QIIME + Phyloseq + MicrobiotaProcess (v1)
gene_x 0 like s 1093 view s
Tags: R, processing, metagenomics, 16S, pipeline
# https://bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc//MicrobiotaProcess.html
# -----------------------------------
# ---- prepare the R environment ----
#Rscript MicrobiotaProcess.R
#NOTE: exit R script, then login again R-environment; rm -rf Phyloseq*_cache
#mkdir figures
rmarkdown::render('Phyloseq.Rmd',output_file='Phyloseq.html')
# with #alpha = 2.0, running the following script further!
# -----------------------------
# ---- 3.1. bridges other tools
##https://github.com/YuLab-SMU/MicrobiotaProcess
##https://www.bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc/MicrobiotaProcess.html
##https://chiliubio.github.io/microeco_tutorial/intro.html#framework
##https://yiluheihei.github.io/microbiomeMarker/reference/plot_cladogram.html
#BiocManager::install("MicrobiotaProcess")
#install.packages("microeco")
#install.packages("ggalluvial")
#install.packages("ggh4x")
library(MicrobiotaProcess)
library(microeco)
library(ggalluvial)
library(ggh4x)
library(gghalves)
## Convert the phyloseq object to a MicrobiotaProcess object
#mp <- as.MicrobiotaProcess(ps.ng.tax)
#mt <- phyloseq2microeco(ps.ng.tax) #--> ERROR
#abundance_table <- mt$abun_table
#taxonomy_table <- mt$tax_table
#ps.ng.tax_abund <- phyloseq::filter_taxa(ps.ng.tax, function(x) sum(x > total*0.01) > 0, TRUE)
#ps.ng.tax_most = phyloseq::filter_taxa(ps.ng.tax_rel, function(x) mean(x) > 0.001, TRUE)
##OPTION1 (NOT_USED): take all samples, prepare ps.ng.tax_abund --> mpse_abund
##mpse <- ps.ng.tax %>% as.MPSE()
#mpse_abund <- ps.ng.tax_abund %>% as.MPSE()
##OPTION2 (USED!): take partial samples, prepare ps.ng.tax or ps.ng.tax_abund (2 replacements!)--> ps.ng.tax_sel --> mpse_abund
ps.ng.tax_sel <- ps.ng.tax_abund
##otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax)[,c("1","2","5","6","7", "15","16","17","18","19","20", "29","30","31","32", "40","41","42","43","44","46")]
##NOTE: Only choose Group2, Group4, Group6, Group8
#> ps.ng.tax_sel
#otu_table() OTU Table: [ 37465 taxa and 29 samples ]
#sample_data() Sample Data: [ 29 samples by 10 sample variables ]
#tax_table() Taxonomy Table: [ 37465 taxa by 7 taxonomic ranks ]
#phy_tree() Phylogenetic Tree: [ 37465 tips and 37461 internal nodes ]
#-Group4: "21","22","23","24","25","26","27","28",
#-Group8: , "47","48","49","50","52","53","55"
otu_table(ps.ng.tax_sel) <- otu_table(ps.ng.tax_abund)[,c("8","9","10","12","13","14", "33","34","35","36","37","38","39","51")]
mpse_abund <- ps.ng.tax_sel %>% as.MPSE()
# A MPSE-tibble (MPSE object) abstraction: 2,352 × 20
# NOTE mpse_abund contains 20 variables: OTU, Sample, Abundance, BarcodeSequence, LinkerPrimerSequence, FileInput, Group,
# Sex_age <chr>, pre_post_stroke <chr>, Conc <dbl>, Vol_50ng <dbl>, Vol_PCR <dbl>, Description <chr>,
# Domain <chr>, Phylum <chr>, Class <chr>, Order <chr>, Family <chr>, Genus <chr>, Species <chr>
# -----------------------------------
# ---- 3.2. alpha diversity analysis
# Rarefied species richness + RareAbundance
mpse_abund %<>% mp_rrarefy()
# 'chunks' represent the split number of each sample to calculate alpha
# diversity, default is 400. e.g. If a sample has total 40000
# reads, if chunks is 400, it will be split to 100 sub-samples
# (100, 200, 300,..., 40000), then alpha diversity index was
# calculated based on the sub-samples.
# '.abundance' the column name of abundance, if the '.abundance' is not be
# rarefied calculate rarecurve, user can specific 'force=TRUE'.
mpse_abund %<>%
mp_cal_rarecurve(
.abundance = RareAbundance,
chunks = 400
)
# The RareAbundanceRarecurve column will be added the colData slot
# automatically (default action="add")
#NOTE mpse_abund contains 22 varibles = 20 varibles + RareAbundance <dbl> + RareAbundanceRarecurve <list>
