Clinical metagenomics [Talks for Shenzhen and so on]
gene_x 0 like s 1252 view s
Tags: metagenomics, 16S, RNA-seq
https://www.nature.com/articles/s41576-019-0113-7
Microbiome: The entirety of organisms that colonize individual sites in the human body.
Microarrays: Commonly referred to as ‘chips’, these platforms consist of spots of DNA fragments, antibodies or proteins printed onto surfaces, enabling massive multiplexing of hundreds to thousands of targets.
Reads: In DNA sequencing, reads are inferred sequences of base pairs corresponding to part of or all of a single DNA fragment.
Metagenomic NGS (mNGS): A shotgun sequencing approach in which all genomic content (DNA and/or RNA) of a clinical or environmental sample is sequenced.
Transmission network analysis: The integration of epidemiological, laboratory and genomic data to track patterns of transmission and to infer origin and dates of infection during an outbreak.
Precision medicine: An approach to medical care by which disease treatment and prevention take into account genetic information obtained by genomic or molecular profiling of clinical samples.
Reference standards: In laboratory test development, well-characterized, standardized and validated reference materials or databases that enable measurement of performance characteristics of an assay, including sensitivity, specificity and accuracy.
Latex agglutination: A clinical laboratory test for detection of a specific antibody in which the corresponding antigen is adsorbed on spherical polystyrene latex particles that undergo agglutination in the presence of the antibody.
Seroconversion: The development of detectable antibodies in the blood that are directed against an infectious agent, such as HIV-1, after which the infectious disease can be detected by serological testing for the antibody. 机体的免疫系统在受到抗原(包括细菌、病毒、甚至自身肿瘤细胞等)刺激后会产生抗体,我们从免疫细胞(B细胞)开始产生抗体算起,将血清中无法检测到抗体至能够检测到抗体的这个“转换点”定义为血清转换。
Library: In DNA sequencing, a collection of DNA fragments with known adapter sequences at one or both ends that is derived from a single clinical or environmental sample.
Sanger sequencing: A classical method of DNA sequencing based on selective incorporation of chain-terminating dideoxynucleotides developed by Frederick Sanger and colleagues in 1977; now largely supplanted by next-generation sequencing.
Subtyping (ST): In microbiology, refers to the identification of a specific genetic variant or strain of a microorganism (for example, virus, bacterium or fungus), usually by sequencing all or part of the genome.
Liquid biopsy: The detection of molecular biomarkers from minimally invasive sampling of clinical body fluids, such as DNA sequences in blood, for the purpose of diagnosing disease.
Spike-in: In laboratory test development, refers to the use of a nucleic acid fragment or positive control microorganism that is added to a negative sample matrix (for example, plasma from blood donors) or clinical samples and that serves as an internal control for the assay.
No-template control: In PCR or sequencing reactions, a negative control sample in which the DNA or cDNA is left out, thus monitoring for contamination that could produce false-positive results.
Biorobots: The automated instrumentation in the clinical laboratory that enables parallel processing of many samples at a time.
Point-of-care: Refers to diagnostic testing or other medical procedures that are done near the time and place of patient care (for example, at the bedside, in an emergency department or in a developing-world field laboratory).
Cluster density: On Illumina sequencing systems, a quality control metric that refers to the density of the clonal clusters that are produced, with each cluster corresponding to a single read. An optimal cluster density is needed to maximize the number and accuracy of reads generated from a sequencing run.
Q-score: A quality control metric for DNA sequencing that is logarithmically related to the base calling error probabilities and serves as a measurement of read accuracy.
Proficiency testing: A method for evaluating the performance of individual laboratories for specific laboratory tests using a standard set of unknown samples that permits interlaboratory comparisons.
Nanopore sequencing: A sequencing method in which DNA or RNA molecules are transported through miniature pores by electrophoresis. Sequencing reads are generated by measurement of transient changes in ionic current as the molecule passes through the pore.
Box 1 | Where is the signal — cellular or cell-free DNA? Metagenomic sequencing for clinical diagnostic purposes typically uses a shotgun approach by sequencing all of the DNA and/or RNA in a clinical sample. Clinical samples can vary significantly in their cellularity, ranging from cell-free fluids (that is, plasma, bronchoalveolar lavage fluid or centrifuged cerebrospinal fluid) to tissues. In the next-generation sequencing (NGS) field, there is great interest in the use of liquid biopsies from cell-free DNA (cfDNA) extracted from body fluids, such as plasma, to identify chromosomal or other genetic mutations and thus diagnose malignancies in the presymptomatic phase123. Similarly, cfDNA analysis has been useful for non-invasive prenatal testing applications, such as for the identification of trisomy 21 (ref.124 ). One study has described the potential utility of cfDNA analysis in diagnosing invasive fungal infection in cases where biopsy is not possible57. Another advantage to cfDNA analysis is the higher sensitivity of metagenomic sequencing owing to less cellular background from the human host. However, limitations of cfDNA analysis may include decreased sensitivity for detection of predominantly intracellular pathogens, such as human T cell lymphotropic virus, Rickettsia spp. and Pneumocystis jirovecii, and loss of the ability to interrogate cellular human host responses with RNA sequencing.
Box 2 | Nanopore sequencing
-
Abstract
- Clinical metagenomic next-generation sequencing (mNGS), the comprehensive analysis of microbial and host genetic material (DNA and RNA) in samples from patients, is rapidly moving from research to clinical laboratories.
- This emerging approach is changing how physicians diagnose and treat infectious disease, with applications spanning a wide range of areas, including antimicrobial resistance [x], the microbiome [x], human host gene expression (transcriptomics) [x] and oncology [x].
- Here, we focus on the challenges of implementing mNGS in the clinical laboratory and address potential solutions for maximizing its impact on patient care and public health.
-
Introduction
- The field of clinical microbiology comprises both diagnostic microbiology, the identification of pathogens from clinical samples to guide management and treatment strategies for patients with infection, and public health microbiology, the surveillance and monitoring of infectious disease outbreaks in the community.
- Traditional diagnostic techniques in the microbiology laboratory include growth and isolation of microorganisms in culture, detection of pathogen-specific antibodies (serology) or antigens and molecular identification of microbial nucleic acids (DNA or RNA), most commonly via PCR.
- [Disadvantage] While most molecular assays target only a limited number of pathogens using specific primers or probes, metagenomic approaches characterize all DNA or RNA present in a sample, enabling analysis of the entire microbiome as well as the human host genome or transcriptome in patient samples.
- Metagenomic approaches have been applied for decades to characterize various niches, ranging from marine environments1 to toxic soils2 to arthropod (节肢动物的) disease vectors 3,4 to the human microbiome5,6.
-
These tools have also been used to identify infections in ancient remains7, discover novel viral pathogens 8 [Viral pathogen discovery] and characterize the human virome in both healthy and diseased states9–11 and for forensic applications12.
-
The capacity to detect all potential pathogens — bacteria, viruses, fungi and parasites — in a sample and simultaneously interrogate host responses has great potential utility in the diagnosis of infectious disease.
- Metagenomics for clinical applications derives its roots from the use of microarrays in the early 2000s13,14.
- Some early successes using this technology include the discovery of the SARS coronavirus15, gene profiling of mutations in cancer16 and in-depth microbiome analysis of different sites in the human body17.
- However, it was the advent of next-generation sequencing (NGS) technologies in 2005 that jump-started the metagenomics field18.
- For the first time, millions to billions of reads could be generated in a single run, permitting analysis of the entire genetic content of a clinical or environmental sample.
-
The proliferation of available sequencing instruments and exponential decreases in sequencing costs over the ensuing decade drove the rapid adoption of NGS technology.
-
To date, several studies have provided a glimpse into the promise of NGS in clinical and public health settings.
