RNA-seq skin organoids on GRCh38+chrHsv1 (final)
gene_x 0 like s 1357 view s
Tags: plot, python, R, pipeline, RNA-seq
-
run nextflow rnaseq
#under sage ln -s /home/jhuang/Tools/nf-core-rnaseq-3.12.0/ rnaseq (rnaseq) nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_GRCh38 --genome GRCh38 --with_umi --umitools_extract_method regex --umitools_bc_pattern '^(?P.{12}).*' -profile docker -resume --max_cpus 54 --max_memory 120.GB --max_time 2400.h --save_align_intermeds --save_unaligned --save_reference --aligner star_salmon --pseudo_aligner salmon --umitools_grouping_method unique #Debug the following error: added "--minAssignedFrags 0 \\" to modules/nf-core/salmon/quant/main.nf option "salmon quant" and added "--min_mapped_reads 0" in the nextflow command (rnaseq) nextflow run rnaseq/main.nf --input samplesheet.csv --outdir results_chrHsv1 --fasta chrHsv1_s17.fasta --gtf chrHsv1_s17.gtf --with_umi --umitools_extract_method regex --umitools_bc_pattern '^(?P.{12}).*' --umitools_dedup_stats -profile test_full -resume --max_memory 256.GB --max_time 2400.h --save_reference --aligner star_salmon --gtf_extra_attributes gene_id --gtf_group_features transcript_id --featurecounts_group_type gene_id --featurecounts_feature_type transcript --skip_rseqc --skip_dupradar --skip_preseq --skip_biotype_qc --skip_deseq2_qc --skip_multiqc --min_mapped_reads 0 #TODO: why a lot of reads were removed due to the too_short? -
import data and pca-plot
# Import the required libraries library("AnnotationDbi") library("clusterProfiler") library("ReactomePA") library(gplots) library(tximport) library(DESeq2) library("org.Hs.eg.db") library(dplyr) library(tidyverse) setwd("~/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon") # Define paths to your Salmon output quantification files files <- c("control_r1" = "./control_r1/quant.sf", "control_r2" = "./control_r2/quant.sf", "HSV.d2_r1" = "./HSV.d2_r1/quant.sf", "HSV.d2_r2" = "./HSV.d2_r2/quant.sf", "HSV.d4_r1" = "./HSV.d4_r1/quant.sf", "HSV.d4_r2" = "./HSV.d4_r2/quant.sf", "HSV.d6_r1" = "./HSV.d6_r1/quant.sf", "HSV.d6_r2" = "./HSV.d6_r2/quant.sf", "HSV.d8_r1" = "./HSV.d8_r1/quant.sf", "HSV.d8_r2" = "./HSV.d8_r2/quant.sf") # Import the transcript abundance data with tximport txi <- tximport(files, type = "salmon", txIn = TRUE, txOut = TRUE) # Define the replicates and condition of the samples replicate <- factor(c("r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2", "r1", "r2")) condition <- factor(c("control", "control", "HSV.d2", "HSV.d2", "HSV.d4", "HSV.d4", "HSV.d6", "HSV.d6", "HSV.d8", "HSV.d8")) # Define the colData for DESeq2 colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) # -- transcript-level count data (x2) -- # Create DESeqDataSet object dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) write.csv(counts(dds), file="transcript_counts.csv") # -- gene-level count data (x2) -- # Read in the tx2gene map from salmon_tx2gene.tsv tx2gene <- read.table("salmon_tx2gene.tsv", header=FALSE, stringsAsFactors=FALSE) # Set the column names colnames(tx2gene) <- c("transcript_id", "gene_id", "gene_name") # Remove the gene_name column if not needed tx2gene <- tx2gene[,1:2] # Import and summarize the Salmon data with tximport txi <- tximport(files, type = "salmon", tx2gene = tx2gene, txOut = FALSE) # Continue with the DESeq2 workflow as before... colData <- data.frame(condition=condition, replicate=replicate, row.names=names(files)) dds <- DESeqDataSetFromTximport(txi, colData=colData, design=~condition) #dds <- dds[rowSums(counts(dds) > 3) > 2, ] #60605-->26543 write.csv(counts(dds, normalized=FALSE), file="gene_counts.csv") # -- merge the raw counts of human and microbe -- #cat ~/DATA/Data_Manja_RNAseq_Organoids/results_GRCh38_unique/star_salmon/gene_counts.csv ~/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon/gene_counts.csv > merged_gene_counts.csv #DELETE the second line "","control_r1","control_r2","HSV.d2_r1","HSV.d2_r2","HSV.d4_r1","HSV.d4_r2","HSV.d6_r1","HSV.d6_r2","HSV.d8_r1","HSV.d8_r2" #~/Tools/csv2xls-0.4/csv_to_xls.py merged_gene_counts.csv -d',' -o raw_gene_counts.xls; # -- for merged analysis due to false normalization factors wenn alone analyzed on virus data -- setwd("~/DATA/Data_Manja_RNAseq_Organoids_Merged/") d.raw <- read.csv("merged_gene_counts.csv", header=TRUE, row.names=1) dds <- DESeqDataSetFromMatrix(countData=d.raw, colData=colData, design=~condition+replicate) dim(counts(dds)) head(counts(dds), 10) rld <- rlogTransformation(dds) #We don't need to run DESeq(dds) before estimateSizeFactors(dds). In fact, the typical workflow in DESeq2 is the opposite: we usually run estimateSizeFactors(dds) (and other preprocessing functions) before running the main DESeq(dds) function. #The estimateSizeFactors function is used to calculate size factors for normalization, which corrects for differences in library size (i.e., the number of read counts) between samples. This normalization step is crucial to ensure that differences in gene expression aren't merely due to differences in sequencing depth between samples. #The DESeq function, on the other hand, performs the main differential expression analysis, comparing gene expression between different conditions or groups. #So, the typical workflow is: # - Create the DESeqDataSet object. # - Use estimateSizeFactors to normalize for library size. # - (Optionally, estimate dispersion with estimateDispersions if not using the full DESeq function later.) # - Use DESeq for the differential expression analysis. # - However, it's worth noting that if you run the main DESeq function directly after creating the DESeqDataSet object, it will automatically perform the normalization (using estimateSizeFactors) and dispersion estimation steps for you. In that case, there's no need to run estimateSizeFactors separately before DESeq. # draw simple pca and heatmap library(gplots) library("RColorBrewer") #mat <- assay(rld) #mm <- model.matrix(~condition, colData(rld)) #mat <- limma::removeBatchEffect(mat, batch=rld$batch, design=mm) #assay(rld) <- mat # -- pca -- png("pca.png", 1200, 800) plotPCA(rld, intgroup=c("condition")) dev.off() # -- heatmap -- png("heatmap.png", 1200, 800) distsRL <- dist(t(assay(rld))) mat <- as.matrix(distsRL) hc <- hclust(distsRL) hmcol <- colorRampPalette(brewer.pal(9,"GnBu"))(100) heatmap.2(mat, Rowv=as.dendrogram(hc),symm=TRUE, trace="none",col = rev(hmcol), margin=c(13, 13)) dev.off() -
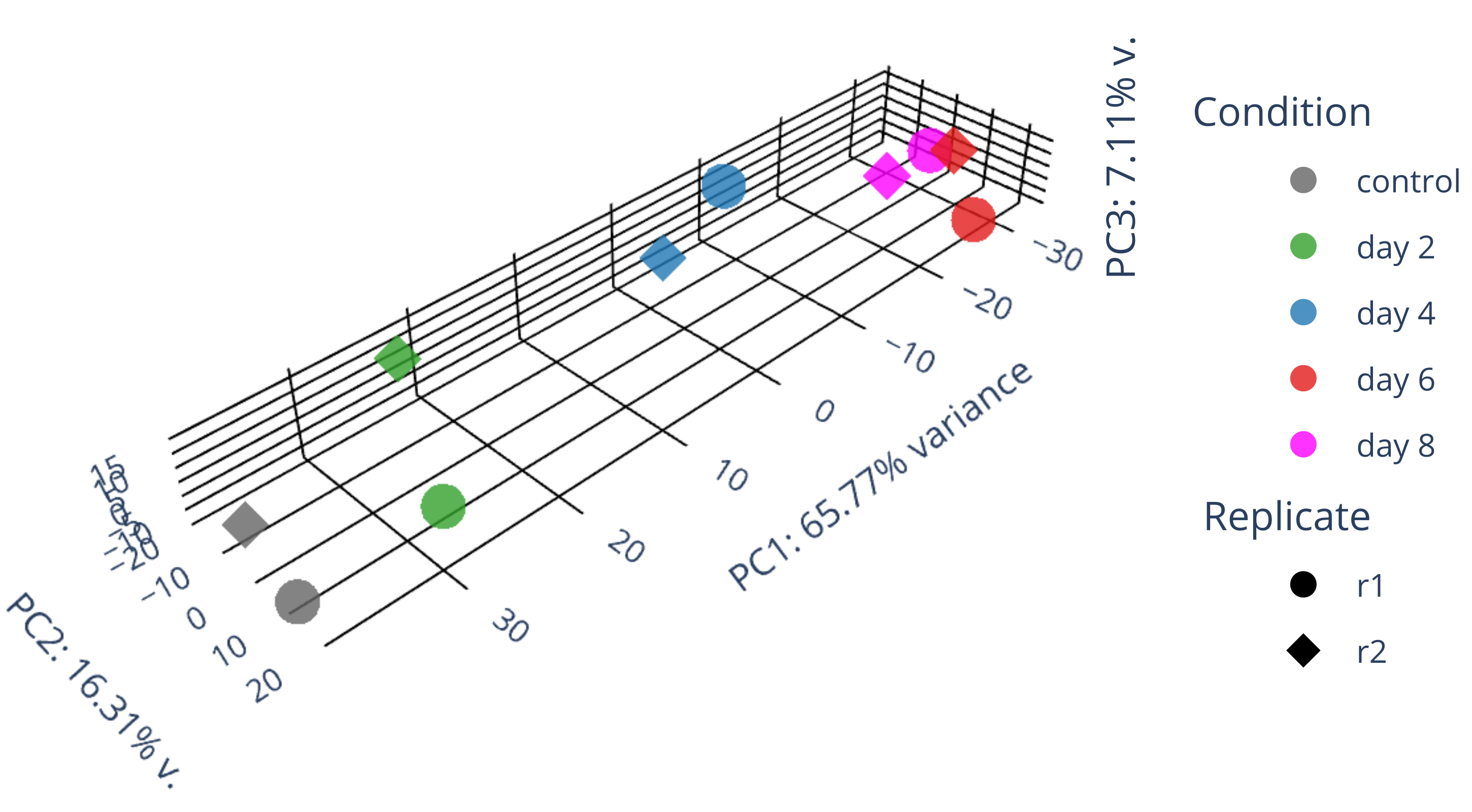
draw 3D PCA plots.
