The location of transposon insertions is identified by sequencing from the transposon insertion site into adjacent regions of the target genome5.
- Saturated mutant libraries can provide very high resolution coverage of the genome allowing the importance of regions within genes to be identified as well as simply a role for the gene itself6,7.
- To identify the repertoire of genes essential for responses to a particular stress condition, the transposon mutant library is grown under that stress condition (test) and in the absence of that stress (control).
- Differences in the prevalence of transposon mutants within the pools are compared between the stress condition of interest and the control6.
- Mutants with an advantage following exposure to the stress condition will proliferate while those that are disadvantaged will become depleted.
- Comparison of the total number of inserts at each site between control and condition experiments indicates loci of interest8.
- The DNA of mutant libraries is sequenced, using a customised protocol, [e.g. transposon-directed insertion site sequencing (TraDIS), transposon sequencing (Tn-seq), high-throughput insertion tracking by deep sequencing (HITS) and insertion sequencing (INSeq)], from the transposon reading into the adjacent genome to identify the location of transposon insertions5,9–11.
- These TIS methods use different transposon-genome junction enrichment techniques including restriction enzyme digestion, (e.g. MmeI), or the use of customised primer annealing followed by high-throughput sequencing of the resulting libraries5,12. These transposon based methods all use short-read sequencing, and the majority of them use either Illumina or Ion torrent sequencing platforms13.
- Sequence reads will not map uniquely to repeat sequences within a reference genome that are longer than the nucleotide sequence reads themselves (Fig. 1)14. * The sequence read length generated by Illumina-based approaches is around 300 base pairs (for a single read), and so reads cannot map uniquely to repeat-sequence motifs that are longer than these reads15.
- Examples of long repeat regions in E. coli include transposase insertion sequences, which are usually over 600 bp, and the several copies of the ribosomal RNA operons, which are over 5 kb16.
- The repeat regions are even a greater challenge in eukaryotic genomes which are much larger and have numerous repeating genetic elements.
- Many repeating elements are of evolutionary importance, and/or have roles in replication, recombination, transcription and genome rearrangement17.
- Thus, transposon insertions into different copies of a repeating element may have different phenotypic effects.
- Therefore, determining the unique locations of transposon insertion sites in repeating elements is essential for formulating meaningful hypotheses from TIS data.
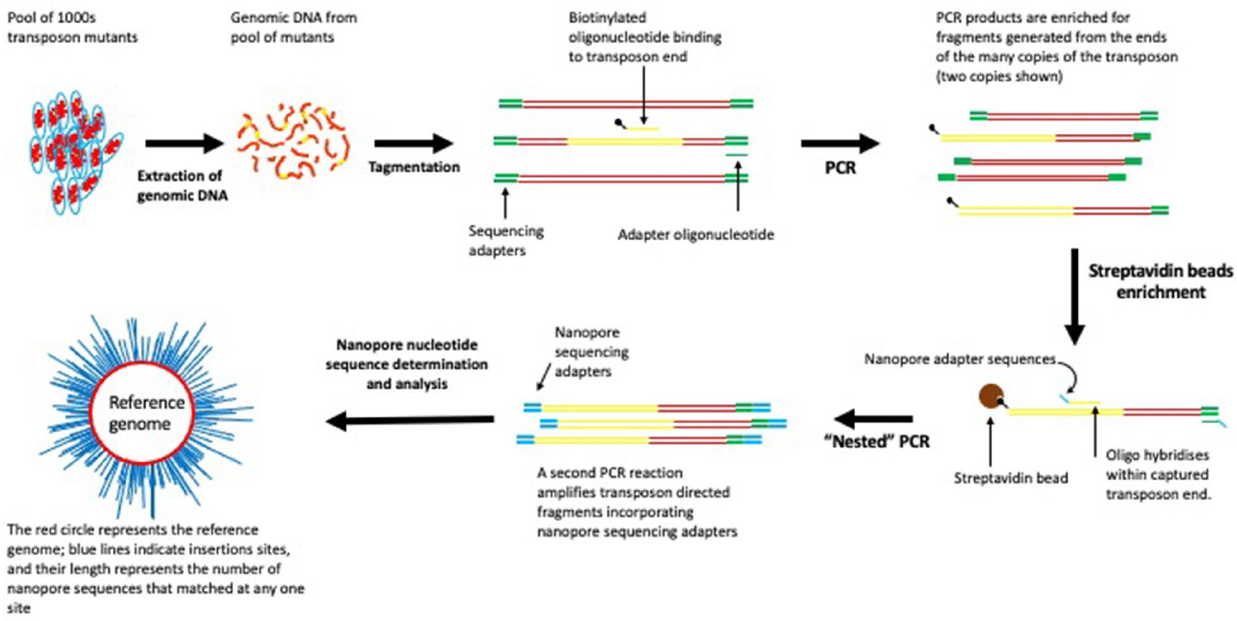
- Here, we describe LoRTIS (Long-Read Transposon Insertion-site Sequencing), which overcomes the limitations resulting from using short-read nucleotide sequences.
