Small RNA processing
gene_x 0 like s 985 view s
Tags:
Small RNA sequencing is a type of RNA-sequencing (RNA-seq) that specifically targets and sequences small RNA molecules in a sample.
RNA-seq is a technique that uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, capturing a snapshot of the transcriptome.
Small RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs), play crucial roles in gene regulation. They typically range from 20 to 30 nucleotides in length.
-
prepare raw data
#mv Data_Ute_smallRNA_3/bundle_v1 Data_Ute_smallRNA_5 ln -s ../Data_Ute_smallRNA_3/bundle_v1 . #OLD MKL_1_wt_1_221216.fastq.gz -> ../221216_NB501882_0404_AHLVNMBGXM/ute/nf796/MKL_1_wt_1_S16_R1_001.fastq.gz #OLD MKL_1_wt_2_221216.fastq.gz -> ../221216_NB501882_0404_AHLVNMBGXM/ute/nf797/MKL_1_wt_2_S17_R1_001.fastq.gz ln -s ./230623_newDemulti_smallRNAs/210817_NB501882_0294_AHW5Y2BGXJ_smallRNA_Ute_newDemulti/2021_nf_ute_smallRNA/nf655/MKL_1_derived_EV_miRNA_S1_R1_001.fastq.gz 2021_August_nf655_MKL-1_EV-miRNA.fastq.gz ln -s ./230623_newDemulti_smallRNAs/210817_NB501882_0294_AHW5Y2BGXJ_smallRNA_Ute_newDemulti/2021_nf_ute_smallRNA/nf657/WaGa_derived_EV_miRNA_S2_R1_001.fastq.gz 2021_August_nf657_WaGa_EV-miRNA.fastq.gz ln -s ./230623_newDemulti_smallRNAs/220617_NB501882_0371_AH7572BGXM_smallRNA_Ute_newDemulti/2022_nf_ute_smallRNA/nf774/0403_WaGa_wt_S1_R1_001.fastq.gz 2022_August_nf774_0403_WaGa_wt.fastq.gz ln -s ./230623_newDemulti_smallRNAs/220617_NB501882_0371_AH7572BGXM_smallRNA_Ute_newDemulti/2022_nf_ute_smallRNA/nf780/0505_MKL1_wt_S2_R1_001.fastq.gz 2022_August_nf780_0505_MKL-1_wt.fastq.gz ln -s ./230623_newDemulti_smallRNAs/221216_NB501882_0404_AHLVNMBGXM_smallRNA_Ute_newDemulti/2022_nf_ute_smallRNA/nf796/MKL-1_wt_1_S1_R1_001.fastq.gz 2022_November_nf796_MKL-1_wt_1.fastq.gz ln -s ./230623_newDemulti_smallRNAs/221216_NB501882_0404_AHLVNMBGXM_smallRNA_Ute_newDemulti/2022_nf_ute_smallRNA/nf797/MKL-1_wt_2_S2_R1_001.fastq.gz 2022_November_nf797_MKL-1_wt_2.fastq.gz ln -s ./230602_NB501882_0428_AHKG53BGXT/demulti_new/nf876/1002_WaGa_sT_Dox_S1_R1_001.fastq.gz 2023_June_nf876_1002_WaGa_sT_Dox.fastq.gz ln -s ./230602_NB501882_0428_AHKG53BGXT/demulti_new/nf887/2312_MKL_1_scr_DMSO_S2_R1_001.fastq.gz 2023_June_nf887_2312_MKL-1_scr_DMSO.fastq.gz -
main run
mkdir our_out # -qc -ra TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -rb 4 #NOT_USED # -mic -mtool Blast -mdb viruses #IGNORING Microbe Module since it is very time-consuming! #jhuang@hamburg:~/DATA/Data_Ute/Data_Ute_smallRNA_5$ java -jar COMPSRA.jar -ref hg38 -qc -ra TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -rb 4 -rh 20 -rt 20 -rr 20 -rlh 8,17 -aln -mt star -ann -ac 1,2,3,4,5,6 -mic -mtool Blast -mdb viruses -in 2021_August_nf655_MKL-1_EV-miRNA.fastq.gz -out ./our_out/ for sample in 2021_August_nf655_MKL-1_EV-miRNA 2021_August_nf657_WaGa_EV-miRNA 2022_August_nf774_0403_WaGa_wt 2022_August_nf780_0505_MKL-1_wt 2022_November_nf796_MKL-1_wt_1 2022_November_nf797_MKL-1_wt_2 2023_June_nf876_1002_WaGa_sT_Dox 2023_June_nf887_2312_MKL-1_scr_DMSO; do mkdir our_out/${sample}/ java -jar COMPSRA.jar -ref hg38 -rh 20 -rt 20 -rr 20 -rlh 8,17 -aln -mt star -ann -ac 1,2,3,4,5,6 -in ${sample}.fastq.gz -out ./our_out/ done -
prepare Data_Ute/Data_Ute_smallRNA_4/sample.list
2021_August_nf655_MKL-1_EV-miRNA 2021_August_nf657_WaGa_EV-miRNA 2022_August_nf774_0403_WaGa_wt 2022_August_nf780_0505_MKL-1_wt 2022_November_nf796_MKL-1_wt_1 2022_November_nf797_MKL-1_wt_2 2023_June_nf876_1002_WaGa_sT_Dox 2023_June_nf887_2312_MKL-1_scr_DMSO -
extract the raw counts and perform statistical test on pre-defined groups.