- For example, NGS was used for the clinical diagnosis of neuroleptospirosis in a 14-year-old critically ill boy with meningoencephalitis19 [-->The enterovirus example in DAMIAN]; this case was the first to demonstrate the utility of metagenomic NGS (mNGS) in providing clinically actionable information, as successful diagnosis prompted appropriate targeted antibiotic treatment and eventual recovery of the patient.
- Examples in public health microbiology include the use of NGS, in combination with transmission network analysis20 [Integration of Sequencing and Epidemiologic Data for Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infections in a Tertiary-Care Hospital], to investigate outbreaks of the Escherichia coli strain O104:H4 (ref. 21) and for surveillance of antimicrobial resistance in the food supply by bacterial whole-genome sequencing22.
- Increasingly, big data provided by mNGS is being leveraged for clinical purposes, including characterization of antibiotic resistance directly from clinical samples23 and analysis of human host response (transcriptomic) data to predict causes of infection and evaluate disease risk24,25.
-
Thus, mNGS can be a key driver for precision diagnosis [What is the exact definition of precision diagnosis?] of infectious diseases, advancing precision medicine [precision diagnosis-->precision medicine] efforts to personalize patient care in this field.
-
Despite the potential and recent successes of metagenomics, clinical diagnostic applications have lagged behind research advances owing to a number of factors.
- [Factor 1] A complex interplay of microbial and host factors influences human health, as exemplified by the role of the microbiome in modulating host immune responses26, and it is often unclear whether a detected microorganism is a contaminant, colonizer or bona fide [真实地] pathogen.
- [Factor 2] Additionally, universal reference standards and proven approaches to demonstrate test validation, reproducibility and quality assurance for clinical metagenomic assays are lacking.
-
Considerations of cost, reimbursement, turnaround time, regulatory considerations and, perhaps most importantly, clinical utility also remain major hurdles for the routine implementation of clinical mNGS in patient care settings 27.
-
We review here the various applications of mNGS currently being exploited in clinical and public health settings.
- We discuss the challenges involved in the adoption of mNGS in the clinical laboratory, including validation and regulatory considerations that extend beyond its initial development in research laboratories, and propose steps to overcome these challenges.
- Finally, we envisage future directions for the field of clinical metagenomics and anticipate what will be achievable in the next 5 years.
-
Applications of clinical metagenomics
- To date, applications of clinical metagenomics have included infectious disease diagnostics for a variety of syndromes and sample types, microbiome analyses in both diseased and healthy states, characterization of the human host response to infection by transcriptomics and the identification of tumour-associated viruses and their genomic integration sites (Fig. 1; Table 1).
- Aside from infectious disease diagnostics, adoption of mNGS in clinical laboratories has been slow, and most applications have yet to be incorporated into routine clinical practice.
- Nonetheless, the breadth and potential clinical utility of these applications are likely to transform the field of diagnostic microbiology in the near future.
3.1. [TODO] Make a similar table as Table 1 for my own projects
- Sequencing method Clinical sample type Potential clinical indications Clinical test available? Refs
- Infectious disease diagnosis — targeted analyses: 1 or 2 papers
- Infectious disease diagnosis — untargeted analyses: 1 or 2 papers
- Microbiome analyses: 1 or 2 papers
- Human host response analyses: RNAseq data 1 or 2 papers
- [Optional] Oncological analyses
3.2. Applications of clinical metagenomics | Infectious disease diagnosis | Introduction
- The traditional clinical paradigm for diagnosis of infectious disease in patients, applied for more than a century, involves a physician formulating a differential diagnosis and then ordering a series of tests (generally ‘one bug, one test’) in an attempt to identify the causative agent.
- The spectrum of conventional testing for pathogens in clinical samples ranges from the identification of microorganisms growing in culture (for example, by biochemical phenotype testing or matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry), the detection of organism-specific biomarkers (such as antigen testing by latex agglutination or antibody testing by enzyme-linked immunosorbent assay (ELISA)) or nucleic acid testing by PCR for single agents to multiplexed PCR testing using syndromic panels.
- These panels generally include the most common pathogens associated with a defined clinical syndrome, such as meningitis (脑膜炎) and encephalitis [ensefәˊlaitis], acute respiratory infection, sepsis or diarrhoeal disease 28–31.
- Molecular diagnostic assays provide a fairly cost-effective and rapid (generally <2 hours of turnaround time) means to diagnose the most common infections.
- However, nearly all conventional microbiological tests in current use detect only one or a limited panel of pathogens at a time or require that a microorganism be successfully cultured from a clinical sample.
- By contrast, while NGS assays in current use cannot compare with conventional tests with respect to speed — the sequencing run alone on a standard Illumina instrument takes >18 hours — mNGS enables a broad range of pathogens — viruses, bacteria, fungi and/or parasites — to be identified from culture or directly from clinical samples on the basis of uniquely identifiable DNA and/or RNA sequences32.
- Another key advantage of NGS approaches is that the sequencing data can potentially be leveraged for additional analyses beyond the mere identification of a causative pathogen, such as microbiome characterization and parallel analyses of human host responses through transcriptome profiling by RNA sequencing (RNA-seq).
- Thus, the clinical utility of NGS in diagnosis may be in the most difficult-to-diagnose cases or for immunocompromised patients, in whom the spectrum of potential pathogens is greater.
- Eventually, mNGS may become cost competitive with multiplexed assays or used as an upfront ‘rule out’ assay to exclude infectious aetiologies.
- Of course, detection of nucleic acids, either by multiplex PCR panels or NGS, does not by itself prove that an identified microorganism is the cause of the illness, and findings have to be interpreted in the clinical context.
- In particular, discovery of an atypical or novel infectious agent in clinical samples should be followed up with confirmatory investigations such as orthogonal testing of tissue biopsy samples and demonstration of seroconversion or via the use of cell culture or animal models, as appropriate8, to ascertain its true pathogenic potential.
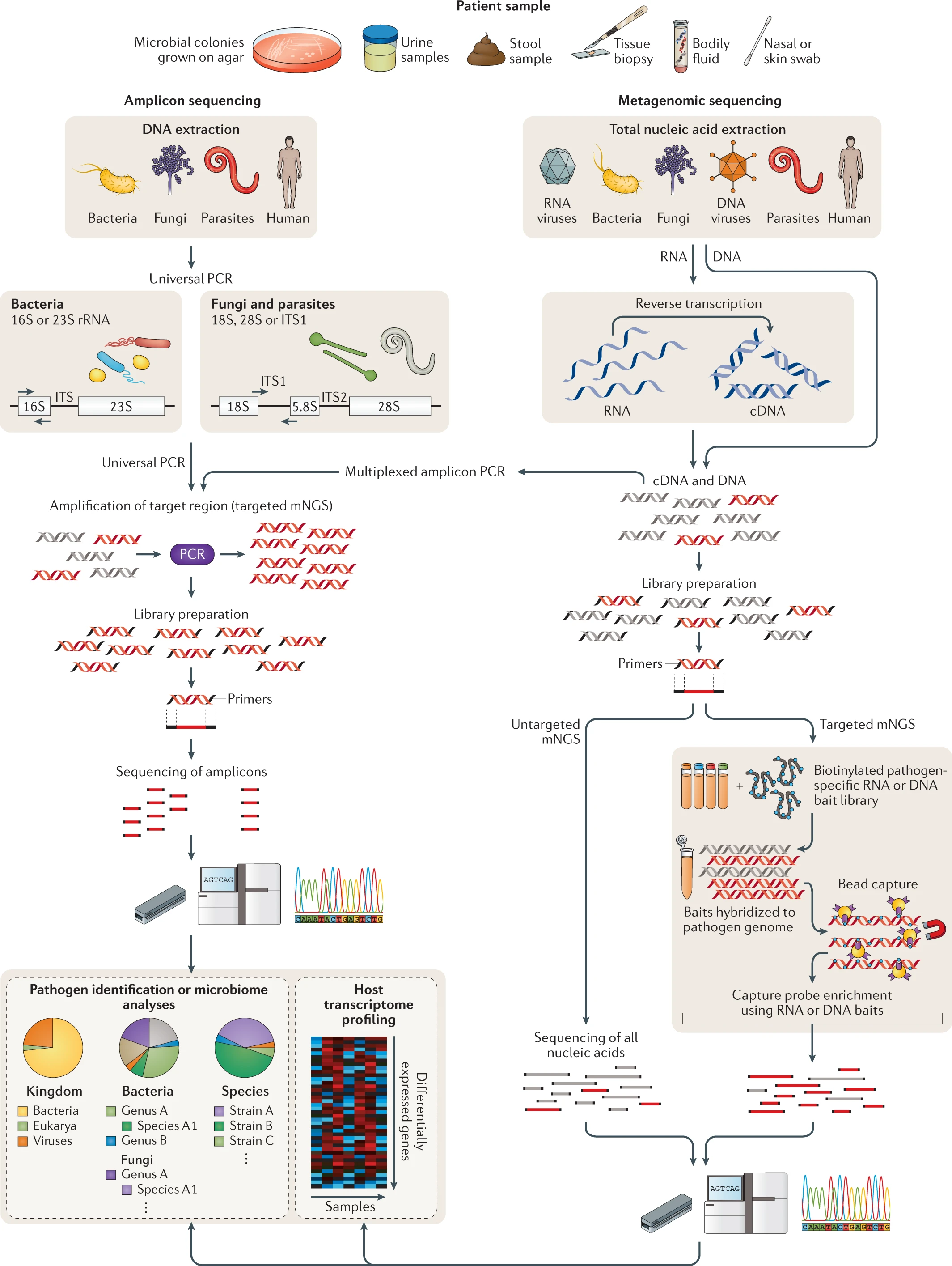
- * NGS of clinical samples as performed in either research or clinical laboratories involves a number of steps, including nucleic acid extraction, enrichment for DNA and/or RNA, library preparation, PCR amplification (if needed), sequencing and bioinformatics analysis (Fig. 2) .
