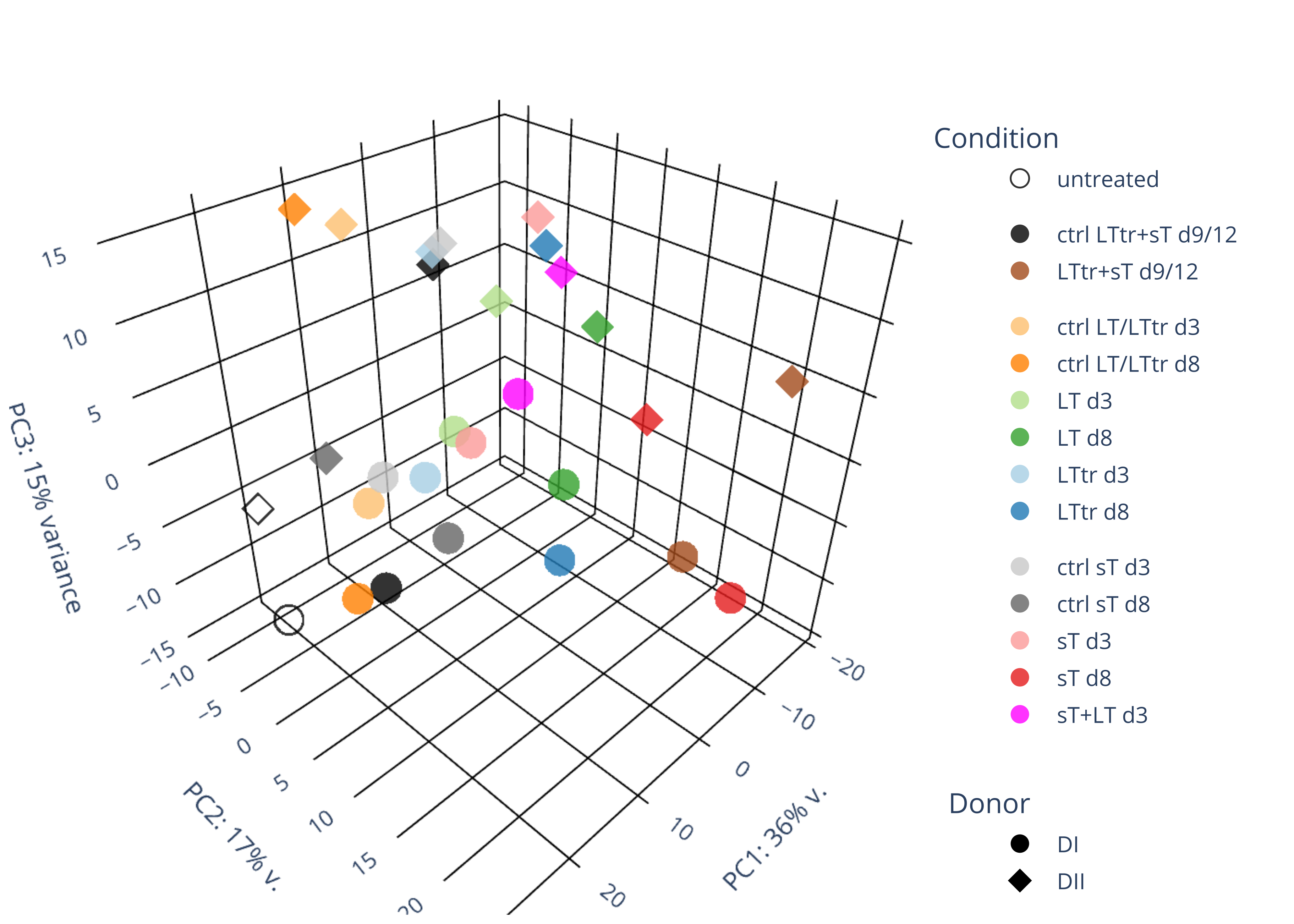
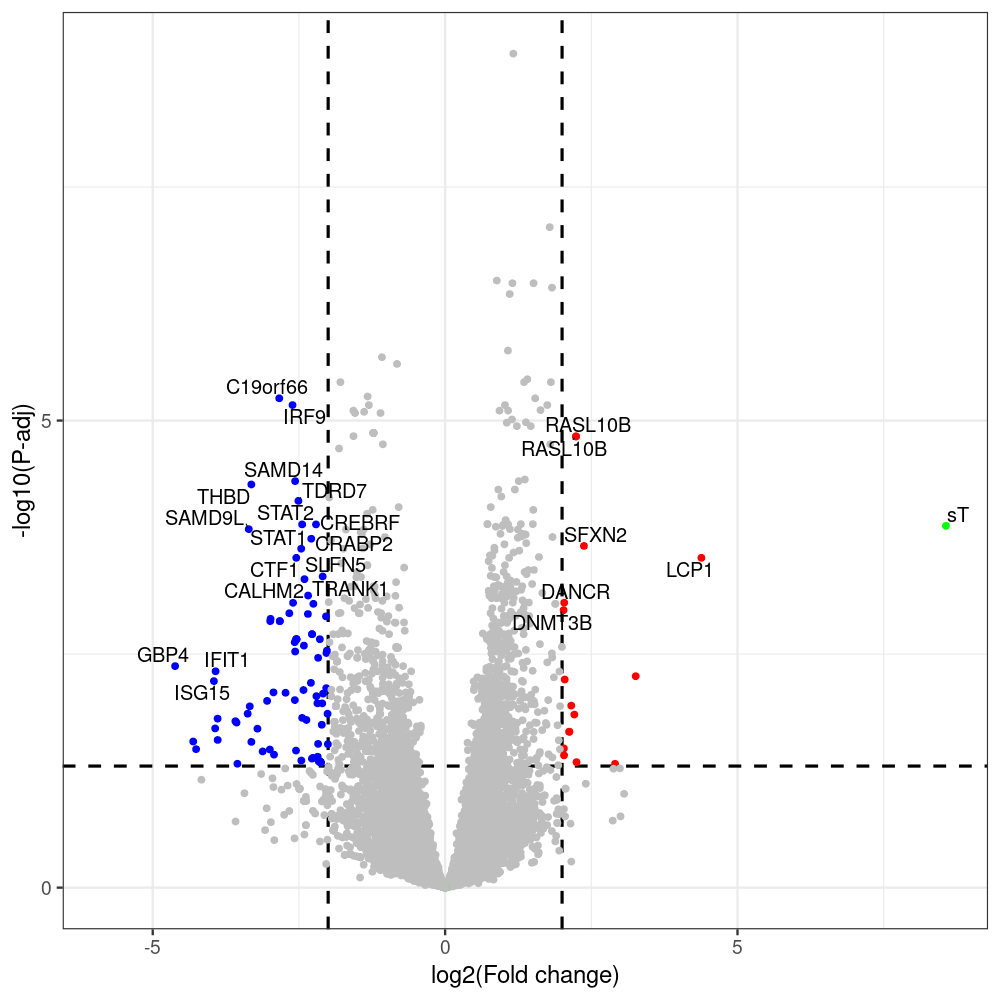
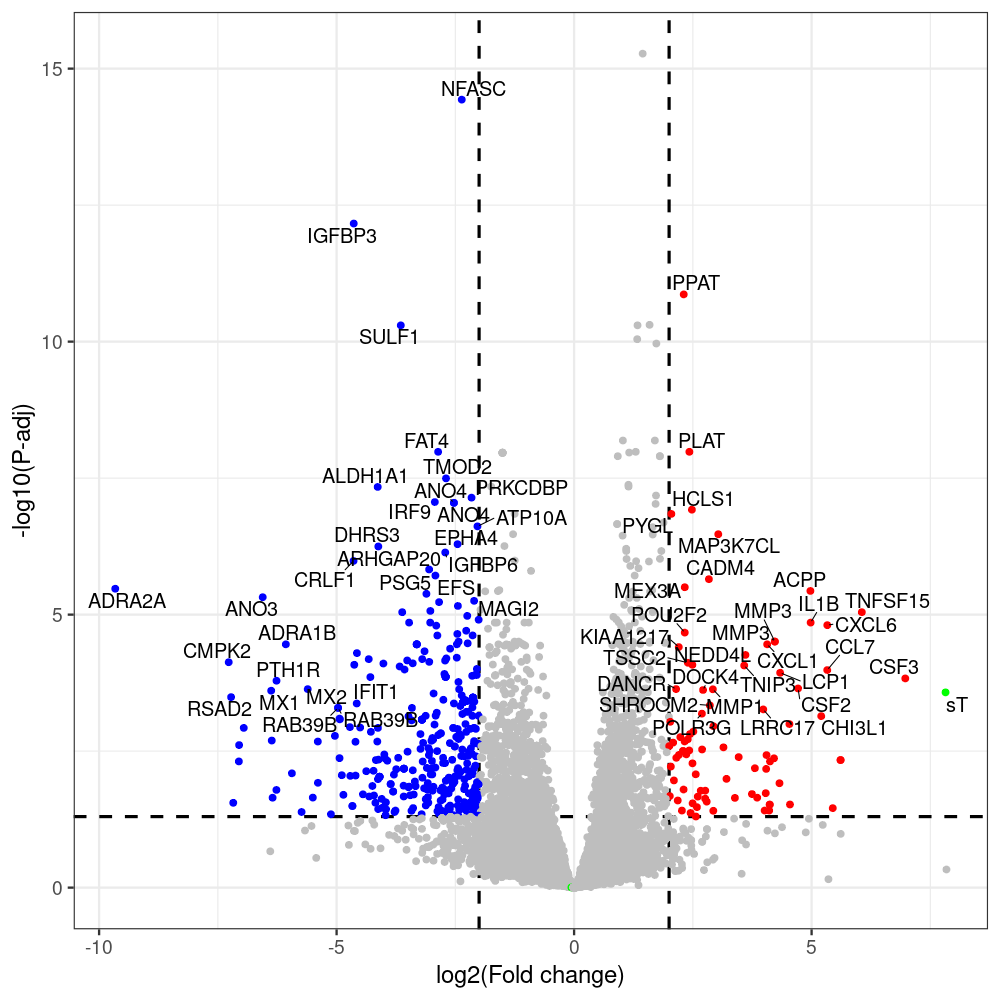
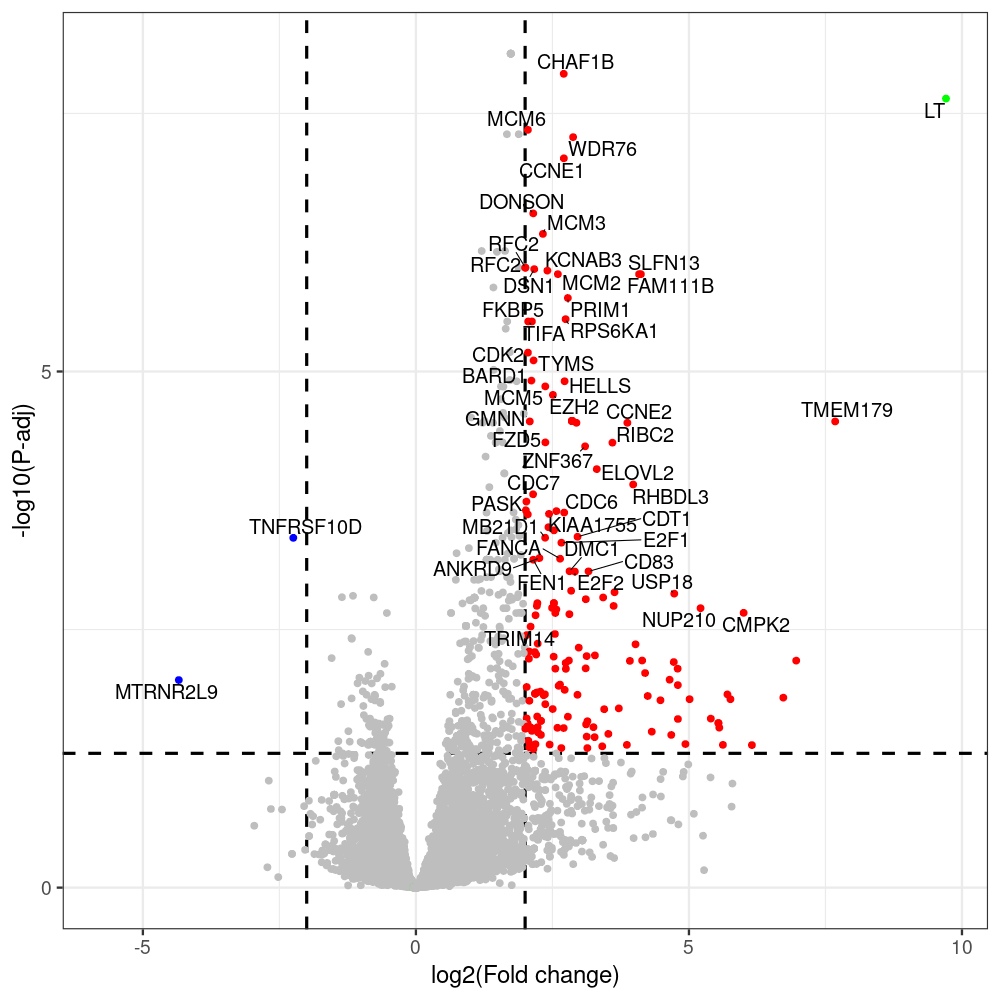
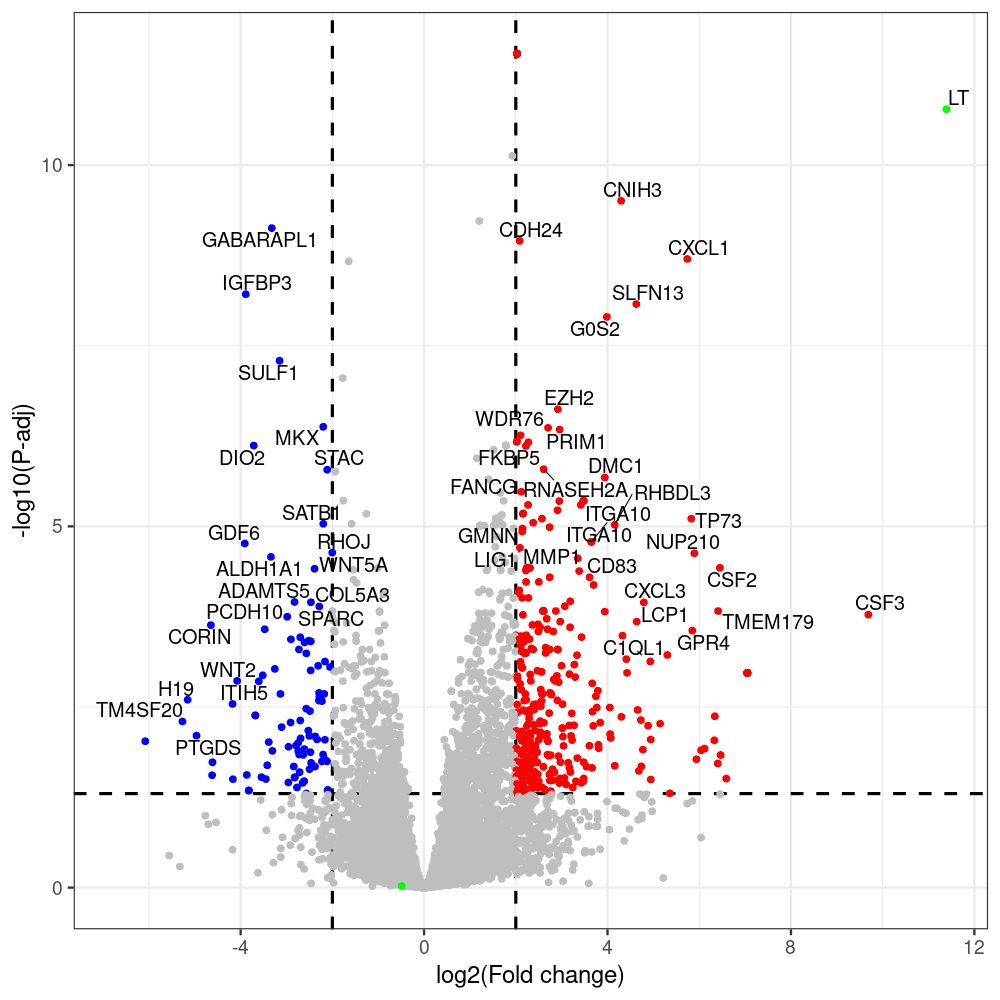
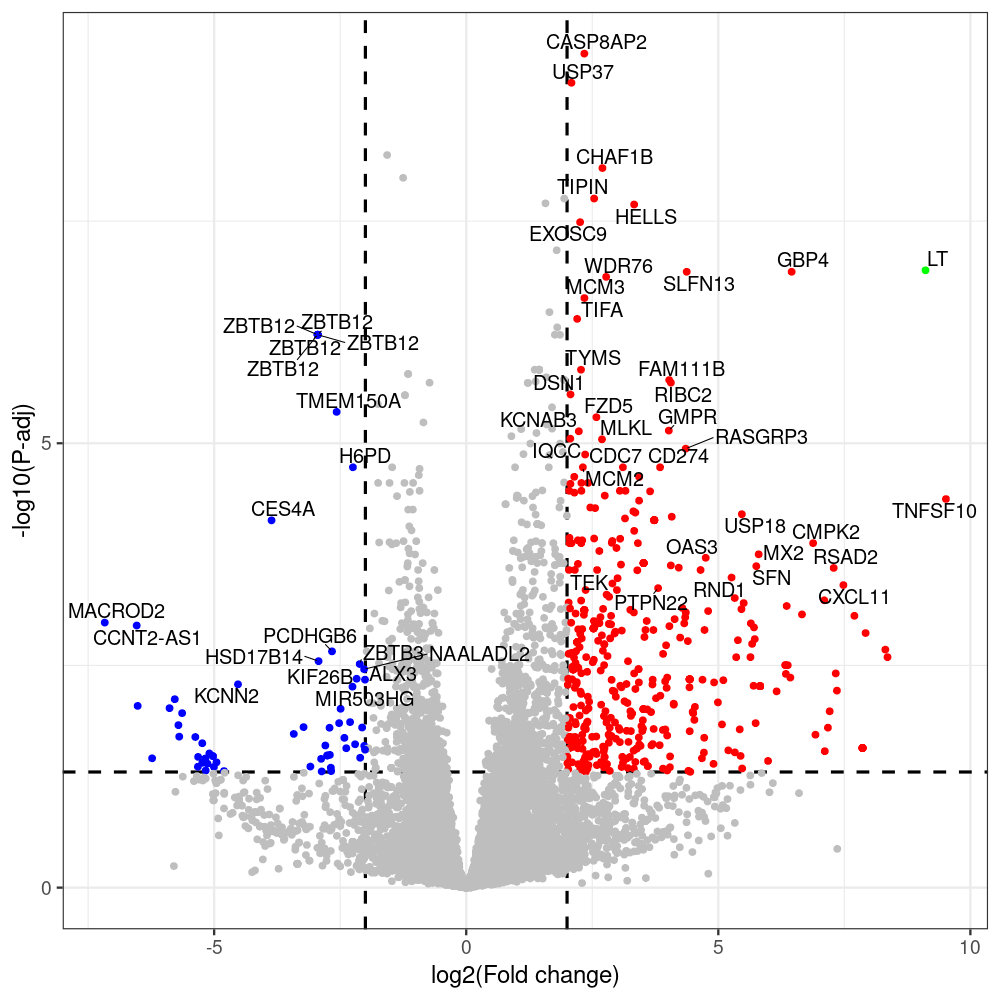
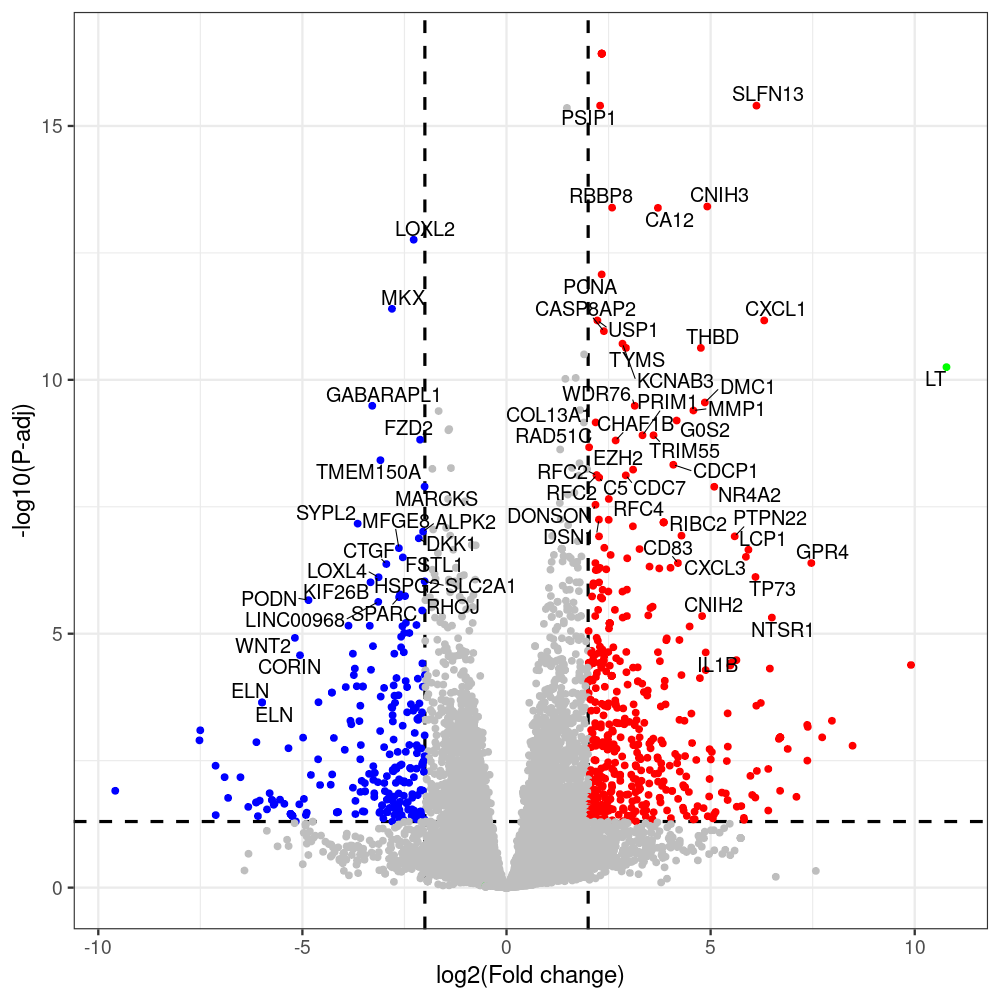
sT manuscript revision: log2FC of sT itself at 3 and 8 dpt
gene_x 0 like s 1317 view s
Tags: plot, R, research
-
nextflow
#under hamm ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq #Debug the following error: added "--minAssignedFrags 0 \\" to modules/nf-core/salmon/quant/main.nf option "salmon quant" and added "--min_mapped_reads 0" in the nextflow command (rnaseq_2021) jhuang@hamm:~/DATA/Data_Denise_sT_RNASeq$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_JN707599 --fasta JN707599.fasta --gtf JN707599.gtf -profile test_full -resume --max_memory 124.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0 (rnaseq_2021) jhuang@hamm:~/DATA/Data_Denise_sT_RNASeq$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_LT --fasta JN707599.fasta --gtf LT.gtf -profile test_full -resume --max_memory 124.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0 (rnaseq_2021) jhuang@hamm:~/DATA/Data_Denise_sT_RNASeq$ /usr/local/bin/nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_sT --fasta JN707599.fasta --gtf sT.gtf -profile test_full -resume --max_memory 124.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0 #DEBUG #V_8_4_1_p602_d8_DonorI,V_8_4_2_p602_d8_DonorI.fastq.gz,,auto #V_8_5_1_p602and604_d3_DonorI,V_8_5_1_602and604_d3_DonorI.fastq.gz,,auto #V_8_5_2_p602and604_d3_DonorII,V_8_5_2_602and604_d3_DonorII.fastq.gz,,auto #mamba install -c bioconda rsem #mamba install fq -
construct DESeqDataSet including host+viral gene expressions
# Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) library("org.Hs.eg.db") library(dplyr) library(tidyverse) setwd("~/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq/results_JN707599/star_salmon") # Define paths to your Salmon output quantification files files <- c("V_8_0_mock_DonorI" = "./V_8_0_mock_DonorI/quant.sf", "V_8_0_mock_DonorII" = "./V_8_0_mock_DonorII/quant.sf", "V_8_1_5_p601_d3_DonorII" = "./V_8_1_5_p601_d3_DonorII/quant.sf", "V_8_1_5_p604_d3_DonorII" = "./V_8_1_5_p604_d3_DonorII/quant.sf", "V_8_1_5_p601_d8_DonorII" = "./V_8_1_5_p601_d8_DonorII/quant.sf", "V_8_1_5_p604_d8_DonorII" = "./V_8_1_5_p604_d8_DonorII/quant.sf", "V_8_1_6_p601_d3_DonorI" = "./V_8_1_6_p601_d3_DonorI/quant.sf", "V_8_1_6_p604_d3_DonorI" = "./V_8_1_6_p604_d3_DonorI/quant.sf", "V_8_1_6_p601_d8_DonorI" = "./V_8_1_6_p601_d8_DonorI/quant.sf", "V_8_1_6_p604_d8_DonorI" = "./V_8_1_6_p604_d8_DonorI/quant.sf", "V_8_2_3_p600_d3_DonorII" = "./V_8_2_3_p600_d3_DonorII/quant.sf", "V_8_2_3_p605_d3_DonorII" = "./V_8_2_3_p605_d3_DonorII/quant.sf", "V_8_2_3_p600_d8_DonorII" = "./V_8_2_3_p600_d8_DonorII/quant.sf", "V_8_2_3_p605_d8_DonorII" = "./V_8_2_3_p605_d8_DonorII/quant.sf", "V_8_2_4_p600_d3_DonorI" = "./V_8_2_4_p600_d3_DonorI/quant.sf", "V_8_2_4_p605_d3_DonorI" = "./V_8_2_4_p605_d3_DonorI/quant.sf", "V_8_2_4_p600_d8_DonorI" = "./V_8_2_4_p600_d8_DonorI/quant.sf", "V_8_2_4_p605_d8_DonorI" = "./V_8_2_4_p605_d8_DonorI/quant.sf", "V_8_4_1_p602_d8_DonorII" = "./V_8_4_1_p602_d8_DonorII/quant.sf", "V_8_4_1_p602_d8_DonorI" = "./V_8_4_1_p602_d8_DonorI/quant.sf", "V_8_3_1_p600and601_d12_DonorI" = "./V_8_3_1_p600and601_d12_DonorI/quant.sf", "V_8_3_1_p604and605_d12_DonorI" = "./V_8_3_1_p604and605_d12_DonorI/quant.sf", "V_8_3_2_p600and601_d9_DonorII" = "./V_8_3_2_p600and601_d9_DonorII/quant.sf", "V_8_3_2_p604and605_d9_DonorII" = "./V_8_3_2_p604and605_d9_DonorII/quant.sf", "V_8_4_2_p602_d3_DonorI" = "./V_8_4_2_p602_d3_DonorI/quant.sf", "V_8_4_2_p602_d3_DonorII" = "./V_8_4_2_p602_d3_DonorII/quant.sf", "V_8_5_1_p602and604_d3_DonorI" = "./V_8_5_1_sTplusLT_d3_Donor1/quant.sf", "V_8_5_2_p602and604_d3_DonorII" = "./V_8_5_2_sTplusLT_d3_Donor2/quant.sf") # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) column_names <- colnames(txi$counts) output_string <- paste(column_names, collapse = ", ") cat(output_string, "\n") #"V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII" col_order <- c("V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII") #reordered.txi <- txi[,col_order] identical(column_names,col_order) condition = as.factor(c("untreated","untreated", "p601_d3","p604_d3", "p601_d8","p604_d8", "p601_d3","p604_d3","p601_d8","p604_d8", "p600_d3","p605_d3","p600_d8", "p605_d8", "p600_d3","p605_d3","p600_d8","p605_d8", "p602_d8","p602_d8", "p600and601_d9d12", "p604and605_d9d12","p600and601_d9d12","p604and605_d9d12", "p602_d3","p602_d3", "p602and604_d3","p602and604_d3")) batch = as.factor(c("200420", "200420", "190927", "190927", "190927", "190927", "190228", "190228", "190228", "190228", "191008", "191008", "191008", "191008", "190228", "190228", "190228", "190228", "200817", "200817", "200420", "200420", "200817", "200817", "210302", "210302", "210504","210504")) ids = as.factor(c("untreated_DonorI","untreated_DonorII", "p601_d3_DonorII","p604_d3_DonorII", "p601_d8_DonorII","p604_d8_DonorII", "p601_d3_DonorI","p604_d3_DonorI","p601_d8_DonorI","p604_d8_DonorI", "p600_d3_DonorII","p605_d3_DonorII","p600_d8_DonorII", "p605_d8_DonorII", "p600_d3_DonorI","p605_d3_DonorI","p600_d8_DonorI","p605_d8_DonorI", "p602_d8_DonorII","p602_d8_DonorI", "p600and601_d12_DonorI", "p604and605_d12_DonorI","p600and601_d9_DonorII","p604and605_d9_DonorII", "p602_d3_DonorI","p602_d3_DonorII", "p602and604_d3_DonorI","p602and604_d3_DonorII")) donor = as.factor(c("DonorI","DonorII", "DonorII","DonorII", "DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorII","DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorI", "DonorI", "DonorI","DonorII","DonorII", "DonorI","DonorII", "DonorI","DonorII")) # Define the colData for DESeq2 #colData = data.frame(row.names=column_names, condition=condition, donor=donor, batch=batch, ids=ids) colData <- data.frame(condition=condition, donor=donor, row.names=names(files)) # -- transcript-level count data (for virus) -- # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition+donor) write.csv(counts(dds), file="transcript_counts.csv") # -- gene-level count data (for virus) -- # Read in the tx2gene map from salmon_tx2gene.tsv tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition+donor) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #60605-->26543 write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv") # -- merge the raw counts of human and microbe -- setwd("~/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq/results/featureCounts") d.raw<- read.delim2("merged_gene_counts_2_f3_f5.txt",sep="\t", header=TRUE, row.names=1) colnames(d.raw)<- c("V_8_0_mock_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_5_p604_d3_DonorII","V_8_1_6_p604_d3_DonorI","V_8_2_3_p605_d8_DonorII","V_8_0_mock_DonorII","V_8_1_5_p601_d8_DonorII","V_8_2_3_p605_d3_DonorII","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d8_DonorII","V_8_2_4_p600_d8_DonorI","V_8_2_4_p600_d3_DonorI","V_8_1_5_p604_d8_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_6_p601_d3_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_4_p605_d3_DonorI","V_8_4_1_p602_d8_DonorI","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_2_4_p605_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_4_1_p602_d8_DonorII","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII", "V_8_5_1_p602and604_d3_DonorI", "V_8_5_2_p602and604_d3_DonorII") #26364 col_order <- c("V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII", "V_8_1_5_p604_d3_DonorII", "V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII", "V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI", "V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII", "V_8_2_3_p605_d8_DonorII", "V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI", "V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI", "V_8_3_1_p600and601_d12_DonorI", "V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII", "V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII", "V_8_5_1_p602and604_d3_DonorI", "V_8_5_2_p602and604_d3_DonorII") reordered.raw <- d.raw[,col_order] write.csv(reordered.raw, file="merged_gene_counts_2_f3_f5_reordered.txt") # Confirm the identification of the headers of two files before merging! #kate ./results/featureCounts/merged_gene_counts_2_f3_f5_reordered.txt #"","V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII" #results_JN707599/star_salmon/gene_counts.csv #"","V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII" #cat ../Data_Denise_RNASeq/results/featureCounts/merged_gene_counts_2_f3_f5_reordered.txt ../Data_Denise_RNASeq/results_JN707599/star_salmon/gene_counts.csv > merged_gene_counts.csv #~/Tools/csv2xls-0.4/csv_to_xls.py merged_gene_counts.csv -d',' -o