- In LoRTIS, as with other TIS methods, we have used the known sequences of the transposon as an anchor point from which to generate DNA fragments by amplification of DNA into the unknown transposon insertion sites for sequencing.
- However, in LoRTIS, by modifying this amplification step, much longer DNA fragments are generated, from which long sequence reads are possible using a MinION nanopore sequencer (Oxford Nanopore)18.
- The resulting long-reads are then aligned with the nucleotide sequence of a reference genome, and the locations identifying the precise positions of the transposon insertions sites determined.
- The longer reads can span repeat sequences and read into adjacent unique sites.
- The long-reads generated by LoRTIS were able to locate transposon insertion sites uniquely within repeating sequence elements of the ribosomal RNA operons, which are approximately 5 kb.
- In addition, we incorporated different nucleotide sequence identifiers (indexes) into replicates to demonstrate that multiple samples can be sequenced on a single flow cell.
- Thus, LoRTIS can identify hundreds of thousands of transposon insertion sites uniquely and overcomes the limitation of short-reads generated by other TIS methods.
Methods: DNA extraction and experimental setup
-
The Escherichia coli strain BW25113 large transposon mutant library described by Yasir et al. was used in this study6. Genomic DNA was extracted using the Quick-DNA Fungal/Bacterial 96 Kit (Zymo Research).
-
LoRTIS and TraDIS-Xpress samples were prepared in duplicate, from DNA extraction to sequencing library preparation. For LoRTIS each replicate was sequenced on a separate flow cell, so Native Barcodes (NBD) were added to each replicate to test the multiplexing capability of the LoRTIS method. NBD details are shown in Supplementary Table 5.
Methods: Preparation of DNA fragments for LoRTIS and TraDIS-Xpress
-
For LoRTIS sequencing library preparation,1 μg of genomic DNA was tagmented using an Illumina compatible MuSeek library preparation kit (Thermo Fisher Scientific) and the tagmented DNA was cleaned using 0.5× volume of AMPure XP magnetic beads (Beckman-Coulter). LongAmp Taq DNA polymerase (New England Biolabs, UK) was used with biotinylated oligonucleotide primers (Supplementary Table 5) that hybridise specifically within the transposon to amplify DNA fragments containing transposon insertion junctions (Supplementary Table 6 PCR1a). A second primer that anneals to MuSeek adapters (‘IonTMu-02’) was used to generate biotinylated double-stranded PCR products containing transposon-genome junctions (Supplementary Table 6 PCR1b). These PCR products were enriched using a Dynabeads™ kilobaseBINDER™ kit (Thermo Fisher Scientific). A nested PCR reaction was carried out using NEB LongAmp Taq polymerase to generate DNA fragments with Oxford Nanopore multiplexing adapters that lacked biotin (Supplementary Table 6 PCR2). The PCR products from this reaction were purified using 0.5× volume of AMPure XP magnetic beads (Beckman-Coulter) and were end-repaired, then nanopore sequencing adapters were ligated according to the Oxford Nanopore protocol.
-
For nucleotide sequencing by LoRTIS, the resulting DNA fragments were loaded onto an Oxford Nanopore MinION flowcell and sequenced for up to 48 h (Fig. 7). Using the LoRTIS protocol, nanopore long-read nucleotide sequence data were generated from two separate runs on an Oxford Nanopore flow cell to generate two replicate data sets that could be compared for reproducibility.
Methods: Bioinformatics
-
FAST5 format data from MinION was processed using Guppy Basecalling software (version 3.6.0) running in High Accuracy Calling (HAC) mode and using GPUs on the Quadram Institute cloud infrastructure. The resulting sequence data in FASTQ format were demultiplexed using QCat (version 1.1.0) and those reads containing transposon nucleotide sequences were identified, trimmed and retained, again using QCat. The nucleotide sequence reads were then located to the BW25113 reference genome (CP009273) using Bio-TraDIS (version 1.4.1)27 and Bio-LoRTIS software (version 0.0.5)28 in a similar way as for short-read data, except that steps to remove the transposon sequences were skipped and Minimap2 (version 2.17-r941)29 was substituted in the place of SMALT. Results from BioTradis, including plot files, were outputted in the same format as for short-read data.
-
The insertion patterns at candidate loci were inspected visually using Artemis (version 18.1.0) which was also used to capture images for figures30. Gene essentiality was determined using tradis_essentiality.R from BioTradis, and scatter charts were plotted to determine reproducibility using Microsoft Excel after normalising for numbers of reads in each replicate. The correlation between TraDIS-Xpress and LoRTIS data was calculated using Spearman's rank correlation coefficient.
TraDIS bioinformatics toolkit:
#https://www.biorxiv.org/content/10.1101/2022.05.26.493556v1.full.pdf
Nano-Bio-TraDIS: https://github.com/quadram-institute-bioscience/LoRTIS
http://sanger-pathogens.github.io/Bio-Tradis/
Downdetector: https://downdetector.com/status/openai/
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
- MCV病毒中的LT与sT蛋白功能
- Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Whole Genome Sequencing: Pricing and Services from Dante Labs and Other Leading Providers
重新审视诊断:微生物细胞游离DNA测序:解决与植入物相关的心血管感染中的未解决挑战