- Any body fluid or tissue yielding sufficient nucleic acid is amenable to NGS analysis, which can either be targeted, that is, enriching individual genes or genomic regions, or untargeted, as is the case for metagenomic ‘shotgun’ approaches (Fig. 2).
- The details for the specific steps vary by laboratory and are described extensively elsewhere33–37.
3.3. * Applications of clinical metagenomics | Infectious disease diagnosis | Targeted NGS analyses
- 多重引物PCR(Multiplexed amplicon PCR)和通用PCR(Universal PCR)是两种不同的聚合酶链反应(PCR)技术,它们在设计和应用上有所区别:
- 多重引物PCR:
- 多重引物PCR是一种可以同时扩增多个目标DNA片段的PCR技术。在这种方法中,多对特异性引物在一个反应体系中使用,允许同时检测多个基因序列。
- 这种方法常用于病原体检测、遗传标记分析和多基因疾病的研究。例如,它可以在一个实验中同时检测多种病毒或细菌的存在。
- 多重引物PCR需要精确设计引物,以避免引物之间的交叉反应和非特异性扩增。
- 通用PCR:
- 通用PCR指使用一对引物针对广泛的物种或多个样本中的相同基因区域进行扩增的PCR技术。这对引物通常针对高度保守的DNA序列,使其能够扩增来自不同物种的相似序列。
- 通用PCR常用于物种鉴定、系统发育分析和环境样本的微生物多样性研究。例如,通过通用PCR可以从多个生物样品中扩增相同的微生物16S rRNA基因区域,以分析不同样品中的微生物组成。
- 通用PCR较为简单,但其特异性较低,可能会扩增到非目标序列。
- 总的来说,多重引物PCR适用于需要同时检测多个特定目标的情况,而通用PCR更适合于需要从广泛样本中扩增相同基因区域的应用。两者各有优势和局限性,选择哪种技术取决于实验的具体需求和目的。
- Universal primers refer to the primers used in universal PCR. Universal PCR is a type of polymerase chain reaction (PCR) where the same set of primers is used to amplify DNA from different species or sources. These primers are designed to target conserved regions of DNA that are similar across various organisms, making them useful for a wide range of applications, including taxonomic classification, phylogenetic studies, and environmental biodiversity assessments. The universal nature of these primers allows for the amplification of a specific gene or DNA segment from a broad array of species within a single PCR assay.
- Targeted approaches have the benefit of increasing the number and proportion of pathogen reads in the sequence data.
- This step can increase the detection sensitivity for microorganisms being targeted, although it limits the breadth of potential pathogens that can be identified.
- * An example of a targeted approach is the use of highly conserved primers for universal PCR amplification and detection of all microorganisms corresponding to a specific type from clinical samples, such as 16S ribosomal RNA (rRNA) gene amplification for bacteria38,39 and 18S rRNA and internal transcribed spacer (ITS) gene amplification for fungi40 (Fig. 2).
- Previously, such approaches were followed by Sanger sequencing of the resulting PCR amplicon to identify the pathogen and make a diagnosis; now, this step is commonly accomplished using NGS.
- Universal PCR for detection of bacteria and fungi has now been adopted in many hospital laboratories and has increased the number and proportion of infectious diagnoses39,41, although the technique is limited by the breadth of detection (that is, bacteria or fungi only or even a more limited range of targets, such as mycobacteria only, depending on the primer sets used) and by concerns regarding sensitivity42.
- * Another example of a targeted NGS approach is the design of primers tiled across the genome to facilitate PCR amplification and amplicon NGS for recovery of viral genomes directly from clinical samples43 .
- This method has been used to track the evolution and spread of Zika virus (ZIKV) in the Americas44–46 and of Ebola virus in West Africa47, with some demonstrations of real-time monitoring having an impact on public health interventions.
- * Another targeted approach is capture probe enrichment, whereby metagenomic libraries are subjected to (使遭受) hybridization using capture ‘bait’ probes48.
- These probes are generally 30–120 bp in length, and the number of probes can vary from less than 50 to more than 2 million 49–52.
- Although this enrichment method has been shown to increase the sensitivity of metagenomic detection in research settings, especially for viruses, it has yet to be used routinely for clinical diagnosis.
- A promising application of this approach may be the enrichment of clinical samples for characterization of antibiotic resistance23, a considerable problem in hospitals and the primary focus of the US National Action Plan for Combating Antibiotic-Resistant Bacteria53.
- * However, drawbacks of capture probe enrichment, compared with untargeted approaches for infectious disease diagnosis, include a bias towards targeted microorganisms, added steps, increased costs and long hybridization times (24–48 hours) as a result of the additional processing needed for maximal efficiency.
3.4. Applications of clinical metagenomics | Infectious disease diagnosis | Untargeted metagenomic NGS analyses
- Untargeted shotgun mNGS analyses forego (放弃) the use of specific primers (namely using Universal Primer in Amplicon sequencing) or probes (namely using baits in targeted mNGS) 54.
- Instead, the entirety of the DNA and/or RNA (after reverse transcription to cDNA) is sequenced.
- *(Can refer to the project of Holger and Anna) With pure cultures of bacteria or fungi, mNGS reads can be assembled into partial or complete genomes.
- (We have the method 3.3. targeted capture probe for the application here) These genome sequences are then used for subtyping and/or monitoring hospital outbreaks in support of infection control and/or public health surveillance efforts. - For example, a seminal study described theuse of whole-genome sequencing of multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae to track the origin and evolution of a hospital outbreak55.
- (We have the method 3.3. targeted capture probe for the application here) This study demonstrated for the first time the high-resolution mapping of likely transmission events in a hospital, some of which were unexpected on the basis of initial epidemiological data, and also identified putative resistance mutations in emerging resistant strains.
- The integration of genomic and epidemiological datayielded actionable insights that would have been useful for curbing transmission.