raw_gene_counts.xls; setwd("~/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq_Merged/") d.raw <- read.csv("merged_gene_counts.csv", header=TRUE, row.names=1) dds <- DESeqDataSetFromMatrix(countData=d.raw, colData=colData, design=~condition+donor) dim(counts(dds)) head(counts(dds), 10) rld <- rlogTransformation(dds) # draw simple pca and heatmap library(gplots) library("RColorBrewer") #mat <- assay(rld) #mm <- model.matrix(~condition, colData(rld)) #mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm) #assay(rld) <- mat # -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
(corrected of last step) construct DESeqDataSet including host+viral gene expressions
column_names <- colnames(d.raw) col_order <- c("V_8_0_mock_DonorI","V_8_0_mock_DonorII","V_8_1_5_p601_d3_DonorII","V_8_1_5_p604_d3_DonorII","V_8_1_5_p601_d8_DonorII","V_8_1_5_p604_d8_DonorII","V_8_1_6_p601_d3_DonorI","V_8_1_6_p604_d3_DonorI","V_8_1_6_p601_d8_DonorI","V_8_1_6_p604_d8_DonorI","V_8_2_3_p600_d3_DonorII","V_8_2_3_p605_d3_DonorII","V_8_2_3_p600_d8_DonorII","V_8_2_3_p605_d8_DonorII","V_8_2_4_p600_d3_DonorI","V_8_2_4_p605_d3_DonorI","V_8_2_4_p600_d8_DonorI","V_8_2_4_p605_d8_DonorI","V_8_4_1_p602_d8_DonorII","V_8_4_1_p602_d8_DonorI","V_8_3_1_p600and601_d12_DonorI","V_8_3_1_p604and605_d12_DonorI","V_8_3_2_p600and601_d9_DonorII","V_8_3_2_p604and605_d9_DonorII","V_8_4_2_p602_d3_DonorI","V_8_4_2_p602_d3_DonorII","V_8_5_1_p602and604_d3_DonorI","V_8_5_2_p602and604_d3_DonorII") identical(column_names,col_order) condition = as.factor(c("untreated","untreated", "p601_d3","p604_d3", "p601_d8","p604_d8", "p601_d3","p604_d3","p601_d8","p604_d8", "p600_d3","p605_d3","p600_d8", "p605_d8", "p600_d3","p605_d3","p600_d8","p605_d8", "p602_d8","p602_d8", "p600and601_d9d12", "p604and605_d9d12","p600and601_d9d12","p604and605_d9d12", "p602_d3","p602_d3", "p602and604_d3","p602and604_d3")) batch = as.factor(c("200420", "200420", "190927", "190927", "190927", "190927", "190228", "190228", "190228", "190228", "191008", "191008", "191008", "191008", "190228", "190228", "190228", "190228", "200817", "200817", "200420", "200420", "200817", "200817", "210302", "210302", "210504","210504")) ids = as.factor(c("untreated_DonorI","untreated_DonorII", "p601_d3_DonorII","p604_d3_DonorII", "p601_d8_DonorII","p604_d8_DonorII", "p601_d3_DonorI","p604_d3_DonorI","p601_d8_DonorI","p604_d8_DonorI", "p600_d3_DonorII","p605_d3_DonorII","p600_d8_DonorII", "p605_d8_DonorII", "p600_d3_DonorI","p605_d3_DonorI","p600_d8_DonorI","p605_d8_DonorI", "p602_d8_DonorII","p602_d8_DonorI", "p600and601_d12_DonorI", "p604and605_d12_DonorI","p600and601_d9_DonorII","p604and605_d9_DonorII", "p602_d3_DonorI","p602_d3_DonorII", "p602and604_d3_DonorI","p602and604_d3_DonorII")) donor = as.factor(c("DonorI","DonorII", "DonorII","DonorII", "DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorII","DonorII","DonorII", "DonorI","DonorI","DonorI","DonorI", "DonorII","DonorI", "DonorI", "DonorI","DonorII","DonorII", "DonorI","DonorII", "DonorI","DonorII")) colData <- data.frame(condition=condition, donor=donor, row.names=column_names) setwd("~/DATA/Data_Denise_sT_PUBLISHING/Data_Denise_RNASeq_Merged_Corrected/") d.raw <- read.csv("merged_gene_counts_corrected.csv", header=TRUE, row.names=1) dds <- DESeqDataSetFromMatrix(countData=d.raw, colData=colData, design=~condition+donor) dim(counts(dds)) head(counts(dds), 10) rld <- rlogTransformation(dds) # draw simple pca and heatmap library(gplots) library("RColorBrewer") #mat <- assay(rld) #mm <- model.matrix(~condition, colData(rld)) #mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm) #assay(rld) <- mat # -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
select the differentially expressed genes
#untreated_DonorI,untreated #untreated_DonorII,untreated #p601_d3_DonorII,mCherry control #p604_d3_DonorII,sT #p601_d8_DonorII,mCherry control #p604_d8_DonorII,sT #p601_d3_DonorI,mCherry control #p604_d3_DonorI,sT #p601_d8_DonorI,mCherry control #p604_d8_DonorI,sT # #p600_d3_DonorII,GFP control #p605_d3_DonorII,LTtr #p600_d8_DonorII,GFP control #p605_d8_DonorII,LTtr #p600_d3_DonorI,GFP control #p605_d3_DonorI,LTtr #p600_d8_DonorI,GFP control #p605_d8_DonorI,LTtr #p602_d3_DonorI,LT #p602_d3_DonorII,LT #p602_d8_DonorII,LT #p602_d8_DonorI,LT # #p600and601_d12_DonorI,GFP+mCherry control #p604and605_d12_DonorI,sT+LTtr #p600and601_d9_DonorII,GFP+mCherry control #p604and605_d9_DonorII,sT+LTtr # #p602and604_d3_DonorI,sT+LT #p602and604_d3_DonorII,sT+LT #CONSOLE: mkdir /home/jhuang/DATA/Data_Denise_sT_immune_evasion_PUBLISHING/Data_Denise_RNASeq_Merged/degenes #---- relevel to control ---- dds$condition <- relevel(dds$condition, "p601_d3") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("p604_d3_vs_p601_d3") dds$condition <- relevel(dds$condition, "p600_d3") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("p605_d3_vs_p600_d3") dds$condition <- relevel(dds$condition, "p600_d3") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("p602_d3_vs_p600_d3") dds$condition <- relevel(dds$condition, "p601_d8") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("p604_d8_vs_p601_d8") dds$condition <- relevel(dds$condition, "p600_d8") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("p605_d8_vs_p600_d8") dds$condition <- relevel(dds$condition, "p600_d8") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("p602_d8_vs_p600_d8") library(biomaRt) listEnsembl() listMarts() #--> total 69, 27 GRCh38.p7 and 39 GRCm38.p4 #ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="104") #GRCh37.p13 ensembl <- useEnsembl("ensembl","hsapiens_gene_ensembl", host="https://grch37.ensembl.org") datasets <- listDatasets(ensembl) # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #> dim(geness) [1] 21938 9 using GRCh38.p7 #> dim(geness) [1] 23517 9 using GRCh37.p13 #for (i in clist) { #i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'external_gene_name', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$geneSymbol = rownames(res) #identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="external_gene_name", by.y="geneSymbol") dim(geness_res) #22044 15 rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("LT", "sT", "VP1", "VP2", "VP3") virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "JN707599" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) viral <- merged_df_sorted[merged_df_sorted$external_gene_name %in% c("LT", "sT", "VP1", "VP2", "VP3"), ] write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) write.csv(as.data.frame(viral), file = paste(i, "viral_annotated.txt", sep="-")) #} # -- 5. draw graphics -- library(ggrepel) #geness_res <- read.csv(file = "p605_d8_vs_p600_d8-all_annotated.txt", sep=",", row.names=1) #i<-"p605_d8_vs_p600_d8" geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("LT", "sT", "VP1", "VP2", "VP3") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:200], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:200])) # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=5) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=5) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() #Under DIR degenes under KONSOLE and delete "start_position","end_position","strand" and so on. #~/Tools/csv2xls-0.4/csv_to_xls.py viral_gene_counts.csv p604_d8_vs_p601_d8-viral_annotated.txt p605_d8_vs_p600_d8-viral_annotated.txt p602_d8_vs_p600_d8-viral_annotated.txt p604_d3_vs_p601_d3-viral_annotated.txt p605_d3_vs_p600_d3-viral_annotated.txt p602_d3_vs_p600_d3-viral_annotated.txt -d$',' -o viral_raw_counts_and_comparisons.xls; #dpt: days post-transfection
点赞本文的读者
还没有人对此文章表态
本文有评论
nicknames of LT, LTtr, sT and so on说:
#untreated_DonorI,untreated #untreated_DonorII,untreated #p601_d3_DonorII,mCherry control #p604_d3_DonorII,sT #p601_d8_DonorII,mCherry control #p604_d8_DonorII,sT #p601_d3_DonorI,mCherry control #p604_d3_DonorI,sT #p601_d8_DonorI,mCherry control #p604_d8_DonorI,sT # #p600_d3_DonorII,GFP control #p605_d3_DonorII,LTtr #p600_d8_DonorII,GFP control #p605_d8_DonorII,LTtr #p600_d3_DonorI,GFP control #p605_d3_DonorI,LTtr #p600_d8_DonorI,GFP control #p605_d8_DonorI,LTtr #p602_d3_DonorI,LT #p602_d3_DonorII,LT #p602_d8_DonorII,LT #p602_d8_DonorI,LT # #p600and601_d12_DonorI,GFP+mCherry control #p604and605_d12_DonorI,sT+LTtr #p600and601_d9_DonorII,GFP+mCherry control #p604and605_d9_DonorII,sT+LTtr # #p602and604_d3_DonorI,sT+LT #p602and604_d3_DonorII,sT+LT
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Kraken2 Installation and Usage Guide
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
- 基因组与表型特性研究:一株药物敏感的鲍曼不动杆菌显示出增强的毒力相关特性和应激耐受性
- A. baumannii HKAB-1 Point-by-point response to Reviewers
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
MicrobiotaProcess Group2 vs Group6 (v1)
Bubble plot for 1457∆atlE vs 1457-M10 vs 1457 vs mock