# default will display the confidence interval around smooth.
# se=TRUE
# NOTE that two colors #c("#00A087FF", "#3C5488FF") for .group = pre_post_stroke; four colors c("#1f78b4", "#33a02c", "#e31a1c", "#6a3d9a") for .group = Group;
p1 <- mpse_abund %>%
mp_plot_rarecurve(
.rare = RareAbundanceRarecurve,
.alpha = Observe,
)
p2 <- mpse_abund %>%
mp_plot_rarecurve(
.rare = RareAbundanceRarecurve,
.alpha = Observe,
.group = Group
) +
scale_color_manual(values=c("#1f78b4", "#e31a1c")) +
scale_fill_manual(values=c("#1f78b4", "#e31a1c"), guide="none")
# combine the samples belong to the same groups if plot.group=TRUE
p3 <- mpse_abund %>%
mp_plot_rarecurve(
.rare = RareAbundanceRarecurve,
.alpha = "Observe",
.group = Group,
plot.group = TRUE
) +
scale_color_manual(values=c("#1f78b4", "#e31a1c")) +
scale_fill_manual(values=c("#1f78b4", "#e31a1c"),guide="none")
png("rarefaction_of_samples_or_groups.png", width=1080, height=600)
p1 + p2 + p3
dev.off()
# ------------------------------------------
# 3.3. calculate alpha index and visualization
library(ggplot2)
library(MicrobiotaProcess)
mpse_abund %<>%
mp_cal_alpha(.abundance=RareAbundance)
mpse_abund
#NOTE mpse_abund contains 28 varibles = 22 varibles + Observe <dbl>, Chao1 <dbl>, ACE <dbl>, Shannon <dbl>, Simpson <dbl>, Pielou <dbl>
f1 <- mpse_abund %>%
mp_plot_alpha(
.group=Group,
.alpha=c(Observe, Chao1, ACE, Shannon, Simpson, Pielou)
) +
scale_fill_manual(values=c("#1f78b4", "#e31a1c"), guide="none") +
scale_color_manual(values=c("#1f78b4", "#e31a1c"), guide="none")
f2 <- mpse_abund %>%
mp_plot_alpha(
.alpha=c(Observe, Chao1, ACE, Shannon, Simpson, Pielou)
)
#ps.ng.tax_sel contais only pre samples --> f1 cannot be generated!
png("alpha_diversity_comparison.png", width=1400, height=600)
f1 / f2
dev.off()
# -------------------------------------------
# 3.4. The visualization of taxonomy abundance (Class)
mpse_abund %<>%
mp_cal_abundance( # for each samples
.abundance = RareAbundance
) %>%
mp_cal_abundance( # for each groups
.abundance=RareAbundance,
.group=Group
)
mpse_abund
#NOTE mpse_abund contains 29 varibles = 28 varibles + RelRareAbundanceBySample <dbl>
# visualize the relative abundance of top 20 phyla for each sample.
# .group=time,
p1 <- mpse_abund %>%
mp_plot_abundance(
.abundance=RareAbundance,
taxa.class = Class,
topn = 20,
relative = TRUE
)
# visualize the abundance (rarefied) of top 20 phyla for each sample.
# .group=time,
p2 <- mpse_abund %>%
mp_plot_abundance(
.abundance=RareAbundance,
taxa.class = Class,
topn = 20,
relative = FALSE
)
png("relative_abundance_and_abundance.png", width= 1200, height=600) #NOT PRODUCED!
p1 / p2
dev.off()
#----
h1 <- mpse_abund %>%
mp_plot_abundance(
.abundance = RareAbundance,
.group = Group,
taxa.class = Class,
relative = TRUE,
topn = 20,
geom = 'heatmap',
features.dist = 'euclidean',
features.hclust = 'average',
sample.dist = 'bray',
sample.hclust = 'average'
)
h2 <- mpse_abund %>%
mp_plot_abundance(
.abundance = RareAbundance,
.group = Group,
taxa.class = Class,
relative = FALSE,
topn = 20,
geom = 'heatmap',
features.dist = 'euclidean',
features.hclust = 'average',
sample.dist = 'bray',
sample.hclust = 'average'
)
# the character (scale or theme) of figure can be adjusted by set_scale_theme
# refer to the mp_plot_dist
png("relative_abundance_and_abundance_heatmap.png", width= 1200, height=600)
aplot::plot_list(gglist=list(h1, h2), tag_levels="A")
dev.off()
# visualize the relative abundance of top 20 class for each .group (Group)
p3 <- mpse_abund %>%
mp_plot_abundance(
.abundance=RareAbundance,
.group=Group,
taxa.class = Class,
topn = 20,
plot.group = TRUE
)
# visualize the abundance of top 20 phyla for each .group (time)
p4 <- mpse_abund %>%
mp_plot_abundance(
.abundance=RareAbundance,
.group= Group,
taxa.class = Class,
topn = 20,
relative = FALSE,
plot.group = TRUE
)
png("relative_abundance_and_abundance_groups.png", width= 1000, height=1000)
p3 / p4
dev.off()