library(gplots) library("RColorBrewer") library(ggplot2) data <- plotPCA(rld, intgroup=c("condition", "replicate"), returnData=TRUE) write.csv(data, file="plotPCA_data.csv") #calculate all PCs including PC3 with the following codes library(genefilter) ntop <- 500 rv <- rowVars(assay(rld)) select <- order(rv, decreasing = TRUE)[seq_len(min(ntop, length(rv)))] mat <- t( assay(rld)[select, ] ) pc <- prcomp(mat) pc$x[,1:3] #df_pc <- data.frame(pc$x[,1:3]) df_pc <- data.frame(pc$x) identical(rownames(data), rownames(df_pc)) #-->TRUE data$PC1 <- NULL data$PC2 <- NULL merged_df <- merge(data, df_pc, by = "row.names") #merged_df <- merged_df[, -1] row.names(merged_df) <- merged_df$Row.names merged_df$Row.names <- NULL # remove the "name" column merged_df$name <- NULL merged_df <- merged_df[, c("PC1","PC2","PC3","PC4","PC5","PC6","PC7","PC8","PC9","PC10","group","condition","replicate")] write.csv(merged_df, file="merged_df_10PCs.csv") summary(pc) #0.5333 0.2125 0.06852 #0.8026 0.09042 0.06578 #0.6577 0.1631 0.07106 #Proportion of Variance 0.6577 0.1631 0.07106 draw_3D.py #/usr/bin/convert PCA_3D.png -crop 2900x1600+250+700 PCA_3D_cropped.png # adjust proportion to real values in the following plot import plotly.graph_objects as go import pandas as pd from sklearn.decomposition import PCA import numpy as np # Read in data as a pandas dataframe #df = pd.DataFrame({ # 'PC1': [-13.999925, -12.504291, -12.443057, -13.065235, -17.316215], # 'PC2': [-1.498823, -3.342411, -6.067055, -8.205809, 3.293993], # 'PC3': [-3.335085, 15.207755, -14.725450, 15.078469, -6.917358], # 'condition': ['GFP d3', 'GFP d3', 'GFP d8', 'GFP d8', 'GFP+mCh d9/12'], # 'replicate': ['DI', 'DII', 'DI', 'DII', 'DI'] #}) df = pd.read_csv('merged_df_10PCs.csv', index_col=0, header=0) df['condition'] = df['condition'].replace("control", "control") df['condition'] = df['condition'].replace("HSV.d2", "day 2") df['condition'] = df['condition'].replace("HSV.d4", "day 4") df['condition'] = df['condition'].replace("HSV.d6", "day 6") df['condition'] = df['condition'].replace("HSV.d8", "day 8") # Fit PCA model to reduce data dimensions to 3 pca = PCA(n_components=3) pca.fit(df.iloc[:, :-3]) X_reduced = pca.transform(df.iloc[:, :-3]) # Get variance ratios explained_variance_ratio = pca.explained_variance_ratio_ # Add reduced data back to dataframe df['PC1'] = X_reduced[:, 0] df['PC2'] = X_reduced[:, 1] df['PC3'] = X_reduced[:, 2] # Create PCA plot with 3D scatter fig = go.Figure() ##ff7f00 condition_color_map = { 'control': 'rgb(100, 100, 100)', 'day 2': '#33a02c', 'day 4': '#1f78b4', 'day 6': '#e31a1c', 'day 8': 'magenta' } replicate_symbol_map = {'r1': 'circle', 'r2': 'diamond'} for replicate, replicate_symbol in replicate_symbol_map.items(): for condition, condition_color in condition_color_map.items(): mask = (df['condition'] == condition) & (df['replicate'] == replicate) fig.add_trace(go.Scatter3d(x=df.loc[mask, 'PC1'], y=df.loc[mask, 'PC2'], z=df.loc[mask, 'PC3'], mode='markers', name=f'{condition}' if replicate == 'r1' else None, legendgroup=f'{condition}', showlegend=True if replicate == 'r1' else False, marker=dict(size=6 if replicate_symbol in ['diamond'] else 10, opacity=0.8, color=condition_color, symbol=replicate_symbol))) for replicate, replicate_symbol in replicate_symbol_map.items(): fig.add_trace(go.Scatter3d(x=[None], y=[None], z=[None], mode='markers', name=replicate, legendgroup=f'{replicate}', showlegend=True, marker=dict(size=10, opacity=1, color='black', symbol=replicate_symbol), hoverinfo='none')) # Annotations for the legend blocks #TODO: calculate the PC values. #TODO: adjust the axis length according to the actual size of axis! fig.update_layout( annotations=[ dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False, text='Condition', font=dict(size=15)), dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False, text='Replicate', font=dict(size=15)) ], scene=dict( #aspectmode='cube', #xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 53% v.', scaleratio=0.53), #yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 21% v.', scaleratio=0.21), #zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 7% variance', scaleratio=0.07), aspectmode='manual', aspectratio=dict(x=explained_variance_ratio[0], y=explained_variance_ratio[1], z=explained_variance_ratio[2]), xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC1: 53% v.', range=[min(df['PC1']), max(df['PC1'])]), yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC2: 21% v.', range=[min(df['PC2']), max(df['PC2'])]), zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title='PC3: 7% variance', range=[min(df['PC3']), max(df['PC3'])]), bgcolor='white' ), margin=dict(l=5, r=5, b=5, t=0) # Adjust the margins to prevent clipping of axis titles ) #fig.show() fig.write_image("fig1.svg") fig.update_layout( annotations=[ dict(x=1.1, y=1.0, xref='paper', yref='paper', showarrow=False, text='Condition', font=dict(size=15)), dict(x=1.1, y=0.6, xref='paper', yref='paper', showarrow=False, text='Donor', font=dict(size=15)), dict(x=1.08, y=0.2, xref='paper', yref='paper', showarrow=False, text=f'PC3: {explained_variance_ratio[2]*100:.2f}% v.', font=dict(size=15), textangle=-90) ], scene=dict( aspectmode='manual', aspectratio=dict(x=explained_variance_ratio[0]*2, y=explained_variance_ratio[1]*2, z=explained_variance_ratio[2]*2), #, range=[min(df['PC1']), max(df['PC1'])] #, range=[min(df['PC2']), max(df['PC2'])] #, range=[min(df['PC3']), max(df['PC3'])] xaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=f'PC1: {explained_variance_ratio[0]*100:.2f}% variance'), yaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=f'PC2: {explained_variance_ratio[1]*100:.2f}% v.'), zaxis=dict(gridcolor='black', backgroundcolor='white', zerolinecolor='black', title=''), bgcolor='white' ), margin=dict(l=0, r=0, b=0, t=0) # Adjust the margins to prevent clipping of axis titles ) fig.write_image("PCA_3D.svg") #/usr/bin/convert g235.png -crop 3250x1680+1+750 PCA_3D_.png -
(optional) estimate size factors and dispersion values.