- Untargeted mNGS of clinical samples is perhaps the most promising approach for the comprehensive diagnosis of infections.
- In principle, nearly all pathogens, including viruses, bacteria, fungi and parasites, can be identified in a single assay56.
- mNGS is a needle-in-a-haystack endeavour, as only a small proportion (typically <1%) of reads are non-human, of which only a subset may correspond to potential pathogens.
- A limitation of mNGS is that the sensitivity of the approach is critically dependent on the level of background.
- Tissues, for example, have increased human host background relative to cell-free body fluids, resulting in a reduced number and proportion of microbial reads and hence a decrease in mNGS sensitivity33,36,37.
- Moreover, defining specific microbial profiles that are diagnostic or predictive of disease development can be difficult, especially from nonsterile sites that harbour a complex microbiome, such as respiratory secretions or stool6.
- Nevertheless, several groups have successfully validated mNGS in Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratories for the diagnosis of infections, including meningitis (脑膜炎) or encephalitis (脑炎)36,37, sepsis33,57 and pneumonia58, and these assays are now available for clinical reference testing of patients.
3.5. Applications of clinical metagenomics | Clinical microbiome analyses
- Many researchers now use mNGS instead of targeted sequencing of the 16S rRNA gene for in-depth characterization of the microbiome59.
- There is growing public awareness of the microbiome and its likely involvement in both acute and chronic disease states60.
- However, no microbiome-based tests have been clinically validated for the diagnosis or treatment of disease, in part owing to an incomplete understanding of the complexity of the microbiome and its role in disease pathogenesis.
- One future clinical application of microbiome analysis may be in the management and treatment of Clostridium difficile-associated disease.
- C. difficile is an opportunistic bacterium that can infect the gut, resulting in the production of toxins that can cause diarrhoea, dehydration, sepsis and death.
- C. difficile infection occurs only in the setting of a microbiome that is altered by factors such as exposure to broad-spectrum antibiotics or recent gastrointestinal surgery61.
- The importance of the microbiome in C. difficile infection is underscored by the 80–90% effectiveness of faecal stool transplantation in treating and potentially curing the disease62,63.
- The use of mNGS to characterize the microbiome in multiple studies has facilitated the development of bacterial probiotic mixtures that can be administered as pills for prophylaxis or treatment of C. difficile-associated disease (Fig. 1B).
- Another potential application of the microbiome is in the analysis of bacterial diversity, which can provide clues as to whether a patient’s illness is infectious or non-infectious.
- For example, a study of mNGS for the identification of respiratory pathogens in patients with pneumonia found that individuals with culture-proven infection had significantly less diversity in their respiratory microbiome25.
- Alterations of the microbiome, known as dysbiosis, have also been shown to be related to obesity, diabetes mellitus and inflammatory bowel disease64, and manipulation of the microbiome may be a pathway to treating these pathological conditions.
3.6. Applications of clinical metagenomics | Human host response analyses
- Clinical mNGS typically focuses on microbial reads; however, there is a complementary role for the analysis of gene expression in studying human host responses to infection65 (Fig. 1c).
- mNGS of RNA libraries used for the detection of pathogens such as RNA viruses in clinical samples incidentally produces host gene expression data for transcriptome (RNA-seq) analyses66.
- Although RNA-seq analyses are commonly performed on whole blood or peripheral blood mononuclear cell (PBMC) samples, any body fluid or tissue type is potentially amenable to these analyses.
- Classification of genes by expression profiling using RNA-seq has been used to characterize several infections, including staphylococcal bacteraemia67, Lyme disease68, candidiasis69, tuberculosis (discriminating between latent and active disease risk)70–72 and influenza73–75.
- Machine-learning-based analyses of RNA-seq data have been used for cancer classification76, and translation of these approaches may be promising for infectious diseases.
- Panels containing a limited number of host biomarkers are being developed as diagnostic assays for influenza77, tuberculosis70 and bacterial sepsis 78.
- Although no RNA-seq-based assay has been clinically validated to date for use in patients, the potential clinical impact of RNA-seq analyses is high.
- Interrogation of RNA reads from microorganisms corresponding to active microbial gene expression might enable the discrimination between infection versus colonization 25 and live (viable) versus dead organisms79.
- Moreover, RNA-seq analyses of the human host can be used to identify novel or underappreciated host–microbial interactions directly from clinical samples, as previously shown for patients with Lyme disease68, dengue 80 or malaria81.
- RNA-seq may be particularly useful in clinical cases in which the causative pathogen is only transiently present (such as early Lyme disease82 or arboviral infections, including West Nile virus83 or ZIKV84); analogous to serologic testing, indirect diagnosis of infections may be possible on the basis of a pathogen-specific human host response.
- Analysis of pathogen-specific host responses may also be useful in discriminating the bona fide causative pathogen or pathogens in a complex clinical metagenomic sample, such as a polymicrobial abscess or respiratory fluid25.
- * Yet another promising application of RNA-seq is in discriminating infectious versus non-infectious causes of acute illness25.
- If an illness is judged more likely to be non-infectious (for example, an autoimmune disease) on the basis of the host response, for example, clinicians may be more willing to discontinue antibiotics and treat the patient aggressively with steroids and other immunosuppressive medications.
- As large-scale sequencing data continue to be generated, perhaps driven by routine clinical mNGS testing, secondary mining of human reads might improve the accuracy of clinical diagnoses by incorporating both microbial and host gene expression data.
3.7. Applications of clinical metagenomics | Applications in oncology
- In oncology, whole-genome or directed NGS approaches to identify mutated genes can be used to simultaneously uncover viruses associated with cancer (that is, herpesviruses, papillomaviruses and polyomaviruses) and/or to gather data on virus–host interactions85.
- For example, mNGS was critical in the discovery of Merkel cell polyomavirus (Fig. 1d), now believed to be the cause of Merkel cell carcinoma, a rare skin cancer seen most commonly in elderly patients86.
- To date, the US Food and Drug Administration (FDA) has approved the clinical use of two NGS panels testing for actionable genomic aberrations in tumour samples 87. - Detection of reads corresponding to both integrated and exogenous viruses in these samples would be possible with the addition of specific viral probes to the panel or accomplished incidentally while sequencing the whole tumour genome or exome.
- Additional knowledge of integrated or active viral infections in cancers and their involvement in signalling pathways may inform preventive and therapeutic interventions with targeted antiviral and/or chemotherapeutic drugs88, as evidenced by the decreased risk of hepatitis C virus-associated hepatocellular carcinoma after treatment with direct-acting antiviral agents89.
- In the future, mNGS of cell-free DNA from liquid biopsy samples (for example, plasma) might be leveraged for the simultaneous identification of early cancer and diagnosis of infection in immunocompromised patients (Box 1).
- [OPTIONAL, or short with 2-3 slides, more technically, e.g. 4.1-4.4] Clinical implementation of metagenomic NGS
Implementation of mNGS in the clinical laboratory is a complex endeavour that requires customization of research protocols using a quality management approach consistent with regulatory standards 90. Library prepara tion reagents, sequencing instrumentation and bioin formatics tools are constantly changing in the research environment. However, in the clinical laboratory, assays need to be implemented following standardized (locked- down) protocols. Changes made to any component of the assay need to be validated and shown to have acceptable performance before testing in patients. Periodic updates and repeat validation studies are performed as deemed necessary to incorporate interim technological advances in NGS reagents, protocols and instrumentation. Metagenomic methods for pathogen detection pres ent a particularly challenging scenario for clinical vali dation (Fig. 3), as it is not practical to test an essentially unlimited number of different organisms for the assay to be considered validated. Although the FDA has pro vided general guidelines for clinical validation of NGS infectious disease testing91, there are no definitive reco mmendations for the clinical implementation of mNGS testing, nor is there mention of specific requirements. However, a best-practice approach can be taken that includes failure-mode analysis and evaluations of per formance characteristics using representative organ isms with ongoing assay monitoring and independent confirmation of unexpected results.