# ---------------------------
# 3.5. Beta diversity analysis
# ---------------------------------------------
# 3.5.1 The distance between samples or groups
# standardization
# mp_decostand wraps the decostand of vegan, which provides
# many standardization methods for community ecology.
# default is hellinger, then the abundance processed will
# be stored to the assays slot.
mpse_abund %<>%
mp_decostand(.abundance=Abundance)
mpse_abund
#NOTE mpse_abund contains 30 varibles = 29 varibles + hellinger <dbl>
# calculate the distance between the samples.
# the distance will be generated a nested tibble and added to the
# colData slot.
mpse_abund %<>% mp_cal_dist(.abundance=hellinger, distmethod="bray")
mpse_abund
#NOTE mpse_abund contains 31 varibles = 30 varibles + bray <list>
# mp_plot_dist provides there methods to visualize the distance between the samples or groups
# when .group is not provided, the dot heatmap plot will be return
p1 <- mpse_abund %>% mp_plot_dist(.distmethod = bray)
png("distance_between_samples.png", width= 1000, height=1000)
p1
dev.off()
# when .group is provided, the dot heatmap plot with group information will be return.
p2 <- mpse_abund %>% mp_plot_dist(.distmethod = bray, .group = Group)
# The scale or theme of dot heatmap plot can be adjusted using set_scale_theme function.
p2 %>% set_scale_theme(
x = scale_fill_manual(
values=c("#1f78b4", "#e31a1c"), #c("orange", "deepskyblue"),
guide = guide_legend(
keywidth = 1,
keyheight = 0.5,
title.theme = element_text(size=8),
label.theme = element_text(size=6)
)
),
aes_var = Group # specific the name of variable
) %>%
set_scale_theme(
x = scale_color_gradient(
guide = guide_legend(keywidth = 0.5, keyheight = 0.5)
),
aes_var = bray
) %>%
set_scale_theme(
x = scale_size_continuous(
range = c(0.1, 3),
guide = guide_legend(keywidth = 0.5, keyheight = 0.5)
),
aes_var = bray
)
png("distance_between_samples_with_group_info.png", width= 1000, height=1000)
p2
dev.off()
# when .group is provided and group.test is TRUE, the comparison of different groups will be returned
# Assuming p3 is a ggplot object after mp_plot_dist call
p3 <- mpse_abund %>%
mp_plot_dist(.distmethod = bray, .group = Group, group.test = TRUE, textsize = 6) +
theme(
axis.title.x = element_text(size = 14), # Customize x-axis label face = "bold"
axis.title.y = element_text(size = 14), # Customize y-axis label
axis.text.x = element_text(size = 14), # Customize x-axis ticks
axis.text.y = element_text(size = 14) # Customize y-axis ticks
)
# Save the plot with the new theme settings
png("Comparison_of_Bray_Distances.png", width = 1000, height = 1000)
print(p3) # Ensure that p3 is explicitly printed in the device
dev.off()
# Extract Bray-Curtis Distance Values and save them in a Excel-table.
library(dplyr)
library(tidyr)
library(openxlsx)
# Define the sample numbers vector
sample_numbers <- c("8", "9", "10", "12", "13", "14", "33", "34", "35", "36", "37", "38", "39", "51")
# Consolidate the list of tibbles using the actual sample numbers
bray_data <- bind_rows(
lapply(seq_along(mpse_abund$bray), function(i) {
tibble(
Sample1 = sample_numbers[i], # Use actual sample number
Sample2 = mpse_abund$bray[[i]]$braySampley,
BrayDistance = mpse_abund$bray[[i]]$bray
)
}),
.id = "PairID"
)
# Print the data frame to check the output
print(bray_data)
# Write the data frame to an Excel file
write.xlsx(bray_data, file = "Bray_Curtis_Distances.xlsx")
#DELETE the column "PairID" in Excel file
# -----------------------
# 3.5.2 The PCoA analysis
#install.packages("corrr")
library(corrr)
#install.packages("ggside")
library(ggside)
mpse_abund %<>%
mp_cal_pcoa(.abundance=hellinger, distmethod="bray")