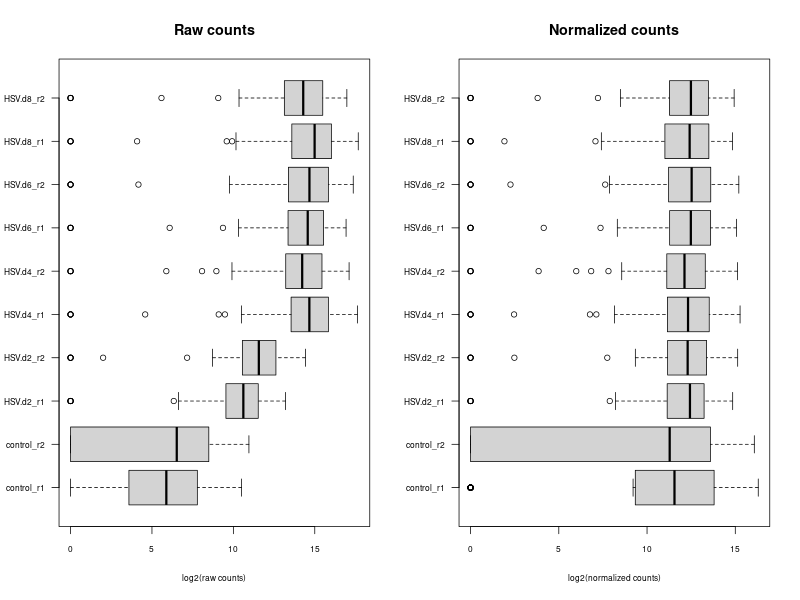
#Size Factors: These are used to normalize the read counts across different samples. The size factor for a sample accounts for differences in sequencing depth (i.e., the total number of reads) and other technical biases between samples. After normalization with size factors, the counts should be comparable across samples. Size factors are usually calculated in a way that they reflect the median or mean ratio of gene expression levels between samples, assuming that most genes are not differentially expressed. #Dispersion: This refers to the variability or spread of gene expression measurements. In RNA-seq data analysis, each gene has its own dispersion value, which reflects how much the counts for that gene vary between different samples, more than what would be expected just due to the Poisson variation inherent in counting. Dispersion is important for accurately modeling the data and for detecting differentially expressed genes. #So in summary, size factors are specific to samples (used to make counts comparable across samples), and dispersion values are specific to genes (reflecting variability in gene expression). sizeFactors(dds) #NULL # Estimate size factors dds <- estimateSizeFactors(dds) # Estimate dispersions dds <- estimateDispersions(dds) #> sizeFactors(dds) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #2.3282468 2.0251928 1.8036883 1.3767551 0.9341929 1.0911693 0.5454526 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #0.4604461 0.5799834 0.6803681 # If alone with virus data, the following BUG occured: #Still NULL --> BUG --> using manual calculation method for sizeFactor calculation! HeLa_TO_r1 HeLa_TO_r2 0.9978755 1.1092227 data.frame(genes = rownames(dds), dispersions = dispersions(dds)) #Given the raw counts, the control_r1 and control_r2 samples seem to have a much lower sequencing depth (total read count) than the other samples. Therefore, when normalization methods are applied, the normalization factors for these control samples will be relatively high, boosting the normalized counts. 1/0.9978755=1.002129023 1/1.1092227= #bamCoverage --bam ../markDuplicates/${sample}Aligned.sortedByCoord.out.bam -o ${sample}_norm.bw --binSize 10 --scaleFactor --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r1Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r1.bw --binSize 10 --scaleFactor 1.002129023 --effectiveGenomeSize 2864785220 bamCoverage --bam ../markDuplicates/HeLa_TO_r2Aligned.sortedByCoord.out.markDups.bam -o HeLa_TO_r2.bw --binSize 10 --scaleFactor 0.901532217 --effectiveGenomeSize 2864785220 raw_counts <- counts(dds) normalized_counts <- counts(dds, normalized=TRUE) #write.table(raw_counts, file="raw_counts.txt", sep="\t", quote=F, col.names=NA) #write.table(normalized_counts, file="normalized_counts.txt", sep="\t", quote=F, col.names=NA) #convert bam to bigwig using deepTools by feeding inverse of DESeq’s size Factor estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) -
compare the normalization methods
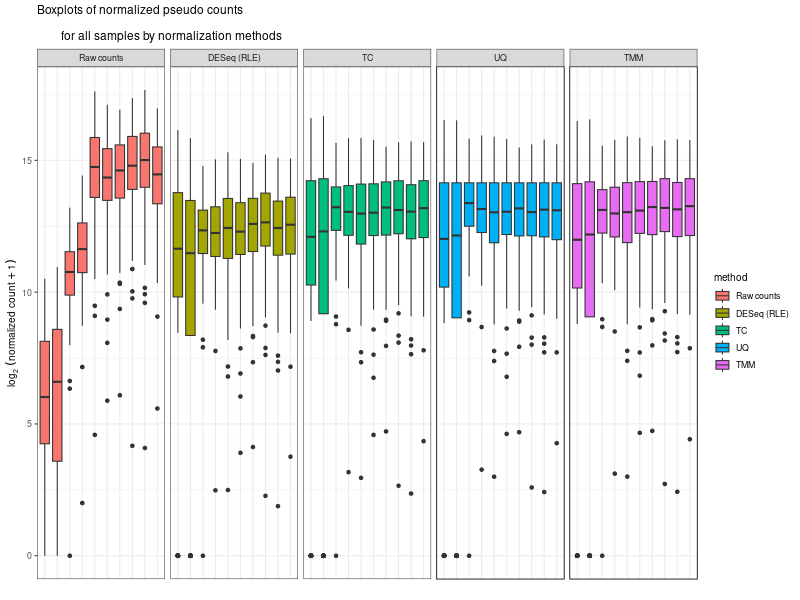
#The provided code indeed simulates the normalization method used by DESeq, which is known as the "Relative Log Expression (RLE)" normalization. The core idea behind this method is to scale the counts in each sample (column) by a size factor derived from the geometric means across all samples. #Here's a step-by-step breakdown of the code: # 1. estimSf function: # - The counts matrix is retrieved from the DESeq dataset. # - For each gene (row), the geometric mean across all samples is computed. # - Counts are divided by their respective gene's geometric mean. # - For each sample (column), the median of these ratios is computed. This median serves as the size factor for the sample. # 2. Once size factors are computed, the counts in the original matrix are then divided by these size factors to get the normalized counts. ### cds is a countDataset estimSf <- function (cds){ # Get the count matrix cts <- counts(cds) # Compute the geometric mean geomMean <- function(x) prod(x)^(1/length(x)) # Compute the geometric mean over the line gm.mean <- apply(cts, 1, geomMean) # Zero values are set to NA (avoid subsequentcdsdivision by 0) gm.mean[gm.mean == 0] <- NA # Divide each line by its corresponding geometric mean # sweep(x, MARGIN, STATS, FUN = "-", check.margin = TRUE, ...) # MARGIN: 1 or 2 (line or columns) # STATS: a vector of length nrow(x) or ncol(x), depending on MARGIN # FUN: the function to be applied cts <- sweep(cts, 1, gm.mean, FUN="/") # Compute the median over the columns med <- apply(cts, 2, median, na.rm=TRUE) # Return the scaling factor return(med) } #https://dputhier.github.io/ASG/practicals/rnaseq_diff_Snf2/rnaseq_diff_Snf2.html #http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#data-transformations-and-visualization #https://hbctraining.github.io/DGE_workshop/lessons/02_DGE_count_normalization.html #https://hbctraining.github.io/DGE_workshop/lessons/04_DGE_DESeq2_analysis.html #https://genviz.org/module-04-expression/0004/02/01/DifferentialExpression/ #DESeq2’s median of ratios [1] #EdgeR’s trimmed mean of M values (TMM) [2] #http://www.nathalievialaneix.eu/doc/html/TP1_normalization.html #very good website! test_normcount <- sweep(raw_counts, 2, sizeFactors(dds), "/") sum(test_normcount != normalized_counts) # round(estimSf(dds),6) is manually calculated sizeFactors of head(estimSf(dds)) all(round(estimSf(dds),6) == round(sizeFactors(dds), 6)) ## Checking the normalization png("normalization_small.png", width=800, height=600) epsilon <- 1 # pseudo-count to avoid problems with log(0) par(mfrow=c(1,2),cex.lab=0.7) boxplot(log2(raw_counts+epsilon), cex.axis=0.7, las=1, xlab="log2(raw counts)", horizontal=TRUE, main="Raw counts") boxplot(log2(normalized_counts+epsilon), cex.axis=0.7, las=1, xlab="log2(normalized counts)", horizontal=TRUE, main="Normalized counts") #boxplot(log10(assays(dds)[["cooks"]]), range=0, las=2) #plotDensity(log2(counts(dds.norm)+epsilon), col=col.pheno.selected, # xlab="log2(counts)", cex.lab=0.7, panel.first=grid()) #plotDensity(log2(counts(dds.norm, normalized=TRUE)+epsilon), col=col.pheno.selected, # xlab="log2(normalized counts)", cex.lab=0.7, panel.first=grid()) dev.off() # since we Gene-level differential expression analysis with DESeq2, the splicing plays no role in the analysis! # 用nanopore 可以 compare transcript length distribution. 有可能Cellline很长,Extracellular vesicles (EVs)很短! #In the folloing code, we compare different normalization methods, however, the function estimateSizeFactors(dge) doesn't work. I want replace the methods with the method implemented above. Please update the code #uding five norm factors from the following five normalization methods. How to replace the last command " sizeFactors(dds) <- dge2" for the five method? library(ggplot2) library(gridExtra) library(reshape2) library(mixOmics) library(RColorBrewer) library(DESeq) library(edgeR) library(VennDiagram) library(devtools) raw_counts_wn <- raw_counts[rowSums(raw_counts) > 0, ] dim(raw_counts_wn) #--Raw counts-- pseudo_counts <- log2(raw_counts_wn + 1) head(pseudo_counts) df_raw <- melt(pseudo_counts, id = rownames(raw_counts_wn)) names(df_raw)[1:2]<- c("id", "sample") df_raw$method <- rep("Raw counts", nrow(df_raw)) head(df_raw) #--DESeq-- cData = data.frame(row.names=colnames(raw_counts_wn), condition=condition) dge<-DESeqDataSetFromMatrix(countData=raw_counts_wn, colData=cData, design=~condition) dge <- estimateSizeFactors(dge) sizeFactors(dge) # Use your function to get the size factors sizeFactorsDGE <- estimSf(dge) isTRUE(all.equal(sizeFactors(dge), sizeFactorsDGE)) # Test the explained method using sweep to simulate the internal process of dge deseq_normcount <- counts(dge, normalized = TRUE) test_normcount <- sweep(raw_counts_wn, 2, sizeFactors(dge), "/") sum(test_normcount != deseq_normcount) pseudo_deseq <- log2(deseq_normcount + 1) df_deseq <- melt(pseudo_deseq, id = rownames(raw_counts_wn)) names(df_deseq)[1:2]<- c("id", "sample") df_deseq$method <- rep("DESeq (RLE)", nrow(df_raw)) #--edgeR-- dge2 <- DGEList(raw_counts_wn) dge2 dge2$samples #--Total count-- pseudo_TC <- log2(cpm(dge2) + 1) df_TC <- melt(pseudo_TC, id = rownames(raw_counts_wn)) names(df_TC)[1:2] <- c ("id", "sample") df_TC$method <- rep("TC", nrow(df_TC)) ##--RPKM-- #gene_lengths_wn <- gene_lengths[rowSums(raw_counts) > 0] #pseudo_RPKM <- log2(rpkm(dge2, gene.length = gene_lengths_wn) + 1) #df_RPKM <- melt(pseudo_RPKM, id = rownames(raw_counts_wn)) #names(df_RPKM)[1:2] <- c ("id", "sample") #df_RPKM$method <- rep("RPKM", nrow(df_RPKM)) #--Upper quartile-- dge2 <- calcNormFactors(dge2, method = "upperquartile") dge2$samples test_normcount <- sweep(dge2$counts, 2, dge2$samples$lib.size*dge2$samples$norm.factors / 10^6, "/") range(as.vector(test_normcount - cpm(dge2))) pseudo_UQ <- log2(cpm(dge2) + 1) df_UQ <- melt(pseudo_UQ, id = rownames(raw_counts_wn)) names(df_UQ)[1:2] <- c ("id", "sample") df_UQ$method <- rep("UQ", nrow(df_UQ)) #--TMM-- dge2 <- calcNormFactors(dge2, method = "TMM") dge2$samples pseudo_TMM <- log2(cpm(dge2) + 1) df_TMM <- melt(pseudo_TMM, id = rownames(raw_counts_wn)) names(df_TMM)[1:2] <- c ("id", "sample") #MODIFIED! df_TMM$method <- rep("TMM", nrow(df_TMM)) #TMM #--Comparison-- png("normalization.png", width=800, height=600) df_allnorm <- rbind(df_raw, df_deseq, df_TC, df_UQ, df_TMM) df_allnorm$method <- factor(df_allnorm$method, levels = c("Raw counts", "DESeq (RLE)", "TC", "UQ", "TMM")) #df_allnorm <- rbind(df_raw, df_TMM) #df_allnorm$method <- factor(df_allnorm$method, levels = c("Raw counts", "DESeq (RLE)")) p <- ggplot(data=df_allnorm, aes(x=sample, y=value, fill=method)) p <- p + geom_boxplot() p <- p + theme_bw() p <- p + ggtitle("Boxplots of normalized pseudo counts\n for all samples by normalization methods") p <- p + facet_grid(. ~ method) p <- p + ylab(expression(log[2] ~ (normalized ~ count + 1))) + xlab("") p <- p + theme(title = element_text(size=10), axis.text.x = element_blank(), axis.ticks.x = element_blank()) print(p) dev.off() #Assign a normalization size factor for downstream analysis from "Raw counts", "DESeq (RLE)", "TC", "UQ", "TMM") #You will need to repeat the DESeq2 analysis steps (like DESeq()) for each set of size factors to see how the results change with different normalization methods. #Note: This approach allows you to apply normalization factors from various methods to DESeq2. But keep in mind that each normalization method was developed with a specific intent and assumptions. Combining normalization methods from one package (like edgeR's TMM) with differential analysis from another package (like DESeq2) might not always be theoretically sound. Always interpret results with caution and preferably in consultation with domain experts. #DESeq (RLE) normalization: sizeFactors(dds) <- sizeFactors(dge) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #0.01996676 0.03367626 0.33493034 0.65395381 4.96825596 4.14671012 4.07416461 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #4.43844964 5.96130192 3.75356239 #Total Count normalization: This is actually just scaling by the total library size. In DESeq2, this would be equivalent to: sizeFactors(dds) <- colSums(raw_counts_wn) #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 # 14594 18992 182152 374750 3404540 2521691 2646741 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 # 3207708 3873254 2430625 identical(colnames(dds), rownames(dge2$samples)) #Upper Quartile (UQ) normalization: uq_factors <- dge2$samples$norm.factors * dge2$samples$lib.size / 10^6 names(uq_factors) <- rownames(dge2$samples) sizeFactors(dds) <- uq_factors #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #0.01578156 0.02060450 0.19559938 0.39108853 3.27858657 2.38221969 2.61381808 # HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #3.03441572 3.65683812 2.30405047 #TMM normalization: tmm_factors <- dge2$samples$norm.factors names(tmm_factors) <- rownames(dge2$samples) sizeFactors(dds) <- tmm_factors #> tmm_factors #control_r1 control_r2 HSV.d2_r1 HSV.d2_r2 HSV.d4_r1 HSV.d4_r2 HSV.d6_r1 #1.0813729 1.0849043 1.0738250 1.0435985 0.9630043 0.9446914 0.9875610 #HSV.d6_r2 HSV.d8_r1 HSV.d8_r2 #0.9459763 0.9441256 0.9479251 #Given the raw counts, the control_r1 and control_r2 samples seem to have a much lower sequencing depth (total read count) than the other samples. Therefore, when normalization methods such as TMM (Trimmed Mean of M-values) or UQ (Upper Quartile normalization) are applied, the normalization factors for these control samples will be relatively high, boosting the normalized counts. #To better understand the situation: # - Check the sequencing depth of each sample. Sum the raw counts across all genes for each sample to get the total read count. If the control samples have a substantially lower total read count, then normalization methods will try to adjust for this discrepancy. # - Consider the normalization method: Different normalization methods might provide slightly different results. TMM normalization, for example, tries to adjust for the compositional differences between samples. It's common to observe larger normalization factors for samples with a lower total read count. # - Visualize the data: MA plots, box plots, or density plots of the raw and normalized counts can help understand the distribution of counts and the effect of normalization. # ---- Adapt the following code to print the normalized_counts using tmm_factors ---- # RLE size factors rle_factors <- sizeFactors(dge) # Print side by side data.frame(RLE = rle_factors, TMM = tmm_factors) sizeFactors(dds) <- rle_factors normalized_counts_rle <- counts(dds, normalized=TRUE) sizeFactors(dds) <- tmm_factors #is it possible in the following command, the calculate de novo normalization factors and ignore the given tmm normalization factors? normalized_counts_tmm <- counts(dds, normalized=TRUE) difference_matrix <- normalized_counts_tmm - normalized_counts_rle all(difference_matrix == 0) -
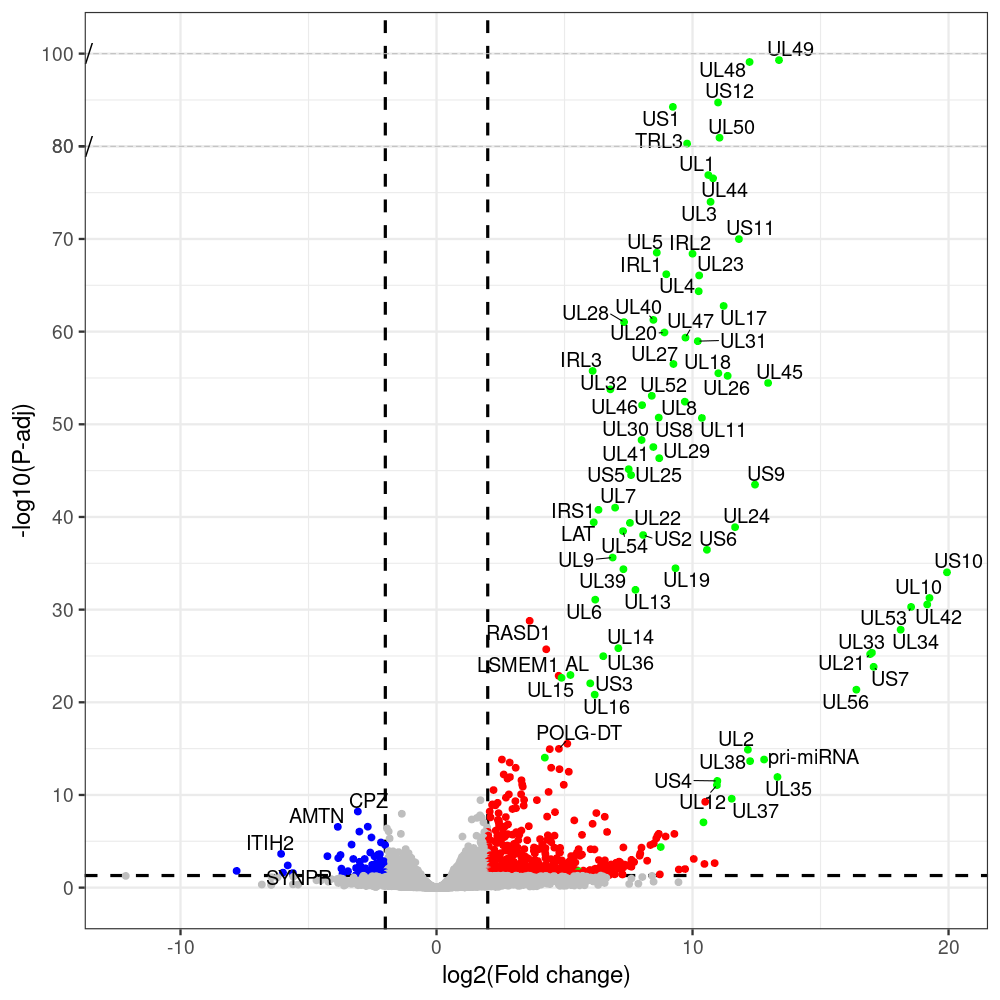
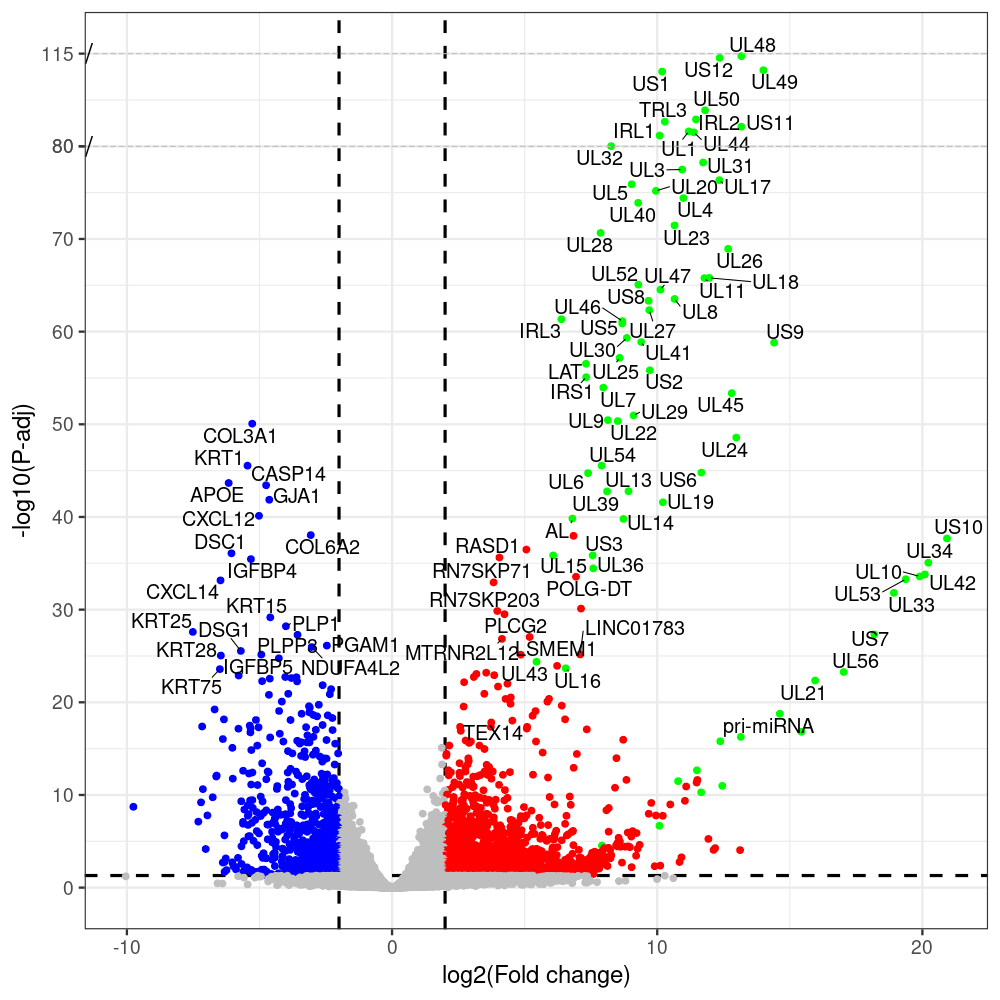
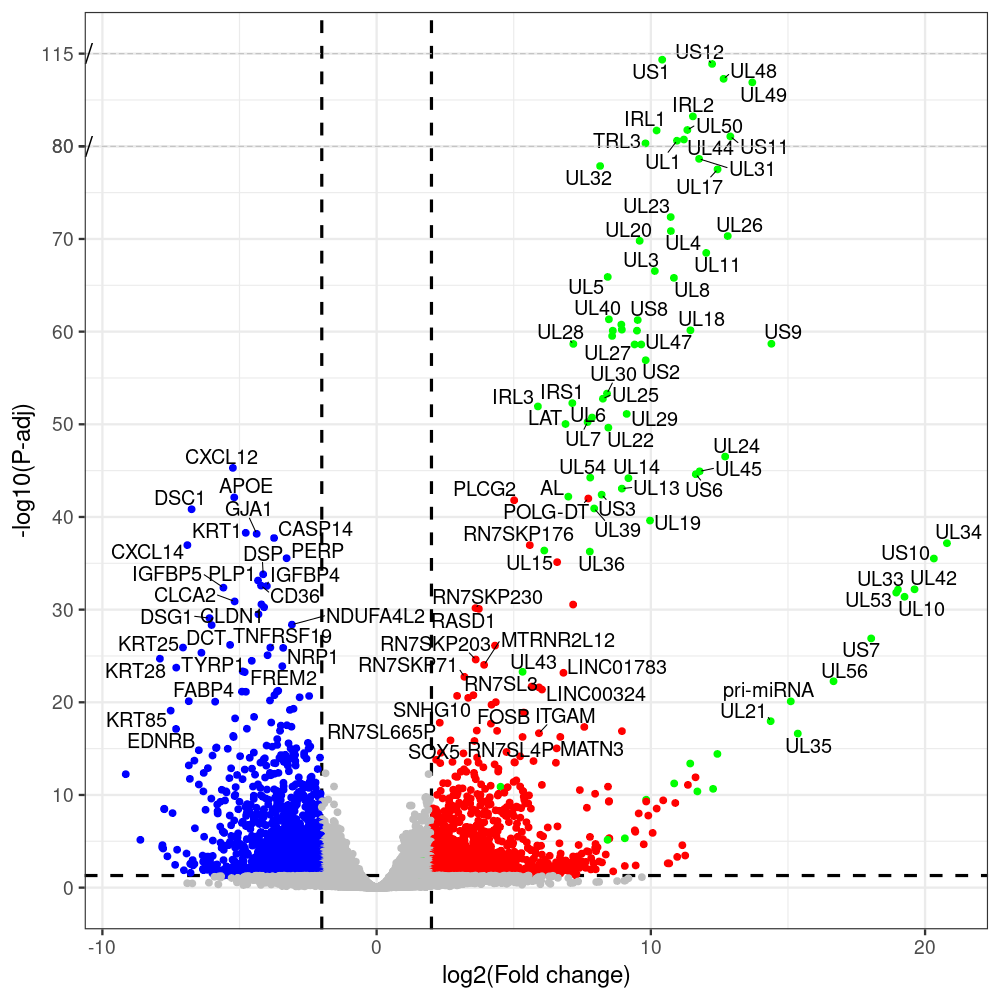
select the differentially expressed genes
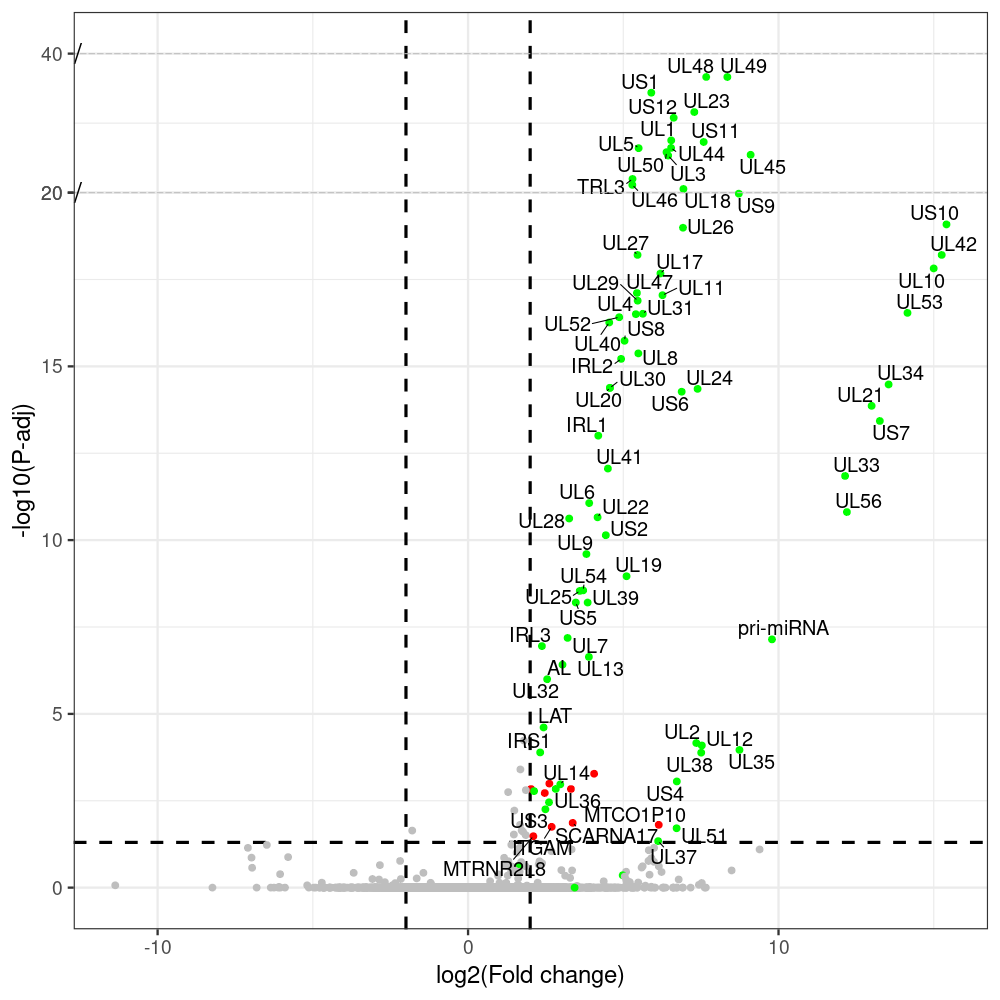
#https://galaxyproject.eu/posts/2020/08/22/three-steps-to-galaxify-your-tool/ #https://www.biostars.org/p/282295/ #https://www.biostars.org/p/335751/ #> condition # [1] control control HSV.d2 HSV.d2 HSV.d4 HSV.d4 HSV.d6 HSV.d6 HSV.d8 HSV.d8 #Levels: control HSV.d2 HSV.d4 HSV.d6 HSV.d8 #CONSOLE: mkdir /home/jhuang/DATA/Data_Manja_RNAseq_Organoids_Virus/results_chrHsv1_downstream/star_salmon/degenes setwd("/home/jhuang/DATA/Data_Manja_RNAseq_Organoids_Merged") #---- relevel to control ---- dds$condition <- relevel(dds$condition, "control") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d2_vs_control","HSV.d4_vs_control", "HSV.d6_vs_control", "HSV.d8_vs_control") dds$condition <- relevel(dds$condition, "HSV.d2") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d4_vs_HSV.d2", "HSV.d6_vs_HSV.d2", "HSV.d8_vs_HSV.d2") dds$condition <- relevel(dds$condition, "HSV.d4") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d6_vs_HSV.d4", "HSV.d8_vs_HSV.d4") dds$condition <- relevel(dds$condition, "HSV.d6") dds = DESeq(dds, betaPrior=FALSE) resultsNames(dds) clist <- c("HSV.d8_vs_HSV.d6") library(biomaRt) listEnsembl() listMarts() #--> total 69, 27 GRCh38.p7 and 39 GRCm38.p4 ensembl <- useEnsembl(biomart = "ensembl", dataset = "hsapiens_gene_ensembl", version="104") datasets <- listDatasets(ensembl) # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(20, 40) compressed_range <- c(20.0, 24.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 20, by=5) y_breaks_compressed <- c(20.0, 24.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 20, by=5) y_labels_compressed <- c(20, 40) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- HSV.d8_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # -- HSV.d6_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # -- HSV.d4_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 100) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 100) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # -- HSV.d2_vs_control.png -- # Define the original and compressed ranges original_range <- c(20, 40) compressed_range <- c(20.0, 24.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 20, by=5) y_breaks_compressed <- c(20.0, 24.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 20, by=5) y_labels_compressed <- c(20, 40) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) #under DIR degenes under KONSOLE for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "~/Tools/csv2xls-0.4/csv_to_xls.py ${i}-all_annotated.txt ${i}-up_annotated.txt ${i}-down_annotated.txt -d$',' -o ${i}.xls;"; done -
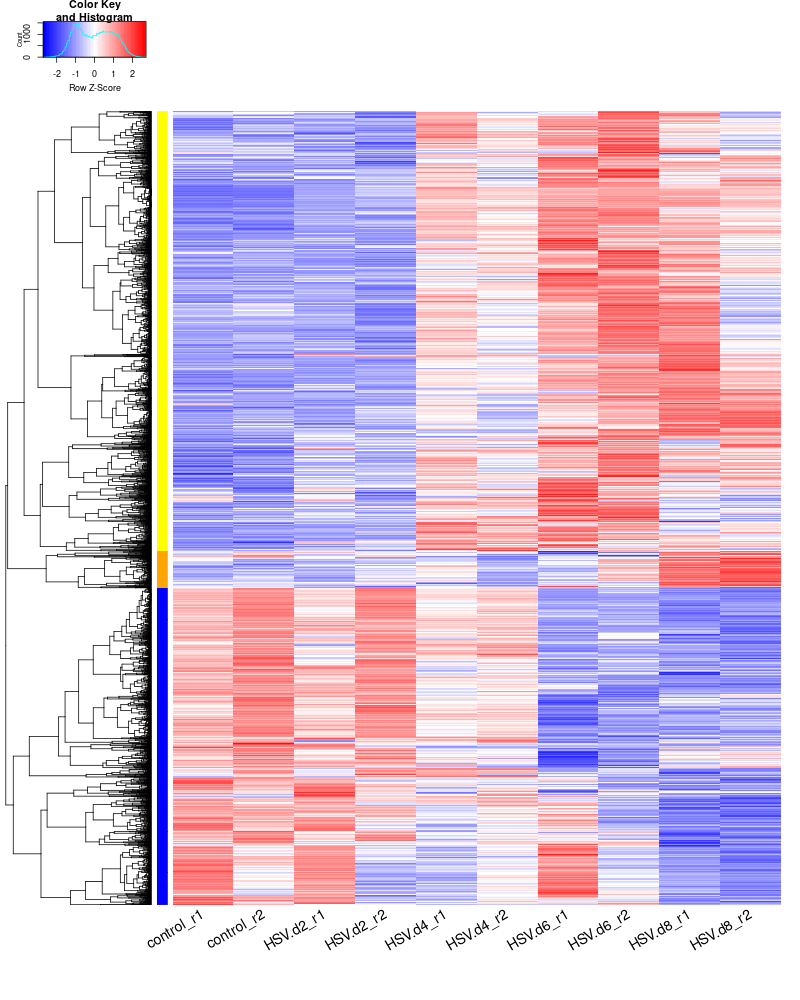
clustering the genes and draw heatmap
for i in HSV.d2_vs_control HSV.d4_vs_control HSV.d6_vs_control HSV.d8_vs_control HSV.d4_vs_HSV.d2 HSV.d6_vs_HSV.d2 HSV.d8_vs_HSV.d2 HSV.d6_vs_HSV.d4 HSV.d8_vs_HSV.d4 HSV.d8_vs_HSV.d6; do echo "cut -d',' -f1-1 ${i}-up_annotated.txt > ${i}-up.id"; echo "cut -d',' -f1-1 ${i}-down_annotated.txt > ${i}-down.id"; done cat *.id | sort -u > ids #add Gene_Id in the first line, delete the "" GOI <- read.csv("ids")$Gene_Id #4647 RNASeq.NoCellLine <- assay(rld) #install.packages("gplots") library("gplots") #clustering methods: "ward.D", "ward.D2", "single", "complete", "average" (= UPGMA), "mcquitty" (= WPGMA), "median" (= WPGMC) or "centroid" (= UPGMC). pearson or spearman datamat = RNASeq.NoCellLine[GOI, ] #datamat = RNASeq.NoCellLine write.csv(as.data.frame(datamat), file ="gene_expressions.txt") constant_rows <- apply(datamat, 1, function(row) var(row) == 0) if(any(constant_rows)) { cat("Removing", sum(constant_rows), "constant rows.\n") datamat <- datamat[!constant_rows, ] } hr <- hclust(as.dist(1-cor(t(datamat), method="pearson")), method="complete") hc <- hclust(as.dist(1-cor(datamat, method="spearman")), method="complete") mycl = cutree(hr, h=max(hr$height)/1.05) mycol = c("YELLOW", "BLUE", "ORANGE", "MAGENTA", "CYAN", "RED", "GREEN", "MAROON", "LIGHTBLUE", "PINK", "MAGENTA", "LIGHTCYAN", "LIGHTRED", "LIGHTGREEN"); mycol = mycol[as.vector(mycl)] #png("DEGs_heatmap.png", width=900, height=800) #cex.lab=10, labRow="", png("DEGs_heatmap.png", width=800, height=1000) heatmap.2(as.matrix(datamat),Rowv=as.dendrogram(hr),Colv = NA, dendrogram = 'row',labRow="", scale='row',trace='none',col=bluered(75), cexCol=1.8, RowSideColors = mycol, margins=c(10,2), cexRow=1.5, srtCol=30, lhei = c(1, 8), lwid=c(2, 8)) #rownames(datamat) #heatmap.2(datamat, Rowv=as.dendrogram(hr), col=bluered(75), scale="row", RowSideColors=mycol, trace="none", margin=c(5,5), sepwidth=c(0,0), dendrogram = 'row', Colv = 'false', density.info='none', labRow="", srtCol=30, lhei=c(0.1,2)) dev.off() #### cluster members ##### write.csv(names(subset(mycl, mycl == '1')),file='cluster1_YELLOW.txt') write.csv(names(subset(mycl, mycl == '2')),file='cluster2_DARKBLUE.txt') write.csv(names(subset(mycl, mycl == '3')),file='cluster3_DARKORANGE.txt') write.csv(names(subset(mycl, mycl == '4')),file='cluster4.txt') #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.txt -d',' -o DEGs_heatmap_cluster_members.xls ~/Tools/csv2xls-0.4/csv_to_xls.py \ significant_gene_expressions.txt \ -d',' -o DEGs_heatmap_expression_data.xls; #### cluster members (advanced) ##### subset_1<-names(subset(mycl, mycl == '1')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_1, ]) #2575 subset_2<-names(subset(mycl, mycl == '2')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_2, ]) #1855 subset_3<-names(subset(mycl, mycl == '3')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_3, ]) #217 subset_4<-names(subset(mycl, mycl == '4')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_4, ]) # subset_5<-names(subset(mycl, mycl == '5')) data <- as.data.frame(datamat[rownames(datamat) %in% subset_5, ]) # # Initialize an empty data frame for the annotated data annotated_data <- data.frame() # Determine total number of genes total_genes <- length(rownames(data)) # Loop through each gene to annotate for (i in 1:total_genes) { gene <- rownames(data)[i] result <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = gene, mart = ensembl) # If multiple rows are returned, take the first one if (nrow(result) > 1) { result <- result[1, ] } # Check if the result is empty if (nrow(result) == 0) { result <- data.frame(ensembl_gene_id = gene, external_gene_name = NA, gene_biotype = NA, entrezgene_id = NA, chromosome_name = NA, start_position = NA, end_position = NA, strand = NA, description = NA) } # Transpose expression values expression_values <- t(data.frame(t(data[gene, ]))) colnames(expression_values) <- colnames(data) # Combine gene information and expression data combined_result <- cbind(result, expression_values) # Append to the final dataframe annotated_data <- rbind(annotated_data, combined_result) # Print progress every 100 genes if (i %% 100 == 0) { cat(sprintf("Processed gene %d out of %d\n", i, total_genes)) } } # Save the annotated data to a new CSV file #write.csv(annotated_data, "cluster1_YELLOW.csv", row.names=FALSE) write.csv(annotated_data, "cluster2_DARKBLUE.csv", row.names=FALSE) write.csv(annotated_data, "cluster3_DARKORANGE.csv", row.names=FALSE) write.csv(annotated_data, "cluster4_DARKMAGENTA.csv", row.names=FALSE) write.csv(annotated_data, "cluster5_DARKCYAN.csv", row.names=FALSE) #~/Tools/csv2xls-0.4/csv_to_xls.py cluster*.csv -d',' -o DEGs_heatmap_clusters.xls -
code of differential gene analysis for clist <- c("HSV.d2_vs_control","HSV.d4_vs_control", "HSV.d6_vs_control", "HSV.d8_vs_control")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d2_vs_control.png -- # Define the original and compressed ranges original_range <- c(20, 40) compressed_range <- c(20.0, 24.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 20, by=5) y_breaks_compressed <- c(20.0, 24.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 20, by=5) y_labels_compressed <- c(20, 40) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d4_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 100) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 100) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[3] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d6_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[4] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # -- HSV.d8_vs_control.png -- # Define the original and compressed ranges original_range <- c(80, 115) compressed_range <- c(80.0, 90.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 80, by=10) y_breaks_compressed <- c(80.0, 90.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 80, by=10) y_labels_compressed <- c(80, 115) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() -
code of differential gene analysis for clist <- c("HSV.d4_vs_HSV.d2", "HSV.d6_vs_HSV.d2", "HSV.d8_vs_HSV.d2")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(15, 20) compressed_range <- c(15.0, 20.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 15, by=5) y_breaks_compressed <- c(15.0, 20.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 15, by=5) y_labels_compressed <- c(15, 20) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=10) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=10) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[3] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=10) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=10) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() -
code of differential gene analysis for clist <- c("HSV.d6_vs_HSV.d4", "HSV.d8_vs_HSV.d4")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(15, 20) compressed_range <- c(15.0, 20.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 15, by=5) y_breaks_compressed <- c(15.0, 20.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 15, by=5) y_labels_compressed <- c(15, 20) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() # -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=5) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=5) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off() -
code of differential gene analysis for clist <- c("HSV.d8_vs_HSV.d6")
# -- 1. export res_df containing both human and virus genes -- #for (i in clist) { i<-clist[1] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] res_df <- as.data.frame(res) write.csv(as.data.frame(res_df[order(res_df$pvalue),]), file = paste(i, "all.txt", sep="-")) up <- subset(res_df, padj<=0.05 & log2FoldChange>=2) down <- subset(res_df, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down.txt", sep="-")) #} # -- 2. annatete human genes 'geness_res', note that the virus genes in that is ignored in the process since they are not in the database -- #for (i in clist) { #i<-clist[2] contrast = paste("condition", i, sep="_") res = results(dds, name=contrast) res <- res[!is.na(res$log2FoldChange),] #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("ENTREZID","EXONID","GENEBIOTYPE","GENEID","GENENAME","PROTEINDOMAINSOURCE","PROTEINID","SEQNAME","SEQSTRAND","SYMBOL","TXBIOTYPE","TXID","TXNAME","UNIPROTID")) #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "ENTREZID", "SYMBOL", "GENENAME","GENEBIOTYPE","TXBIOTYPE","SEQSTRAND","UNIPROTID")) # In the ENSEMBL-database, GENEID is ENSEMBL-ID. #geness <- AnnotationDbi::select(edb86, keys = rownames(res), keytype = "GENEID", columns = c("GENEID", "SYMBOL", "GENEBIOTYPE")) # "ENTREZID", "TXID","TXBIOTYPE","TXSEQSTART","TXSEQEND" #geness <- geness[!duplicated(geness$GENEID), ] #using getBM replacing AnnotationDbi::select #filters = 'ensembl_gene_id' means the records should always have a valid ensembl_gene_ids. geness <- getBM(attributes = c('ensembl_gene_id', 'external_gene_name', 'gene_biotype', 'entrezgene_id', 'chromosome_name', 'start_position', 'end_position', 'strand', 'description'), filters = 'ensembl_gene_id', values = rownames(res), mart = ensembl) geness_uniq <- distinct(geness, ensembl_gene_id, .keep_all= TRUE) #merge by column by common colunmn name, in the case "GENEID" res$ENSEMBL = rownames(res) identical(rownames(res), rownames(geness_uniq)) res_df <- as.data.frame(res) geness_res <- merge(geness_uniq, res_df, by.x="ensembl_gene_id", by.y="ENSEMBL") dim(geness_res) rownames(geness_res) <- geness_res$ensembl_gene_id geness_res$ensembl_gene_id <- NULL #} # -- 3. prepare annatete virus genes -- virus_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") # replace with the actual list of column names virus_rows <- res_df[rownames(res_df) %in% virus_genes, ] virus_rows$external_gene_name <- rownames(virus_rows) virus_rows$chromosome_name <- "chrHsv1" # Define default values based on data type default_values <- list( character = NULL, numeric = 0, integer = 0L, logical = FALSE ) # Ensure that virus_rows has the same columns as geness_res for (col in colnames(geness_res)) { if (!col %in% colnames(virus_rows)) { data_type <- class(geness_res[[col]])[1] default_value <- default_values[[data_type]] virus_rows[[col]] <- rep(default_value, nrow(virus_rows)) } } missing_cols <- setdiff(colnames(geness_res), colnames(virus_rows)) for (col in missing_cols) { virus_rows[[col]] <- NA # Or another default value as appropriate } # Reorder columns in virus_rows to match the order in geness_res virus_rows <- virus_rows[, colnames(geness_res), drop = FALSE] # -- 4. merge them together -- #for (i in clist) { merged_df <- rbind(geness_res, virus_rows) merged_df_sorted <- as.data.frame(merged_df[order(merged_df$padj),]) write.csv(merged_df_sorted, file = paste(i, "all_annotated.txt", sep="-")) up <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange>=2) down <- subset(merged_df_sorted, padj<=0.05 & log2FoldChange<=-2) write.csv(as.data.frame(up[order(up$log2FoldChange,decreasing=TRUE),]), file = paste(i, "up_annotated.txt", sep="-")) write.csv(as.data.frame(down[order(abs(down$log2FoldChange),decreasing=TRUE),]), file = paste(i, "down_annotated.txt", sep="-")) #} # -- 5. draw graphics -- #geness_res <- read.csv(file = "HSV.d2_vs_control-all_annotated.txt", sep=",", row.names=1) geness_res <- merged_df_sorted # Color setting geness_res$Color <- ifelse(geness_res$padj > 0.05 | abs(geness_res$log2FoldChange) < 2, "gray", ifelse(geness_res$log2FoldChange > 0, "red", "blue")) # Predefined genes colored in green predefined_genes <- c("AL", "IRL1", "IRL2", "IRL3", "IRS1", "LAT", "ORF-O/P", "Ori-S", "pri-miRNA", "TRL1", "TRL2", "TRL3", "TRS1", "UL1", "UL10", "UL11", "UL12", "UL13", "UL14", "UL15", "UL16", "UL17", "UL18", "UL19", "UL2", "UL20", "UL21", "UL22", "UL23", "UL24", "UL25", "UL26", "UL27", "UL28", "UL29", "UL3", "UL30", "UL31", "UL32", "UL33", "UL34", "UL35", "UL36", "UL37", "UL38", "UL39", "UL4", "UL40", "UL41", "UL42", "UL43", "UL44", "UL45", "UL46", "UL47", "UL48", "UL49", "UL5", "UL50", "UL51", "UL52", "UL53", "UL54", "UL55", "UL56", "UL6", "UL7", "UL8", "UL9", "US1", "US10", "US11", "US12", "US2", "US3", "US4", "US5", "US6", "US7", "US8", "US9") geness_res$Color[geness_res$external_gene_name %in% predefined_genes] <- "green" geness_res$invert_Padj <- (-log10(geness_res$padj)) * sign(geness_res$log2FoldChange) top_g <- unique(c(geness_res[order(geness_res$invert_Padj, decreasing = TRUE), 'external_gene_name'][1:100], geness_res[order(geness_res$invert_Padj, decreasing = FALSE), 'external_gene_name'][1:100])) # Define the original and compressed ranges original_range <- c(40, 50) compressed_range <- c(40.0, 50.0) # Calculate breaks for the y-axis y_breaks_below <- seq(0, 40, by=5) y_breaks_compressed <- c(40.0, 50.0) y_breaks_above <- c() y_breaks <- c(y_breaks_below, y_breaks_compressed, y_breaks_above) y_labels_below <- seq(0, 40, by=5) y_labels_compressed <- c(40, 50) y_labels_above <- c() y_labels <- c(y_labels_below, y_labels_compressed, y_labels_above) # Adjust the p-values based on the ranges geness_res$adjusted_pvalue <- with(geness_res, ifelse(-log10(padj) > original_range[1] & -log10(padj) <= original_range[2], ((-log10(padj) - original_range[1]) / (original_range[2] - original_range[1])) * (compressed_range[2] - compressed_range[1]) + compressed_range[1], ifelse(-log10(padj) > original_range[2], -log10(padj) - (original_range[2] - original_range[1]) + (compressed_range[2] - compressed_range[1]), -log10(padj)))) # Create the plot png(paste(i, "png", sep="."), width=1000, height=1000) ggplot(geness_res, aes(x = log2FoldChange, y = adjusted_pvalue, color = Color, label = external_gene_name)) + geom_vline(xintercept = c(2, -2), lty = "dashed", size = 1.5) + geom_hline(yintercept = -log10(0.05), lty = "dashed", size = 1.5) + geom_point(size = 3) + labs(x = "log2(Fold change)", y = "-log10(P-adj)", color = "Significance") + scale_color_identity() + geom_text_repel(data = subset(geness_res, external_gene_name %in% top_g & padj < 0.05 & (abs(log2FoldChange) >= 2)), size = 7, point.padding = 0.15, color = "black", min.segment.length = .1, box.padding = .2, lwd = 2) + theme_bw(base_size = 24) + theme(legend.position = "bottom") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[1], ymax = compressed_range[1], linetype = "dashed", color = "grey") + #annotate("rect", xmin = -Inf, xmax = Inf, ymin = compressed_range[2], ymax = compressed_range[2], linetype = "dashed", color = "grey") + #annotate("text", x = -Inf, y = compressed_range[1], label = "/", hjust = 0, size = 10) + #annotate("text", x = -Inf, y = compressed_range[2], label = "/", hjust = 0, size = 10) + scale_y_continuous(breaks = sort(y_breaks), labels = sort(y_labels)) dev.off()
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- Kraken2 Installation and Usage Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
- 基因组与表型特性研究:一株药物敏感的鲍曼不动杆菌显示出增强的毒力相关特性和应激耐受性
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
RNAseq Analysis LT-K331A-LTtr-control
MicrobiotaProcess Group2 vs Group6 (v1)
Small RNA sequencing processing in the example of smallRNA_7