4.1. Clinical implementation of metagenomic NGS | Sensitivity and enrichment or depletion methods
Sensitivity and enrichment or depletion methods
A key limitation of mNGS is its decreased sensitivity with
high background, either predominantly from the human
host (for example, in tissue biopsies) or the microbiome
(for example, in stool). The background can be clini
cally relevant as the pathogen load in infections, such as
Shigella flexneri in stool from patients with diarrhoea92 or
ZIKV in plasma from patients with vector-borne febrile
illness93, can be very low (<103 copies per ml).
Host depletion methods for RNA libraries have been
developed and shown to be effective, including DNase I
treatment after extraction to remove residual human
background DNA94; the use of RNA probes followed
by RNase H treatment95; antibodies against human and
mitochondrial rRNA (the most abundant host RNA
types in clinical samples)96; and/or CRISPR–Cas9-based
approaches, such as depletion of abundant sequences by
hybridization97.
Unfortunately, there are no comparably effective
parallel methods for DNA libraries. Limited enrich
ment in the 3–5 times range can be achieved with
the use of antibodies against methylated human host
DNA98, which enriches microbial reads owing to the
lack of methylated DNA in most pathogen genomes.
Differential lysis of human cells followed by degrada
tion of background DNA with DNase I — thus retain
ing and enriching for nucleic acid from organisms with
cell walls, which include some bacteria and fungi — has
been shown to provide substantial microbial enrichment
of up to 1,000 times94,99,100. However, the performance of
differential lysis methods can be limited by a number
of factors. These limitations include potential decreased
sensitivity for microorganisms without cell walls, such
as Mycoplasma spp. or parasites; a possible paradoxi
cal increase in exogenous background contamination
by use of additional reagents101; and the inability to
detect free nucleic acid from dead organisms that are
lysed in vivo by human host immune cells or antibiotic
treatment. The importance of retaining the ability for
cell-free DNA detection from culture-negative samples
from dead organisms is also why incorporation of a
propidium monoazide treatment step to select for DNA
from live organisms may not be clinically useful as an
enrichment method for mNGS102 . In general, both the
differential lysis and propidium monoazide approaches
would also be cumbersome to implement in a highly
reproducible fashion, which is needed for clinical
laboratory validation.
To some extent, the human host background limi
tation may be overcome with brute force, made possi
ble by the increasing capacities of available sequencers.
For instance, an astrovirus was detected in a child with
encephalitis by ultradeep sequencing of brain tissue,
yielding only 1,612 reads out of ~134 million (0.0012%)
sequences103. Yet another approach to improve sensitiv
ity is to leverage a hybrid method for enrichment, such
as metagenomic sequencing with spiked primers46 .
Combining targeted with untargeted sequencing, the
method uses variably sized panels (100–10,000) of short
primers that are added (‘spiked’) into reaction mixtures
to enrich for specific target organisms while retaining
the breadth of metagenomic sequencing for off-target
organisms. When spiked at the reverse transcription
step, a panel of ZIKV-specific primers was found to
increase the number of ZIKV reads by more than ten
fold without appreciably decreasing broad metagenomic
sensitivity for other pathogens, enabling whole-genome
viral sequencing to characterize ZIKV spread from
Brazil into Central America and Mexico46.
4.2. Clinical implementation of metagenomic NGS | Laboratory workflow considerations
The complexity of mNGS analysis requires highly
trained personnel and extreme care in sample handling
to avoid errors and cross-contamination. Even miniscule
amounts of exogenous DNA or RNA introduced during
sample collection, aliquoting, nucleic acid extraction,
library preparation or pooling can yield a detectable
signal from contaminating reads. In addition, labora
tory surfaces, consumables and reagents are not DNA
free. A database of background microorganisms com
monly detected in mNGS data and arising from nor
mal flora or laboratory contamination101,104 typically
needs to be maintained for accurate mNGS analyses.
Microorganisms on this list are either not reported or
will require higher thresholds for reporting if they are
clinically significant organisms.
Clinical laboratory operations are characterized by
a defined workflow with scheduled staffing levels and
are less amenable to on-demand testing than those of
research laboratories. As samples are typically handled in
batches, the frequency of batch analysis is a major deter
minant of overall turnaround time. Unless fully auto
mated sample-handling systems are readily available,
wet lab manipulations for mNGS require considerable
hands-on time to perform, as well as clinical staff who
are highly trained in molecular biology procedures.
There are ergonomic concerns with repetitive tasks
such as pipetting, as well as potential for inadvertent
sample mix-up or omission of critical steps in the work
flow. Maintaining high quality during complex mNGS
procedures can be stressful to staff, as slight deviations in
sample handling can lead to major changes in the results
generated. Separating the assay workflow into multiple
discrete steps to be performed by rotating shifts can be
helpful to avoid laboratory errors.
4.3. Clinical implementation of metagenomic NGS | Reference standards
Well-characterized reference standards and controls areneeded to ensure mNGS assay quality and stability overtime. Most available metagenomic reference materialsare highly customized to specific applications (for example, ZymoBIOMICS Microbial Community Standardfor microbiome analyses and bacterial and fungal metagenomics105) and/or focused on a more limited spectrum of organisms (for example, the National Instituteof Standards and Technology (NIST) reference materialsfor mixed microbial DNA detection, which contain onlybacteria106). Thus, these materials may not be applicableto untargeted mNGS analyses.
Custom mixtures consisting of a pool of microorganisms (mock microbial communities) or theirnucleic acids can be developed as external controls toestablish limits of detection for mNGS testing. Internalspike-in control standards are available for other NGS
applications such as transcriptome analysis by RNA-seq, with External RNA Controls Consortium (ERCC)RNA standards composed of synthetic RNA oligonucleotides spanning a range of nucleotide lengths andconcentrations 107. The complete set or a portion ofthe ERCC RNA standards (or their DNA equivalents)can be used as spike-in internal controls to controlfor assay inhibition and to quantify titres of detectedpathogens by standard curve analysis108. Nonetheless,
the lack of universally accepted reference standards formNGS makes it difficult to compare assay performancesbetween different laboratories. There is a critical needfor standardized reference organisms and genomicmaterials to facilitate such comparisons and to defineoptimal analysis methods.
4.4. Clinical implementation of metagenomic NGS | Bioinformatics challenges
User-friendly bioinformatics software for analysis ofmNGS data is not currently available. Thus, customizedbioinformatics pipelines for analysis of clinical mNGSdata56,109–111 still require highly trained programming staffto develop, validate and maintain the pipeline for clinicaluse. The laboratory can either host computational servers locally or move the bioinformatics analysis and datastorage to cloud platforms. In either case, hardware andsoftware setups can be complex, and adequate measuresmust be in place to protect confidential patient sequence
data and information, especially in the cloud environment.Storage requirements for sequencing data can quicklybecome quite large, and the clinical laboratory must decideon the quantity, location and duration of data storage.
Bioinformatics pipelines for mNGS analysis use anumber of different algorithms, usually developed forthe research setting and constantly updated by software developers. As for wet lab procedures, it is usuallynecessary to make custom modifications to the pipelinesoftware and then lock down both the software and reference databases for the purposes of clinical validation112.
A typical bioinformatics pipeline consists of a series of
analysis steps from raw input FASTQ files including
quality and low-complexity filtering, adaptor trimming,
human host subtraction, microorganism identification
by alignment to reference databases, optional sequence
assembly and taxonomic classification of individual
reads and/or contiguous sequences (contigs) at levels
such as family, genus and species (Fig. 4). Each step in
the pipeline must be carefully assessed for accuracy and
completeness of data processing, with consideration for
propagation of errors. Sensitivity analyses should be
performed with the inclusion of both in silico data and
data generated from clinical samples. Customized data
sets can be prepared to mimic input sequence data and
expand the range of microorganisms detected through in
silico analysis37. The use of standardized reference mate
rials and NGS data sets is also helpful in comparative
evaluation of different bioinformatics pipelines105.