# The dimensions of ordination analysis will be added the colData slot (default).
mpse_abund
mpse_abund %>% print(width=380, n=2)
#NOTE mpse_abund contains 34 varibles = 31 varibles + `PCo1 (30.16%)` <dbl>, `PCo2 (15.75%)` <dbl>, `PCo3 (10.53%)` <dbl>
#BUG why 36 variables in mpse_abund %>% print(width=380, n=1) [RareAbundanceBySample <list>, RareAbundanceByGroup <list>]
#> methods(class=class(mpse_abund))
# [1] [ [[<- [<-
# [4] $ $<- arrange
# [7] as_tibble as.data.frame as.phyloseq
#[10] coerce coerce<- colData<-
#[13] distinct filter group_by
#[16] left_join mp_adonis mp_aggregate_clade
#[19] mp_aggregate mp_anosim mp_balance_clade
#[22] mp_cal_abundance mp_cal_alpha mp_cal_cca
#[25] mp_cal_clust mp_cal_dca mp_cal_dist
#[28] mp_cal_nmds mp_cal_pca mp_cal_pcoa
#[31] mp_cal_pd_metric mp_cal_rarecurve mp_cal_rda
#[34] mp_cal_upset mp_cal_venn mp_decostand
#[37] mp_diff_analysis mp_diff_clade mp_envfit
#[40] mp_extract_abundance mp_extract_assays mp_extract_dist
#[43] mp_extract_feature mp_extract_internal_attr mp_extract_rarecurve
#[46] mp_extract_refseq mp_extract_sample mp_extract_taxonomy
#[49] mp_extract_tree mp_filter_taxa mp_mantel
#[52] mp_mrpp mp_plot_abundance mp_plot_alpha
#[55] mp_plot_diff_boxplot mp_plot_diff_res mp_plot_dist
#[58] mp_plot_ord mp_plot_rarecurve mp_plot_upset
#[61] mp_plot_venn mp_rrarefy mp_select_as_tip
#[64] mp_stat_taxa mutate otutree
#[67] otutree<- print pull
#[70] refsequence refsequence<- rename
#[73] rownames<- select show
# [ reached getOption("max.print") -- omitted 6 entries ]
#see '?methods' for accessing help and source code
# We also can perform adonis or anosim to check whether it is significant to the dissimilarities of groups.
mpse_abund %<>%
mp_adonis(.abundance=hellinger, .formula=~Group, distmethod="bray", permutations=9999, action="add")
mpse_abund %>% mp_extract_internal_attr(name=adonis)
#NOTE mpse_abund contains 34 varibles, no new variable, it has been saved in mpse_abund and can be extracted with "mpse_abund %>% mp_extract_internal_attr(name='adonis')"
#The result of adonis has been saved to the internal attribute !
#It can be extracted using this-object %>% mp_extract_internal_attr(name='adonis')
#The object contained internal attribute: PCoA ADONIS
#Permutation test for adonis under reduced model
#Terms added sequentially (first to last)
#Permutation: free
#Number of permutations: 9999
#
#vegan::adonis2(formula = .formula, data = sampleda, permutations = permutations, method = distmethod)
# Df SumOfSqs R2 F Pr(>F)
#Group 1 0.23448 0.22659 3.5158 5e-04 ***
#Residual 12 0.80032 0.77341
#Total 13 1.03480 1.00000
#---
#Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
# ("1","2","5","6","7", "15","16","17","18","19","20", "29","30","31","32", "40","41","42","43","44","46")
#div.df2[div.df2 == "Group1"] <- "aged.post"
#div.df2[div.df2 == "Group3"] <- "young.post"
#div.df2[div.df2 == "Group5"] <- "aged.post"
#div.df2[div.df2 == "Group7"] <- "young.post"
# ("8","9","10","12","13","14", "21","22","23","24","25","26","27","28", "33","34","35","36","37","38","39","51", "47","48","49","50","52","53","55")
#div.df2[div.df2 == "Group2"] <- "aged.pre"
#div.df2[div.df2 == "Group4"] <- "young.pre"
#div.df2[div.df2 == "Group6"] <- "aged.pre"
#div.df2[div.df2 == "Group8"] <- "young.pre"
#Group1: f.aged and post
#Group2: f.aged and pre
#Group3: f.young and post
#Group4: f.young and pre
#Group5: m.aged and post
#Group6: m.aged and pre
#Group7: m.young and post
#Group8: m.young and pre
#[,c("1","2","5","6","7", "8","9","10","12","13","14")]
#[,c("15","16","17","18","19","20", "21","22","23","24","25","26","27","28")]
#[,c("29","30","31","32", "33","34","35","36","37","38","39","51")]
#[,c("40","41","42","43","44","46", "47","48","49","50","52","53","55")]
#For the first set:
#a6cee3: This is a light blue color, somewhat pastel and soft.
