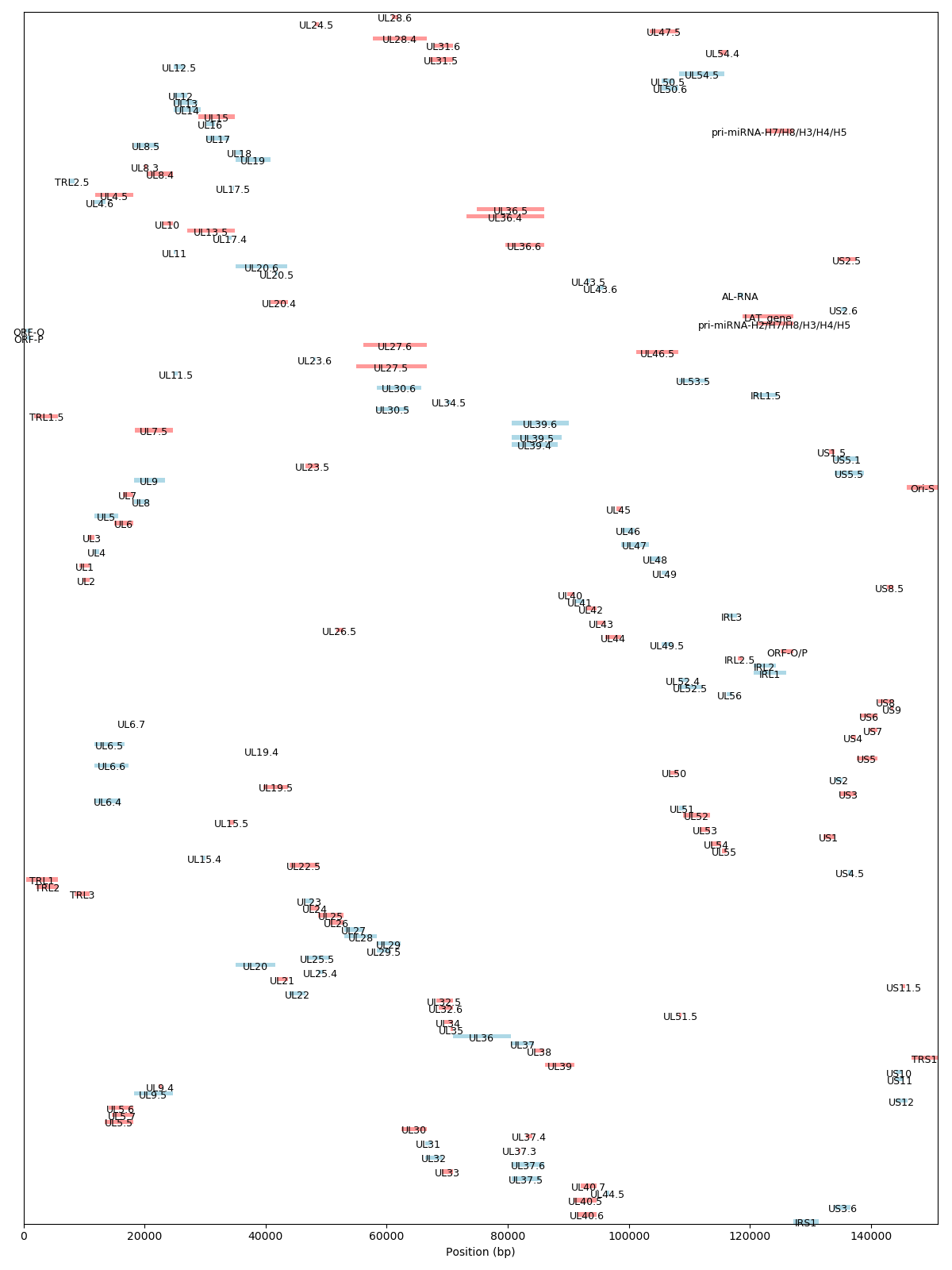
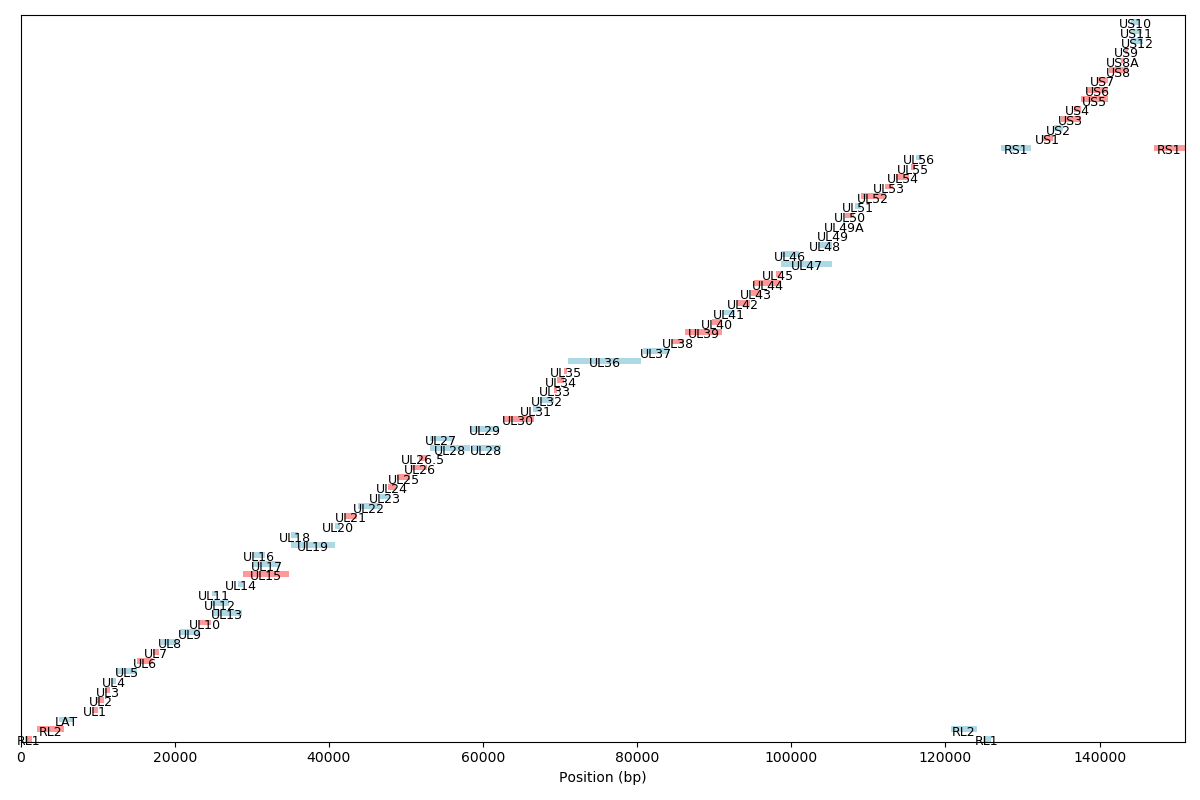
Genomic Organization of Herpes Simplex Virus type 1 (HSV-1 s17)
gene_x 0 like s 1227 view s
Tags: plot, python, RNA-seq
HSV (Herpes Simplex Virus) contains two types of repeated sequences, which are:
-
IRS (Internal Repeat, Short): This refers to the short repeated sequences located between the unique long (UL) and unique short (US) regions of the HSV genome. There are two copies of IRS in the genome flanking the US region.
-
TRL (Terminal Repeat, Long) and IRL (Internal Repeat, Long): These are the long repeated sequences in the HSV genome. The TRL sequences are found at the very ends (terminals) of the linear HSV genome, while the IRL sequences are found internally, flanking the UL region.
The organization of the HSV genome can be summarized as: TRL - UL - IRL - US - IRS - US (in reverse orientation) - IRL
Here's a brief breakdown:
-
UL (Unique Long): This is a unique sequence region found once in the genome.
-
US (Unique Short): This is another unique sequence region but it is shorter than UL and is found flanked by IRS sequences.
-
IRS (Internal Repeat Short): These are short repeated sequences that flank the US region.
-
TRL (Terminal Repeat Long) and IRL (Internal Repeat Long): The long repeated sequences found at the genome's terminals and internally flanking the UL region.
These repeated sequences play crucial roles in the HSV life cycle, especially during the processes of recombination, genome replication, and the switch between latency and active replication.
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
def read_gtf(filename):
with open(filename, 'r') as file:
lines = file.readlines()
features = []
for line in lines:
if not line.startswith("#"):
split_line = line.strip().split("\t")
feature_type = split_line[2]
start = int(split_line[3])
end = int(split_line[4])
try:
gene_name = [x for x in split_line[8].split(";") if "gene_id" in x][0].split('"')[1]
except:
gene_name = "unknown"
strand = split_line[6]
features.append((feature_type, start, end, gene_name, strand))
return features
def plot_features(features, genome_id):
fig, ax = plt.subplots(figsize=(12, 16))
y_offset = 0
y_increment = 1
y_positions = {}
for feature_type, start, end, gene_name, strand in features:
if feature_type == "gene":
if gene_name not in y_positions:
y_positions[gene_name] = y_offset
y_offset += y_increment
y_pos = y_positions[gene_name]
color = "lightblue" if strand == "-" else (1, 0.6, 0.6) # Using RGB
rect = mpatches.Rectangle([start, y_pos], end-start, 0.6, ec="none", fc=color)
ax.add_patch(rect)
if strand == "+":
ax.text((start + end) / 2, y_pos, gene_name, ha='center', va='center', fontsize=9)
else:
ax.text((start + end) / 2, y_pos, gene_name, ha='center', va='center', fontsize=9)
ax.set_xlim(0, max([f[2] for f in features]))
ax.set_ylim(0, y_offset)
ax.set_yticks([])
ax.set_xlabel("Position (bp)")
ax.set_title(f"") #f"Genomic Organization of {genome_id}"
plt.tight_layout()
plt.savefig(f"{genome_id}_genomic_organization.png")
plt.show()
if __name__ == "__main__":
genome_id = "chrHsv1_s17"
features = read_gtf("chrHsv1_s17.gtf")
plot_features(features, genome_id)
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences
- 阳光房漏水怎么办?丁基胶带才是最佳密封选择
- DAMIAN Post-processing for Flavivirus and FSME
- All tools and services of BV-BRC
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
RNA-seq skin organoids on GRCh38+chrHsv1 (final)
YopQ Secretion Boxplot and Fitting Function