Additionally, public databases for microbial reference
genomes are being continuously updated, and laborato
ries need to keep track of the exact versions used in addi
tion to dealing with potential misannotations and other
database errors. Larger and more complete databases
containing publicly deposited sequences such as the
National Center for Biotechnology Information (NCBI)
Nucleotide database are more comprehensive but also
contain more errors than curated, more limited data
bases such as FDA-ARGOS91,113 or the FDA Reference
Viral Database (RVDB) 114. A combined approach that
incorporates annotated sequences from multiple data
bases may enable greater confidence in the sensitivity
and specificity of microorganism identification.
Performance validation and verification for bioinfor
matics analysis constitute a time-consuming endeavour
and include analysis of control and patient data sets and
comparisons, with orthogonal clinical testing to deter
mine the accuracy of the final result36. Establishing
thresholds enables separation of true-positive matches
from the background, and these thresholds can incor
porate metrics such as the number of sequence reads
aligning to the detected microorganism, normalized to
reads per million, external no-template control samples or
internal spike-in material; the number of nonoverlapping
genomic regions covered; and the read abundance in clin
ical samples relative to negative control samples (to avoid
reporting of contaminant organisms). Receiver–operator
curve (ROC) analysis is a useful tool to determine opti
mal threshold values for a training set of clinical samples
with known results, with verification of pre-established
thresholds using an independent validation set36.
As in the wet lab workflow, analysis software and ref
erence databases should ideally be locked down before
validation and clinical use. Many laboratories maintain
both production and up-to-date development versions
of the clinical reference database (for example, the NCBI
nucleotide database is updated every 2 weeks), with the
production database being updated at regular, prespec
ified intervals. Standardized data sets should be used to
verify the database after any update and to ensure that
assay results are accurate and reproducible, as errors
can be introduced from newly deposited sequences and
clinical metadata.
4.5. Clinical implementation of metagenomic NGS | Cost considerations
Although there have been substantial cost reductions in
the generation of sequence data, the overall per-sample
reagent cost for sequencing remains fairly high. Most lab
oratories lack the robotic equipment and established
automated protocols to multiplex large numbers
of patient samples in a single run. Thus, the majority of
library preparation methods for mNGS are performed
manually and hence incur considerable staff time. The
additional resources needed to run and maintain a
bioinformatics analysis pipeline are also considerable,
and steps taken to ensure regulatory oversight can add
notably to costs as well. This leads to an overall cost
of several hundreds to thousands of dollars per sam
ple analysed, which is higher than that for many other
clinical tests.
Technical improvements in hardware are needed
for mNGS sample processing to increase throughput
and to reduce costs. As NGS procedures become more
standardized, there has been a drive towards increasing
automation with the use of liquid-handling biorobots115.
Typically, two biorobots are needed for clinical mNGS
for both the pre-amplification and post-amplification
steps to avoid PCR amplicon cross-contamination.
Increased multiplexing is also possible with the greatly
enhanced output from the latest generation of sequenc
ers, such as the Illumina NovaSeq instruments. However,
a potential limitation with running larger numbers of
samples per run is longer overall turnaround times for
clinical use owing to the requirement for batch pro
cessing as well as sample workflow and computational
analysis considerations. Additionally, high-throughput
processing of clinical samples for NGS may only be
possible in reference laboratories. The development of
microfluidic devices for NGS sample library preparation,
such as VolTRAX116, could eventually enable clinicians
to use mNGS more widely in hospital laboratories or
point-of-care settings.
4.6. Clinical implementation of metagenomic NGS | Regulatory considerations
Clinical laboratories are highly regulated, and general
laboratory and testing requirements apply to all mole
cular diagnostic assays reported for patient care 90 .
Quality control is paramount, and methods must be
developed to ensure analytic accuracy throughout the
assay workflow. Important quality control steps can
include initial sample quality checks, library param
eters (concentration and size distribution), sequence
data generation (cluster density and Q-score), recovery of
internal controls and performance of external controls.
Validation data generated during assay development and
implementation should be recorded and made availa
ble to laboratory inspectors (for laboratory-developed
tests) or submitted to regulatory agencies, such as the
FDA in the USA or the European Medicines Agency
(EMA) in Europe, for approval.
Ongoing monitoring is particularly important for
mNGS assays to verify acceptable performance over
time and to investigate atypical findings36. Monitoring is
accomplished using sample internal controls, intra-run
control samples, swipe tests for contamination and perio
dic proficiency testing. Unexpected or unusual results are
further investigated by reviewing patients’ clinical charts
or by confirmatory laboratory testing using orthogonal
methods. Identification of microorganisms that have
not been identified before in the laboratory should be
independently confirmed, usually through clinical ref
erence or public health laboratory testing. Atypical or
novel organisms should be assessed for their clinical
significance, and these findings should be reported and
discussed with health-care providers, with consideration
for their potential pathogenicity and for further testing
and treatment options. Clinical microbial sequencing
boards, modelled after tumour boards in oncology, can
be convened via real-time teleconferencing to discuss
mNGS results with treatment providers in clinical con
text (Fig. 3). Detection of microorganisms with public
health implications such as Sin Nombre hantavirus or
Ebola virus should be reported, as appropriate, to the
relevant public health agencies.
-
Conclusions and future perspectives