#b2df8a: A soft, pale green, similar to a light lime.
#fb9a99: A soft pink, slightly peachy or salmon-like.
#cab2d6: A pale purple, reminiscent of lavender or a light mauve.
#For the second set:
#1f78b4: This is a strong, vivid blue, close to cobalt or a medium-dark blue.
#33a02c: A medium forest green, vibrant and leafy.
#e31a1c: A bright red, very vivid, similar to fire engine red.
#6a3d9a: This would be described as a deep purple, akin to a dark lavender or plum.
p1 <- mpse_abund %>%
mp_plot_ord(
.ord = pcoa,
.group = Group,
.color = Group,
.size = 2.4,
.alpha = 1,
ellipse = TRUE,
show.legend = FALSE # don't display the legend of stat_ellipse
) +
scale_fill_manual(
#values = c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#cab2d6", "#6a3d9a"),
#values = c("#a6cee3", "#b2df8a", "#fb9a99", "#cab2d6"),
values = c("#1f78b4", "#e31a1c"),
guide = guide_legend(keywidth=1.6, keyheight=1.6, label.theme=element_text(size=12))
) +
scale_color_manual(
#values=c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#cab2d6", "#6a3d9a"),
#values = c("#a6cee3", "#b2df8a", "#fb9a99", "#cab2d6"),
values = c("#1f78b4", "#e31a1c"),
guide = guide_legend(keywidth=1.6, keyheight=1.6, label.theme=element_text(size=12))
)
#scale_fill_manual(values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500")) +
#scale_color_manual(values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"))
#scale_fill_manual(values=c("#00A087FF", "#3C5488FF")) +
#scale_color_manual(values=c("#00A087FF", "#3C5488FF"))
#png("PCoA.png", width= 1000, height=1000)
#svg("PCoA.svg", width= 11, height=10)
#svg("PCoA_.svg", width=10, height=10)
#svg("PCoA.svg")
pdf("PCoA_Group2_vs_Group6.pdf")
p1
dev.off()
#FF0000: Red
#000000: Black
#0000FF: Blue
#C0C0C0: Silver
#00FF00: Lime (often referred to simply as Green in web colors)
#FFFF00: Yellow
#00FFFF: Aqua (also known as Cyan)
#FFA500: Orange
# The size of point also can be mapped to other variables such as Observe, or Shannon
# Then the alpha diversity and beta diversity will be displayed simultaneously.
p2 <- mpse_abund %>%
mp_plot_ord(
.ord = pcoa,
.group = Group,
.color = Group,
.size = Shannon,
.alpha = Observe,
ellipse = TRUE,
show.legend = FALSE # don't display the legend of stat_ellipse
) +
scale_fill_manual(
values = c("#1f78b4", "#e31a1c"), #only needs four colors.
#values = c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"),
guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8))
) +
scale_color_manual(
values = c("#1f78b4", "#e31a1c"), #only needs four colors.
#values=c("#FF0000", "#000000", "#0000FF", "#C0C0C0", "#00FF00", "#FFFF00", "#00FFFF", "#FFA500"),
guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8))
) +
scale_size_continuous(
range=c(0.5, 3),
guide = guide_legend(keywidth=0.6, keyheight=0.6, label.theme=element_text(size=8))
)
pdf("PCoA2_Group2_vs_Group6.pdf")
p2
dev.off()
# ------------------------------------------
# 3.5.3 Hierarchical cluster (tree) analysis
#input should contain hellinger!
mpse_abund %<>%
mp_cal_clust(
.abundance = hellinger,
distmethod = "bray",
hclustmethod = "average", # (UPGAE)
action = "add" # action is used to control which result will be returned
)
mpse_abund
mpse_abund %>% print(width=380, n=2)
#NOTE mpse_abund contains 34 varibles, no new variable, the column bray has been new calculated!