#The following results calculate raw counts (Note: If you only want to merge the count files, you can use -fm -fms.) java -jar COMPSRA.jar -ref hg38 -fun -fm -fms 1-8 -fdclass 1,2,3,4,5 -fdann -pro COMPSRA_MERGE -inf ./sample.list -out ./our_out/ #The following command calculate statistical test after defining case and control. java -jar COMPSRA.jar -ref hg38 -fun -fd -fdclass 1,2,3,4,5,6 -fdcase 1-2 -fdctrl 3-6 -fdnorm cpm -fdtest mwu -fdann -pro COMPSRA_DEG -inf ./sample.list -out ./our_out/ sed -i -e 's/_August/-08/g' COMPSRA_MERGE_0_miRNA.txt sed -i -e 's/_November/-11/g' COMPSRA_MERGE_0_miRNA.txt sed -i -e 's/_June/-06/g' COMPSRA_MERGE_0_miRNA.txt sed -i -e 's/_STAR_Aligned_miRNA.txt//g' COMPSRA_MERGE_0_miRNA.txt #sed -i -e 's/_piRNA.txt//g' COMPSRA_MERGE_0_piRNA.txt #sed -i -e 's/_tRNA.txt//g' COMPSRA_MERGE_0_tRNA.txt #sed -i -e 's/_snoRNA.txt//g' COMPSRA_MERGE_0_snoRNA.txt #sed -i -e 's/_snRNA.txt//g' COMPSRA_DEG_0_snRNA.txt sed -i -e 's/_August/-08/g' COMPSRA_DEG_0_miRNA.txt sed -i -e 's/_November/-11/g' COMPSRA_DEG_0_miRNA.txt sed -i -e 's/_June/-06/g' COMPSRA_DEG_0_miRNA.txt sed -i -e 's/_STAR_Aligned_miRNA.txt//g' COMPSRA_DEG_0_miRNA.txt import pandas as pd df = pd.read_csv('COMPSRA_MERGE_0_miRNA.txt', sep='\t', index_col=0) df = df.drop(columns=['Unnamed: 9']) # Assuming df is your DataFrame df.loc['Sum'] = df.sum(numeric_only=True) df.to_csv('COMPSRA_MERGE_0_miRNA_.txt', sep='\t') df = pd.read_csv('COMPSRA_MERGE_0_piRNA.txt', sep='\t', index_col=0) df = df.drop(columns=['Unnamed: 9']) df.loc['Sum'] = df.sum(numeric_only=True) df.to_csv('COMPSRA_MERGE_0_piRNA_.txt', sep='\t') df = pd.read_csv('COMPSRA_MERGE_0_snoRNA.txt', sep='\t', index_col=0) df = df.drop(columns=['Unnamed: 9']) df.loc['Sum'] = df.sum(numeric_only=True) df.to_csv('COMPSRA_MERGE_0_snoRNA_.txt', sep='\t') df = pd.read_csv('COMPSRA_MERGE_0_snRNA.txt', sep='\t', index_col=0) df = df.drop(columns=['Unnamed: 9']) df.loc['Sum'] = df.sum(numeric_only=True) df.to_csv('COMPSRA_MERGE_0_snRNA_.txt', sep='\t') df = pd.read_csv('COMPSRA_MERGE_0_tRNA.txt', sep='\t', index_col=0) df = df.drop(columns=['Unnamed: 9']) df.loc['Sum'] = df.sum(numeric_only=True) df.to_csv('COMPSRA_MERGE_0_tRNA_.txt', sep='\t') #samtools flagstat **.bam #47217410 + 0 in total (QC-passed reads + QC-failed reads) #45166321 + 0 mapped (95.66% : N/A) #2051089 + 0 in total (QC-passed reads + QC-failed reads) #TODO: check the microRNA in the paper mentioned in which position? #Single publications on EVs as transport vehicles for specific miRNAs in the pathogenesis of Merkel cell carcinoma have also been reported, such as miR-375 and its function in proliferation ~/Tools/csv2xls-0.4/csv_to_xls.py COMPSRA_MERGE_0_miRNA_.txt \ COMPSRA_MERGE_0_piRNA_.txt \ COMPSRA_MERGE_0_tRNA_.txt \ COMPSRA_MERGE_0_snoRNA_.txt \ COMPSRA_MERGE_0_snRNA_.txt \ -d$'\t' -o raw_counts.xls; # sorting the DEG table, change the sheet labels to 'miRNA_between_columns_B-C_and_columns_D-G' ~/Tools/csv2xls-0.4/csv_to_xls.py COMPSRA_DEG_0_miRNA.txt -d$'\t' -o normalized_and_significance_test_miRNA.xls; ##merging the row counts and statical values #cut -f1-1 COMPSRA_MERGE_0_snoRNA.txt > f1_MERGE #cut -f1-1 COMPSRA_DEG_0_snoRNA.txt > f1_DEG #cut -f1-1 COMPSRA_MERGE_0_miRNA.txt > f1_MERGE #cut -f1-1 COMPSRA_DEG_0_miRNA.txt > f1_DEG #diff f1_MERGE f1_DEG -
calculate the number of total reads, total human reads and total non-human reads.
for sample in 2021_August_nf655_MKL-1_EV-miRNA 2021_August_nf657_WaGa_EV-miRNA 2022_August_nf774_0403_WaGa_wt 2022_August_nf780_0505_MKL-1_wt 2022_November_nf796_MKL-1_wt_1 2022_November_nf797_MKL-1_wt_2 2023_June_nf876_1002_WaGa_sT_Dox 2023_June_nf887_2312_MKL-1_scr_DMSO; do echo "--------------- ${sample} ---------------" samtools flagstat ./${sample}/${sample}_STAR_Aligned.out.bam samtools flagstat ./${sample}/${sample}_STAR_Aligned_UnMapped.bam done
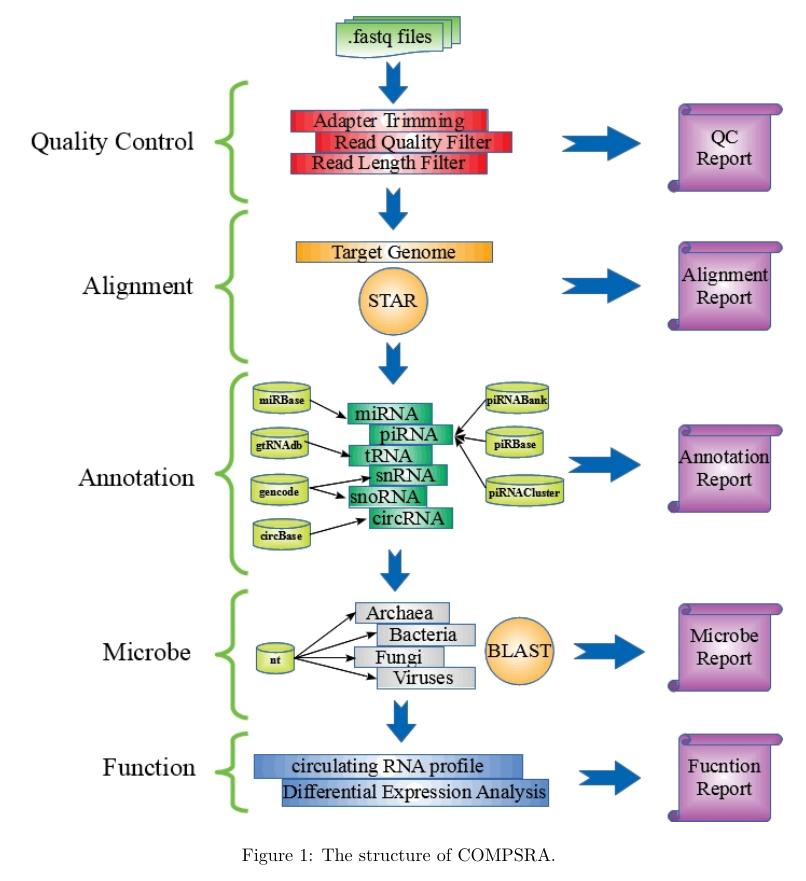
COMPSRA was composed of five functional modules: Quality Control, Alignment, Annotation, Microbe and Function. They are integrated into a pipeline and each module can also process independently.