Technological advancements in library preparation methods, sequence generation and computational bio informatics are enabling quicker and more comprehen sive metagenomic analyses at lower cost. Sequencing technologies and their applications continue to evolve. Real-time sequencing in particular may be a game- changing technology for point-of-care applications in clinical medicine and public health, as laboratories have begun to apply these tools to diagnose atypical infec tions and track pathogen outbreaks, as demonstrated by the recent deployment of real-time nanopore sequencing for remote epidemiological surveillance of Ebola44 and ZIKV44,45, and even for use aboard the International Space Station117 (Box 2). Nonetheless, formidable challenges remain when implementing mNGS for routine patient care. In par ticular, sensitivity for pathogen detection is decreased in clinical samples with a high nucleic acid background or with exceedingly low pathogen titres; this concern is only partially mitigated by increasing sequencing depth per sample as costs continue to drop. As a comprehen sive direct detection method, mNGS may eventually replace culture, antigen detection and PCR methods in clinical microbiology, but indirect approaches such as viral serological testing will continue to play a key part in the diagnostic work-up for infections27, and functional assays such as culture and phenotypic susceptibility test ing will likely always be useful for research studies. In summary, while current limitations suggest that mNGS is unlikely to replace conventional diagnostics in the short term, it can be a complementary, and perhaps essential, test in certain clinical situations. Although the use of mNGS for informing clinical care has been demonstrated in multiple case reports and small case series118, nearly all studies have been retro spective, and clinical utility has yet to be established in a large-scale prospective clinical trial. Prospective clinical studies will be critical to understand when to perform mNGS and how the diagnostic yield compares with that of other methods. For example, the mNGS transcrip tomic approach might enable effective treatment triage, whereby antimicrobials are only needed for patients showing an ‘infectious profile’ of gene expression and those with a ‘non-infectious profile’ can be treated for other causes. In particular, prospective clinical trial and economic data showing the cost-effectiveness of these relatively expensive tests in improving patient outcomes are needed to justify their use. These data will also sup port a pathway towards regulatory approval and clini cal reimbursement. High-quality evidence that clinical metagenomic assays are effective in guiding patient management will require protocols that minimize potential assay and patient selection bias and compare relevant health outcomes using data sets generated from large patient cohorts119,120. We predict that, over the next 5 years, prospective clinical trial data evaluating the clinical utility and cost- effectiveness of mNGS will become available; overall costs and turnaround time for mNGS will continue to drop; other aspects of mNGS beyond mere identifica tion, such as incorporation of human host response and microbiome data, will prove clinically useful; robotic sample handling and microfluidic devices will be devel oped for push-button operation; computational analysis platforms will be more widely available, both locally and on the cloud, obviating the need for dedicated bioinfor matics expertise; and at least a few mNGS-based diag nostic assays for infectious diseases will attain regulatory approval with clinical reimbursement. We will witness the widespread democratization of mNGS as genomic analyses become widely accessible not only to physicians and researchers but also to patients and the public via crowdsourcing initiatives121,122 . Furthermore, in a world with constantly emerging pathogens, we envisage that mNGS-based testing will have a pivotal role in monitor ing and tracking new disease outbreaks. As surveillance networks and rapid diagnostic platforms such as nano pore sequencing are deployed globally, it will be possi ble to detect and contain infectious outbreaks at a much earlier stage, saving lives and lowering costs. In the near future, mNGS will not be a luxury but a necessity in the clinician’s armamentarium as we engage in the perpetual fight against infectious diseases. -
Fig. 1 for chapter 4 | Clinical applications of metagenomic sequencing (USING: Overview of applications of clinical metagenomics:
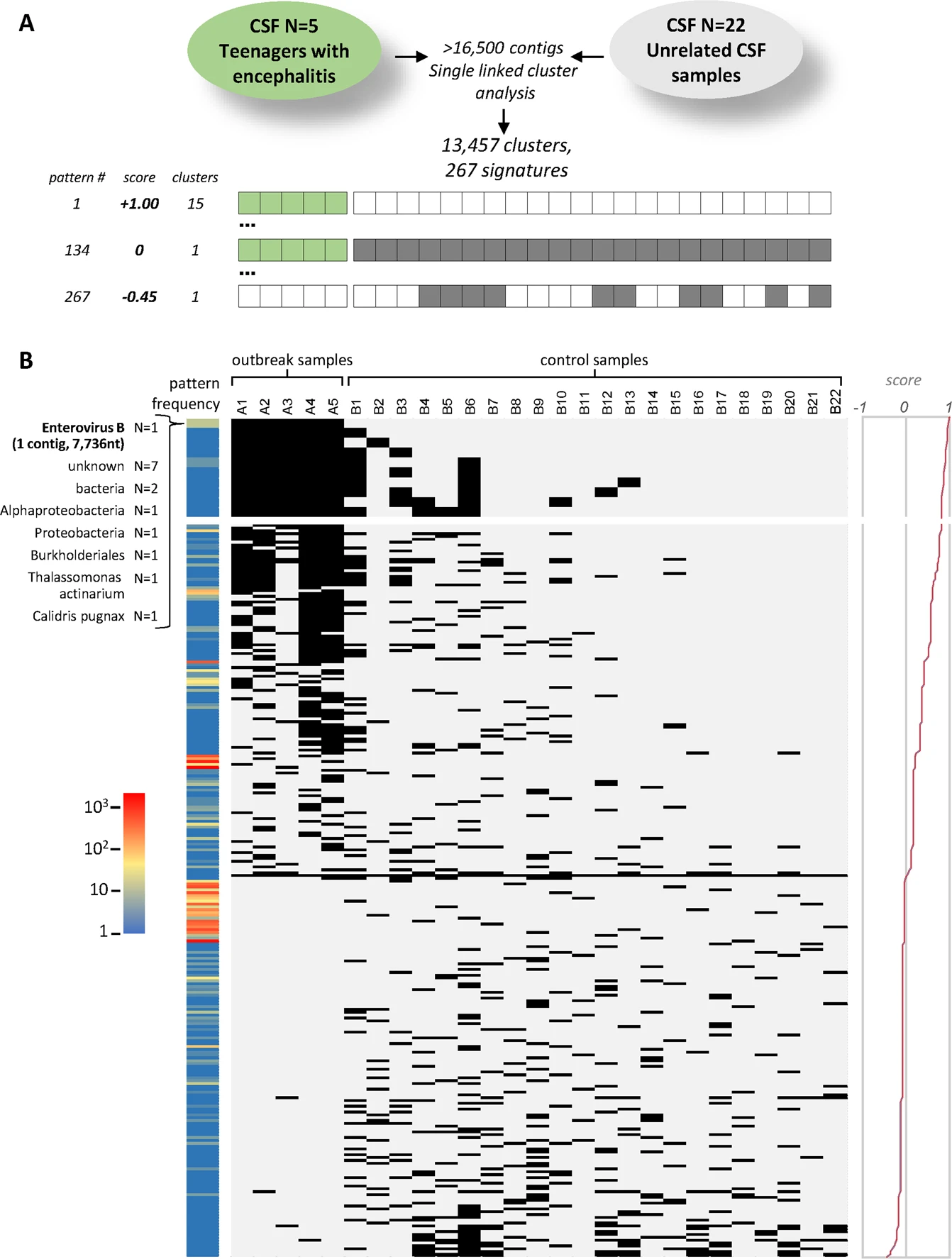
- Infectius disease diagnostics (untargeted analyses using metagnenomic sequencing using DAMIAN): DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples, explain the cohort samples! will be further developed!
- With the methods, we can only assemble a small part or a short contig of virus or bacteria. However, if we want to know if want to compare two different isolates, we need generally the complete sequences of virus, we can use the targeted capture sequencing!
- Infectius disease diagnostics (targeted analyses using capture probe enrichment) Paper: Target capture sequencing reveals a monoclonal outbreak of respiratory syncytial virus B infections among adult hematologic patients
-
Microbiome analyses (Metagenomic sequencing using 16S Amplicon sequencing or Unbiased shotgun metagenomics?) • Unbiased shotgun metagenomics • Amplicon metagenomics Fragment DNA and sequence PCR amplify a gene of interest randomly Tells you what types of organisms there are Bacteria/Archaea (16S rRNA), Microbial Unexpected Viral Euks (18S rRNA), Fungi (ITS), #DELETE "Virus (no Infection good marker) ----> • Targeted analyses using capture probe enrichment"
-
Human host response analyses (RNA sequencing)
-
[TODO]: based on the Figure 2, make shorter slide (Overview.png)
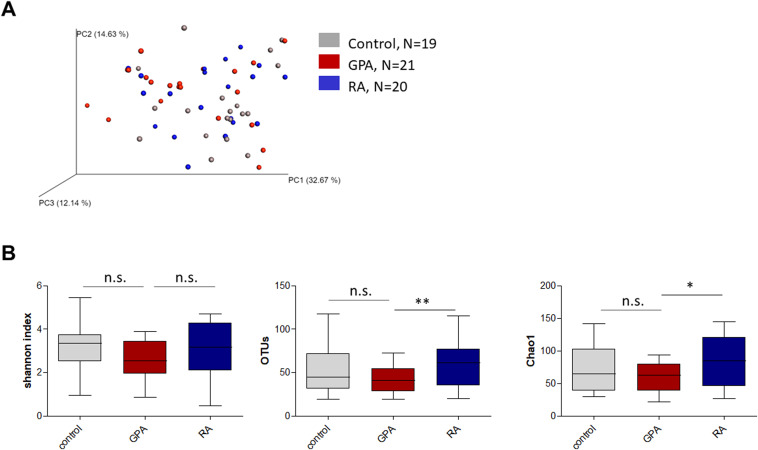
- Project1: mark the keywords in the plots with highlighted color e.g. with green: Amplicon sequencing + Bacteria + microbiome analyses; Changes in the composition of the upper respiratory tract microbial community in granulomatosis with polyangiitis; Fig. 1. Alpha and beta diversity of nasal samples from patients with GPA and RA and healthy controls. (Figure1.jpg)
- Project2: keywords: Metagenomic sequencing + Targeted mNGS + microbiome analyses (monitoring hospital outbreaks); Target capture sequencing reveals a monoclonal outbreak of respiratory syncytial virus B infections among adult hematologic patients
- Project 3: keywords: Metagenomic sequencing + Untargeted mNGS + Pathogen identification; DAMIAN: an open source bioinformatics tool for fast, systematic and cohort based analysis of microorganisms in diagnostic samples in the example enterovirus detection Figure7.png.