# if action = 'add', the result of hierarchical cluster will be added to the MPSE object
# mp_extract_internal_attr can extract it. It is a treedata object, so it can be visualized
# by ggtree.
sample.clust <- mpse_abund %>% mp_extract_internal_attr(name='SampleClust')
#The object contained internal attribute: PCoA ADONIS SampleClust
sample.clust
#--> The associated data tibble abstraction: 27 × 30
library(ggtree)
p <- ggtree(sample.clust) +
geom_tippoint(aes(color=Group)) +
geom_tiplab(as_ylab = TRUE) +
ggplot2::scale_x_continuous(expand=c(0, 0.01))
png("hierarchical_cluster1.png", width= 1000, height=800)
p
dev.off()
#https://bioconductor.org/packages/release/bioc/vignettes/MicrobiotaProcess/inst/doc//MicrobiotaProcess.html
# mapping = aes(x = RelRareAbundanceBySample-->Group,
# y = Sample-->Group,
# fill = Phyla
# ),
library(ggtreeExtra)
library(ggplot2)
# Extract relative abundance of phyla
phyla.tb <- mpse_abund %>%
mp_extract_abundance(taxa.class=Phylum, topn=30)
# The abundance of each samples is nested, it can be flatted using the unnest of tidyr.
phyla.tb %<>% tidyr::unnest(cols=RareAbundanceBySample) %>% dplyr::rename(Phyla="label")
phyla.tb
phyla.tb %>% print(width=380, n=10)
p1 <- p +
geom_fruit(
data=phyla.tb,
geom=geom_col,
mapping = aes(x = RelRareAbundanceBySample,
y = Sample,
fill = Phyla
),
orientation = "y",
#offset = 0.4,
pwidth = 3,
axis.params = list(axis = "x",
title = "The relative abundance of phyla (%)",
title.size = 4,
text.size = 2,
vjust = 1),
grid.params = list()
)
png("hierarchical_cluster2_Phyla.png", width = 1000, height = 800)
p1
dev.off()
# Extract relative abundance of classes
class.tb <- mpse_abund %>%
mp_extract_abundance(taxa.class = Class, topn = 30)
# Flatten and rename the columns
class.tb %<>% tidyr::unnest(cols = RareAbundanceBySample) %>% dplyr::rename(Class = "label")
# View the data frame
class.tb
# Create the plot
p1 <- p +
geom_fruit(
data = class.tb,
geom = geom_col,
mapping = aes(x = RelRareAbundanceBySample,
y = Sample,
fill = Class
),
orientation = "y",
pwidth = 3,
axis.params = list(axis = "x",
title = "The relative abundance of classes (%)",
title.size = 4,
text.size = 2,
vjust = 1),
grid.params = list()
)
# Save the plot to a file #ERROR-->NEED to be DEBUGGED!
png("hierarchical_cluster2_Class.png", width = 1000, height = 800)
print(p1)
dev.off()
# -----------------------
# 3.6 Biomarker discovery
library(ggtree)
library(ggtreeExtra)
library(ggplot2)
library(MicrobiotaProcess)
library(tidytree)
library(ggstar)
library(forcats)
#----BUG: why resulting in 26 taxa != 16 in the end ----
mpse_abund %>% print(width=150)
#mpse_abund %<>%
# mp_cal_abundance( # for each samples
# .abundance = RareAbundance
# ) %>%
# mp_cal_abundance( # for each groups
# .abundance=RareAbundance,
# .group=Group
# )
#mpse_abund
mpse_abund %<>%
mp_diff_analysis(
.abundance = RelRareAbundanceBySample,
.group = Group,
first.test.alpha = 0.01, filter.p="pvalue"
)
# The result is stored to the taxatree or otutree slot, you can use mp_extract_tree to extract the specific slot.
taxa.tree <- mpse_abund %>%
mp_extract_tree(type="taxatree")
taxa.tree
## And the result tibble of different analysis can also be extracted with tidytree (>=0.3.5)
#LDAupper, LDAmean, LDAlower,
taxa.tree %>% select(label, nodeClass, Sign_Group, fdr) #%>% dplyr::filter(!is.na(fdr))
taxa.tree %>% print(width=150, n=200)
# -- replace the pvalue and fdr with pvalue and p-adjusted from DESeq enrichment results --
#TODO: replace the values of pvalue and fdr in taxa.tree, with the values of pvalue and padj from sigtab, if the the tips in taxa.tree could be found in colnames(sigtab).
#tree_data <- get.data(taxa.tree)
#as.treedata(taxa.tree)
#d <- tibble(label = paste0('t', 1:4), trait = rnorm(4))
tree_data <- as_tibble(taxa.tree)
#full_join(x, d, by = 'label') %>% as.treedata
# Modify tree_data by joining with sigtab and updating Sign_Group
sigtab$label <- rownames(sigtab)
#write.xlsx(sigtab, file = "sigtab.xlsx")
sum(sigtab$padj<0.05))
#taxa.tree <- left_join(tree_data, sigtab[, c("label", "log2FoldChange", "pvalue", "padj")], by = 'label') %>% as.treedata
taxa.tree2 <- tree_data %>%
left_join(sigtab[, c("label", "baseMean", "log2FoldChange", "lfcSE", "stat", "pvalue", "padj")], by = "label") %>%
mutate(Sign_Group = case_when(
log2FoldChange > 0 & padj <= 0.05 ~ "Group2",
log2FoldChange < 0 & padj <= 0.05 ~ "Group6",
TRUE ~ NA_character_ # Sets Sign_Group to NA otherwise
)) %>%
as.treedata() # Convert the dataframe to a treedata object
taxa.tree2 %>% print(width=380, n=20)
# ---- print taxa_data2 to Excel, why resulting in 26 records? ----
taxa_data2 <- as_tibble(taxa.tree2)
sum(!is.na(taxa_data2$Sign_Group))
sapply(taxa_data2, class)
# Remove or transform list columns if not needed
taxa_data2_simplified <- taxa_data2 %>%
select(-RareAbundanceBySample, -RareAbundanceByGroup) %>%
mutate(across(where(is.list), ~toString(.))) # Convert lists to character strings if needed
# Replace NA with a placeholder, such as "NA" or another suitable representation
taxa_data2_simplified <- taxa_data2_simplified %>%
mutate(across(everything(), ~ifelse(is.na(.), "NA", .)))
taxonomy_data <- as.data.frame(mp_extract_taxonomy(mpse_abund))
colnames(taxa_data2_simplified)[colnames(taxa_data2_simplified) == "label"] <- "OTU"
combined_data <- left_join(taxa_data2_simplified, taxonomy_data, by = "OTU")
library(writexl)
write_xlsx(combined_data, "taxa_data2.xlsx")
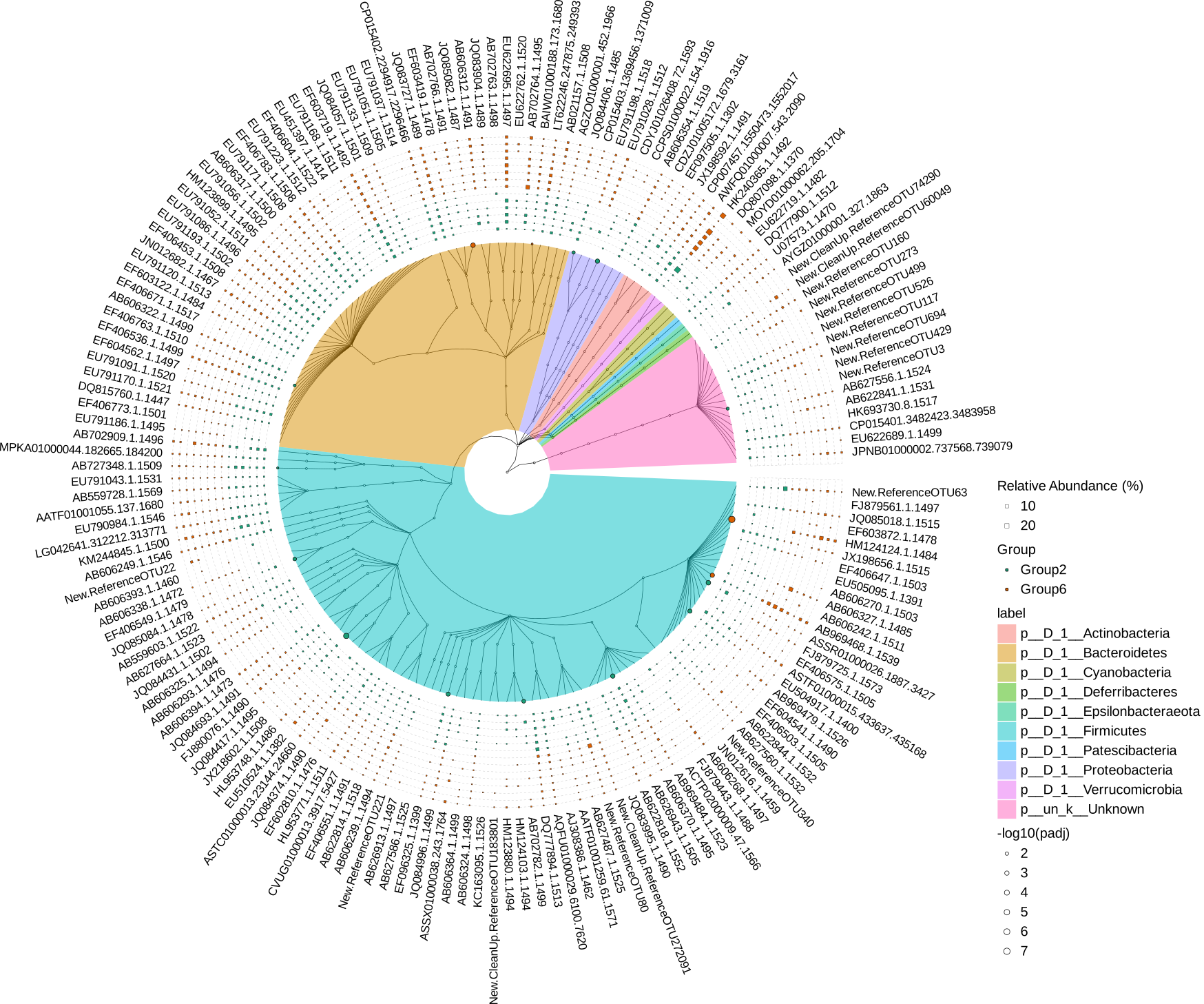
# Since taxa.tree is treedata object, it can be visualized by ggtree and ggtreeExtra
p1 <- ggtree(
taxa.tree2,
layout="radial",
size = 0.3
) +
geom_point(
data = td_filter(!isTip),
fill="white",
size=1,
shape=21
)
# display the high light of phylum clade.
p2 <- p1 +
geom_hilight(
data = td_filter(nodeClass == "Phylum"),
mapping = aes(node = node, fill = label)
)
# display the relative abundance of features(OTU)
p3 <- p2 +
ggnewscale::new_scale("fill") +
geom_fruit(
data = td_unnest(RareAbundanceBySample),
geom = geom_star,
mapping = aes(
x = fct_reorder(Sample, Group, .fun=min),
size = RelRareAbundanceBySample,
fill = Group,
subset = RelRareAbundanceBySample > 0
),
starshape = 13,
starstroke = 0.25,
offset = 0.03,
pwidth = 0.4,
grid.params = list(linetype=2)
) +
scale_size_continuous(
name="Relative Abundance (%)",
range = c(.5, 3)
) +
scale_fill_manual(values=c("#1B9E77", "#D95F02"))
# display the tip labels of taxa tree
p4 <- p3 + geom_tiplab(size=6, offset=4.0)
# display the LDA of significant OTU.
#p5 <- p4 +
# ggnewscale::new_scale("fill") +
# geom_fruit(
# geom = geom_col,
# mapping = aes(
# x = LDAmean,
# fill = Sign_Group,
# subset = !is.na(LDAmean)
# ),
# orientation = "y",
# offset = 0.3,
# pwidth = 0.5,
# axis.params = list(axis = "x",
# title = "Log10(LDA)",
# title.height = 0.01,
# title.size = 2,
# text.size = 1.8,
# vjust = 1),
# grid.params = list(linetype = 2)
# )
# display the significant (FDR-->pvalue-->padj) taxonomy after kruskal.test (default)
#shape = 21,
#scale_size_continuous(range=c(1, 3)) +
p6 <- p4 +
ggnewscale::new_scale("size") +
geom_point(
data=td_filter(!is.na(Sign_Group)),
mapping = aes(size = -log10(padj),
fill = Sign_Group,
),
shape = 21,
) +
scale_size_continuous(range=c(1, 4)) +
scale_fill_manual(values=c("#1B9E77", "#D95F02"))
svg("diff_analysis_Group2_vs_Group6.svg",width=22, height=22)
#png("differently_expressed_otu.png", width=2000, height=2000)
p6 + theme(
legend.key.height = unit(1.0, "cm"),
legend.key.width = unit(1.0, "cm"),
legend.spacing.y = unit(0.01, "cm"),
legend.text = element_text(size = 20),
legend.title = element_text(size = 20)
#legend.position = c(0.99, 0.01)
)
dev.off()
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Kraken2 Installation and Usage Guide
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- 病毒与癌症——挑战、突破与未来方向
- Processing Data_Michelle_RNAseq_2025
- 从共生到致病:金黄色葡萄球菌在鼻腔定植与人工关节感染过程中的转录表达谱
- 外排泵缺失揭示鲍曼不动杆菌 ATCC19606 对氯霉素的菌株特异性转录组适应机制
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Phyloseq + MicrobiotaProcess + PICRUSt2
MicrobiotaProcess Group2 vs Group6 (v1)