-
Quality Control: To deal with fastq files and filter out the adapter sequences and reads with low quality.
- FASTQ files from the small RNA sequencing of biological samples are the default input.
- First, the adapter portions of a read are trimmed along with any randomized bases at ligation junctions that are produced by some small RNA-seq kits (e.g., NEXTflexT M Small RNA-Seq kit). The adapter sequences, typically situated at the 3' (3-prime) end, vary across different kits. Some commonly employed adapter sequences are listed below:
- TruSeq Small RNA (Illumina) TGGAATTCTCGGGTGCCAAGG
- Small RNA Kits V1 (Illumina) TCGTATGCCGTCTTCTGCTTGT
- Small RNA Kits V1.5 (Illumina) ATCTCGTATGCCGTCTTCTGCTTG
- NEXTflex Small RNA Sequencing Kit v3 for Illumina Platforms (Bioo Scientific) TGGAATTCTCGGGTGCCAAGG
- LEXOGEN Small RNA-Seq Library Prep Kit (Illumina) TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC
- The read quality of the remaining sequence is evaluated using its corresponding Phred score.
- Poor quality reads are removed according to quality control parameters set in the command line (-rh 20 –rt 20 –rr 20).
- Users can specify qualified reads of specific length intervals for input into subse-quent modules.
-
Alignment:
- To align the clean reads to the reference genome. COMPSRA uses STAR as its default RNA sequence aligner with default parameters which are customizable on the command line.
- Qualified reads from the QC module output are first mapped to the human genome hg19/hg38, and then aligned reads are quantified and annotated in the Annotation Module.
- Reads that could not be mapped to the human genome are saved into a FASTA file for input into the Microbe Module.
-
Annotation:
- To annotate different kinds of circulating RNAs based on the alignment result.
- COMPSRA currently uses several different small RNA databases for annotating human genome mapped reads and provides all the possible annotations:
- miRBase (Kozomara and Griffiths-Jones, 2011) for miRNA;
- piRNABank (Sai Lakshmi and Agrawal, 2008);
- piRBase (Zhang, et al., 2014) and piRNACluster (Rosenkranz, 2016) for piRNA;
- gtRNAdb (Chan and Lowe, 2016) for tRNA;
- GENCODE release 27 (Harrow, et al., 2012) for snRNA and snoRNA;
- circBase (Glazar, et al., 2014) for circular RNA.
- To conform the different reference human genome versions in these databases, we use an automatic LiftOver created by the UCSC Genome Browser Group.
- All the databases used are already pre-built, enabling speedy annotation.
-
Microbe:
- To predict the possible species of microbes existed in the samples.
- The qualified reads that could not be mapped to the human genome in the Alignment Module are aligned to the nucleotide (nt) database (Coordinators, 2013) from UCSC using BLAST.
- The four major microbial taxons archaea, bacteria, fungi and viruses are supported.
-
Function:
- To perform differential expression analysis and other functional studies to be extended.
- The read count of each RNA molecule that is identified in the Annotation Module is outputted as a tab-delimited text file according to RNA type.
- With more than one sample FASTQ file inputs, the output are further aggregated into a data matrix of RNA molecules as rows and samples as columns showing the read counts of an RNA molecule across different samples.
- The user can mark each sample FASTQ file column as either a case or a control in the command line, and perform a case versus control differential expression analysis for each RNA molecule using the Mann-Whitney rank sum test (Wilcoxon Rank Sum Test) as the default statistical test.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Setup conda environments
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- MicrobiotaProcess_PCA_Group3-4.R processing Data_Karoline_16S_2025
- Phyloseq.Rmd processing Data_Karoline_16S_2025
- Processing RNAseq_2025_WT_vs_ΔIJ_on_ATCC19606
- Stihl Elektro-Heckenschneider
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Sorry, 没有相似文章