- Project 4: keywords: Metagenomic sequencing + Untargeted mNGS + microbiome analyses; (Can refer to the project of Holger and Anna) With pure cultures of bacteria or fungi, mNGS reads can be assembled into partial or complete genomes; Based on the provided description, the project does indeed relate to microbiome analyses; Genomics of Invasive Cutibacterium acnes Isolates from Deep-Seated Infections; C.acnes_Figure1.jpg
- Future project 5: keywords: Metagenomic sequencing + Untargeted mNGS + Host transcriptome profiling: Flowchart3.png.
-
A
- Applications in infectious disease diagnostics include direct identification of microorganisms from primary clinical samples (part Aa);
- antimicrobial resistance prediction by characterization of resistance genes (part Ab);
- detection of species-level or strain-level virulence determinants, such as secretion of specific endotoxins or exotoxins (part Ac);
-
and antiviral resistance prediction (part Ad). As shown for HIV-1, recovery of the complete viral genome from a patient sample by metagenomic next-generation sequencing (mNGS) (part Ad, graph) facilitates sequence analysis to predict susceptibility [sәseptәˊbiliti] or resistance to antiretroviral drugs (part Ad, bar plot); [????] the susceptibility profile for the analysed strain (black bars) predicts resistance to the non-nucleoside reverse transcriptase inhibitor (NNRTI) class of drugs (denoted by an asterisk), as opposed to nucleoside reverse transcriptase inhibitors (NRTIs) or protease inhibitors (PIs).
-
B
-
Microbiome analyses can inform disease prognosis in acute and chronic disease states and underlie the development of probiotic therapies. Coloured bars represent individual microbiota species. A reduction in species diversity is seen in dysbiosis (an unhealthy state), such as present in patients with Clostridium difficile-associated disease. Stool from healthy individuals can be harvested to treat patients with C. difficile infection by faecal stool transplantation or as orally administered encapsulated faecal pills. Alternatively, synthetic stool generated from microbiota species observed in healthy individuals can be used as probiotics to treat patients. In addition to C. difficile infection, chronic diseases such as obesity, inflammatory bowel disease and diabetes mellitus are potential targets for probiotic therapy.
-
C RNA-sequencing-based transcriptomics can improve the diagnosis of infectious and non-infectious conditions on the basis of the human host response. Host transcriptomic profiling by NGS can enable the construction of a classifier metric to discriminate between patients with infection (red bars) from uninfected patients (blue bars) with high accuracy (part Ca).
-
Metric scores above the dotted line indicate infection, whereas scores below the dotted line indicate absence of infection; the overall accuracy of the classifier metric shown is 83%. Cluster heat map analysis identifies individual, differentially expressed host genes associated with infection (genes A–F) versus those associated with no infection (genes G–L) (part Cb).
-
D
- Sequencing of viral tumours or liquid biopsy analyses in oncology can be used for simultaneous pathogen detection and characterization of host genetic mutations.
- mNGS can be used to detect Merkel cell polyomavirus, the virus associated with the development of Merkel cell carcinoma.
- Simultaneous sequencing of host DNA can identify mutations that arise from integration of the viral genome containing the full-length large T antigen (LT) followed by subsequent truncation of the LT antigen (part Da) or truncation of the LT antigen before viral genome integration (part Db).
-
Both of these two mutations lead to cellular transformation that drives tumour proliferation.
-
Although promising, many of these sequencing-based applications have yet to be incorporated into routine clinical practice.
-
Fig. 2 for chapter 4 | Targeted versus untargeted shotgun metagenomic next-generation sequencing approaches (USING: Amplicon sequencing vs Metagenomic sequencing).
A variety of patient samples, as well as cultured microbial colonies, can be analysed using targeted or untargeted metagenomic next-generation sequencing (mNGS) methods for pathogen identification, microbiome analyses and/or host transcriptome profiling. Universal PCR (left) is a targeted mNGS approach that uses primers designed from conserved regions such as the ribosomal RNA (rRNA) genes that are universally conserved among bacteria (16S or 23S rRNA) or fungi and parasites (18S rRNA, 28S rRNA or internal transcribed spacer (ITS)). Other sets of primers can be designed to target a defined set of pathogens and/or genes and used for multiplex reverse transcription PCR or PCR (multiplexed amplicon PCR). NGS library preparation and sequencing of the resultant amplicons enable pathogen identification down to the genus or species level. Metagenomic sequencing (right) entails unbiased shotgun sequencing of all microbial and host nucleic acids present in a clinical sample. Separate DNA and RNA libraries are constructed; the DNA library is used for identification of bacteria, fungi, DNA viruses and parasites, whereas the RNA library is used for identification of RNA viruses and RNA sequencing-based human host transcriptome profiling (heat map, bottom right). As no primers or probes are used in unbiased mNGS, the vast majority of reads corresponds to the human host and, thus, detection of pathogens from metagenomic libraries is a ‘needle-in-a-haystack’ endeavour. An optional capture probe enrichment step using magnetic beads enables targeted mNGS of pathogens and/or genes from metagenomic libraries. All these methods are compatible with sequencing on traditional benchtop instruments such as the Illumina HiSeq and portable nanopore sequencers such as the Oxford Nanopore Technologies MinION. -
Fig. 3 for chapter 5 [OPTIONAL, but the figure contains no content.
However, it is a good figure showing routine of future dignostics, they clinician like it] | Challenges to routine deployment of metagenomic sequencing in the clinical setting. At each step in the process, multiple factors (bullet points) must be taken into account when implementing a clinical metagenomic pipeline for diagnosis of infections to maximize accuracy and clinical relevance. In particular, it is often useful to interpret and discuss the results of metagenomic next-generation-sequencing (mNGS) testing in a clinical context as part of a clinical microbial sequencing board, akin to a tumour board in oncology. EMR, electronic medical record. -
Fig. 4 | A typical metagenomic next-generation sequencing [IGNORING] bioinformatics pipeline.
A next-generation sequencing (NGS) data set, generally in FASTQ or sequence alignment map (SAM) format, is analysed on a computational server, portable laptop or desktop computer or on the cloud. An initial preprocessing step consists of low- quality filtering, low-complexity filtering and adaptor trimming. Computational host subtraction is performed by mapping reads to the host (for example, human) genome and setting aside host reads for subsequent transcriptome (RNA) or genome (DNA) analysis. The remaining unmapped reads are directly aligned to large reference databases, such as the National Center for Biotechnology Information (NCBI) GenBank database or microbial reference sequence or genome collections, or are first assembled de novo into longer contiguous sequences (contigs) followed by alignment to reference databases. After taxonomic classification, in which individual reads or contigs are assigned into specific taxa (for example, species, genus and family), the data can be analysed and visualized in a number of different formats. These include coverage map and pairwise identity plots to determine how much of the microbial genome has been recovered and its similarity to reference genomes in the database; Krona plots to visualize taxonomic diversity in the metagenomic library ; phylogenetic analysis to compare assembled genes, gene regions or genomes to reference sequences; and heat maps to show microorganisms that were detected in the clinical samples. OTU, operational taxonomic unit.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Setup the environment for lumicks-pylake and C_Trap-Multimer-photontrack.ipynb
- 🧬 Cadmium Resistance Gene Analysis in Staphylococcus epidermidis HD46
- MCV病毒中的LT与sT蛋白功能
- Analysis of the RNA binding protein (RBP) motifs for RNA-Seq and miRNAs (v3, simplied)
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
QIIME + Phyloseq + MicrobiotaProcess (v1)
Plotting Alpha Diversities from 16S rRNA Sequencing Data
Phyloseq + MicrobiotaProcess + PICRUSt2
PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics