Calling peaks using findPeaks of HOMER
gene_x 0 like s 2153 view s
Tags: Motif Discovery, NGS, ChIP-seq
https://fonseca.lab.mcgill.ca/resources/20220810_ATAC_Analysis_Demo/guide.html#peaks
- Introduction
- Accessing the data
- Peak calling
- Annotating peak files
- Filtering the data
- Visualizing the data
- Gene set enrichment analysis
- Principal component analysis
- Motif analysis
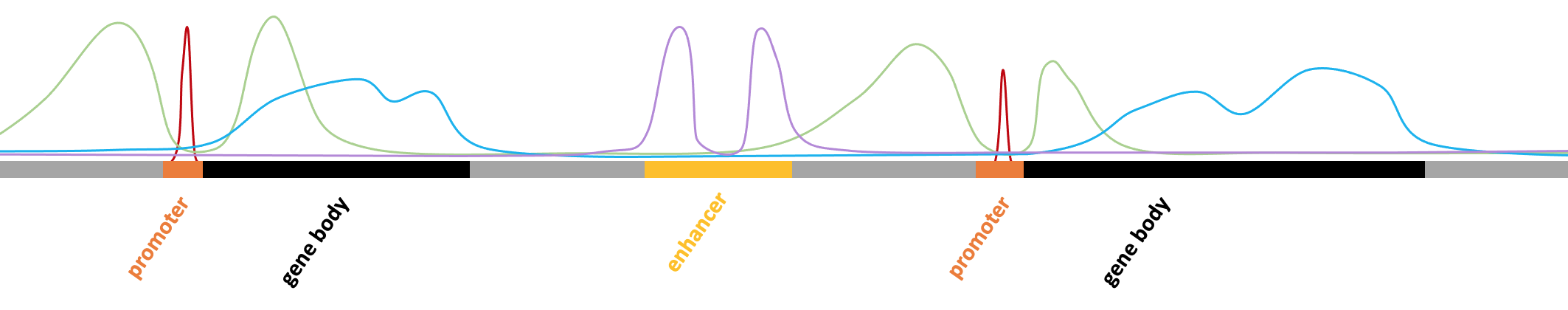
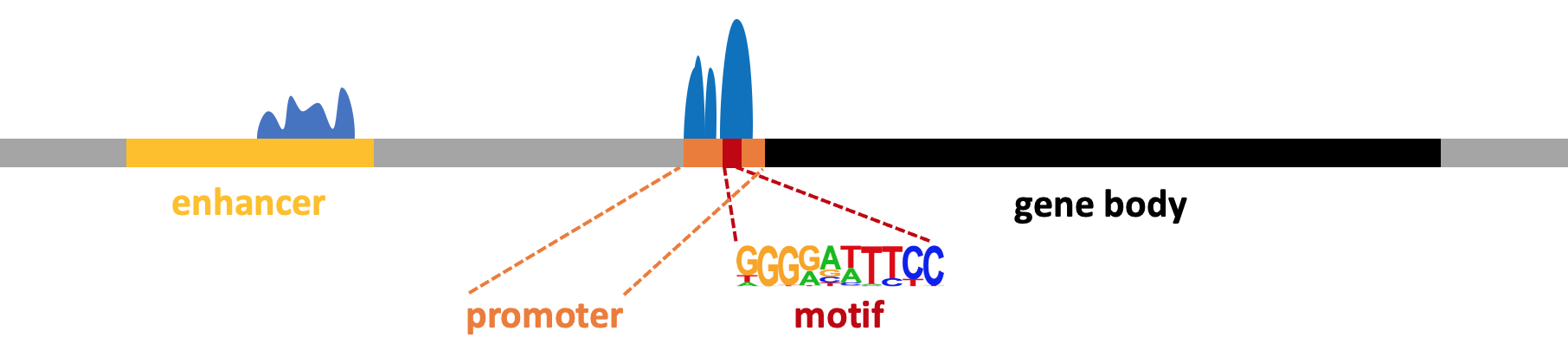
H3K4me3 is associated with active promoters and is commonly found in narrow peaks. Thus, you might consider using the "-style factor" parameter when using Homer's findPeaks command, which is designed for transcription factors and histone modifications with narrow peaks.
Here's a basic usage of findPeaks for H3K4me3:
findPeaks <tag directory> -style factor -o auto -i <input tag directory>
#makeTagDirectory <alignment file>
conda activate myperl
#makeTagDirectory <output directory> <input file> -genome hg38
for sample in p783_ChIP_DonorI p783_ChIP_DonorII p783_input_DonorI p783_input_DonorII; do
makeTagDirectory ${sample} ../picard/${sample}.dedup.sorted.bam -genome hg38
done
The "-style factor" option sets several parameters automatically (like the size of the sliding window, the required local fold enrichment over the background, and others) that are suitable for factor-style peaks.
Remember that peak calling can be somewhat subjective and might require additional parameter tuning and downstream filtering depending on the specific characteristics of your dataset, your experiment, and your research question.
In some cases, you may want to adjust parameters such as the -size (to specify the size of the region used for peak finding), -minDist (to specify the minimum distance between peaks), or others. However, for most typical use cases, the default parameters set by the "-style factor" option should work well for H3K4me3.
H3K27me3 (trimethylation of the 27th lysine residue on the H3 protein) is a histone modification associated with transcriptional repression, and it is often found in broad domains rather than narrow peaks. Thus, you might want to use the "-style histone" parameter when using Homer's findPeaks command, which is designed for histone modifications with broad peaks.
Here's a basic usage of findPeaks for H3K27me3:
findPeaks <tag directory> -style histone -o auto -i <input tag directory>
#<tag directory> should be replaced with the name of your tag directory.
The "-style histone" option sets several parameters automatically that are suitable for histone-style peaks, including larger window sizes and a more stringent fold-enrichment requirement over local background.
Remember that peak calling can be somewhat subjective and might require additional parameter tuning and downstream filtering depending on the specific characteristics of your dataset, your experiment, and your research question.
In some cases, you may want to adjust parameters such as the -size (to specify the size of the region used for peak finding), -minDist (to specify the minimum distance between peaks), or others. However, for most typical use cases, the default parameters set by the "-style histone" option should work well for H3K27me3.
H3K27ac (acetylation of the 27th lysine residue on the H3 protein) is a histone modification typically associated with active promoters and enhancers. Unlike H3K27me3, which tends to occur in broader regions, H3K27ac often forms relatively sharp peaks similar to those of transcription factor binding sites.
Here's a basic usage of findPeaks for H3K27ac:
findPeaks <tag directory> -style histone -o auto -i <input tag directory>
#<tag directory> should be replaced with the name of your tag directory.
The "-style histone" option is designed to work well with many histone modifications, including H3K27ac. This setting automatically adjusts several parameters to values that are typically appropriate for histone modifications.
However, peak calling is not an exact science and can be somewhat subjective. Depending on the specific characteristics of your dataset and your research question, you might need to adjust the parameters. Some parameters that you may want to experiment with include -size (to specify the size of the region used for peak finding), -minDist (to specify the minimum distance between peaks), or -region (to call broader enriched regions rather than sharp peaks).
In some cases, using "-style factor" might be more appropriate for H3K27ac data, given the sharpness of the peaks. You can try both and see which set of peaks makes more sense in your specific experimental context. It's also always a good idea to visualize some of the called peaks in a genome browser to ensure that the peaks look reasonable.
H3K4me1 (monomethylation of the fourth lysine residue on the H3 protein) is a histone modification usually associated with enhancer regions. The peaks of H3K4me1 often span a broad region.
Here's a basic usage of findPeaks for H3K4me1:
findPeaks <tag directory> -style histone -o auto -i <input tag directory>
#<tag directory> should be replaced with the name of your tag directory.
The -style histone option is designed to work well with many histone modifications, including H3K4me1. This setting automatically adjusts several parameters to values that are typically appropriate for histone modifications.
However, peak calling is not an exact science and the "best" parameters can depend on various factors including the specifics of your experiment and your research question. You might need to adjust parameters based on your specific needs. Some parameters that you may want to adjust include -size (to specify the size of the region used for peak finding), -minDist (to specify the minimum distance between peaks), or -region (to call broader enriched regions rather than sharp peaks).
In the case of H3K4me1, the peaks are generally broader than other histone modifications such as H3K4me3. Therefore, you might want to consider increasing the -size parameter or use the -region option to better capture these broad regions of enrichment.
As always, after peak calling, you should visualize some of the called peaks in a genome browser to ensure that they look reasonable and align well with the experimental data.
H3K9me3 is a histone modification often associated with transcriptional repression and heterochromatin, and it tends to form large domains rather than sharp peaks.
Here's a basic usage of findPeaks for H3K9me3:
findPeaks <tag directory> -style histone -o auto -i <input tag directory>
#<tag directory> should be replaced with the name of your tag directory.
The -style histone option is designed to work well with many histone modifications. This setting automatically adjusts several parameters to values that are typically appropriate for histone modifications.
However, for modifications like H3K9me3 that often cover broad domains, you might want to use the -region option to call enriched regions rather than individual peaks. This will capture the broader domains characteristic of H3K9me3:
findPeaks <tag directory> -style histone -region -o auto -i <input tag directory>
Peak calling is not an exact science and the "best" parameters can depend on various factors including the specifics of your experiment and your research question. Therefore, you may need to adjust parameters based on your specific needs.
Finally, it's always good to check the output of your peak calling by visualizing some of the called peaks in a genome browser. This will ensure they align well with the experimental data and look like what you would expect given what is known about the biology of the histone mark.
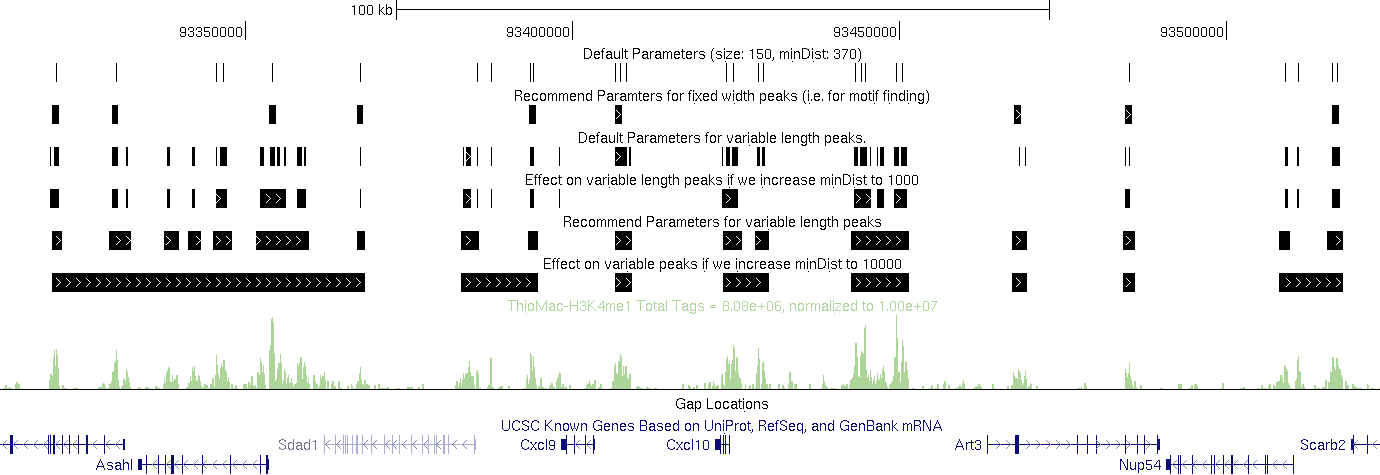
#Default Parameters:
findPeaks Macrophage-H3K4me1/ -i Macrophage-IgG -size 150 -minDist 370 > output.txt (i.e. defaults)
#Recommend Parameters for fixed width peaks (i.e. for motif finding):
findPeaks Macrophage-H3K4me1/ -i Macrophage-IgG -size 1000 -minDist 2500 > output.txt
#Default Parameters for variable length peaks.
findPeaks Macrophage-H3K4me1/ -i Macrophage-IgG -region -size 150 -minDist 370 > output.txt
#Effect on variable length peaks if we increase minDist to 1000.
findPeaks Macrophage-H3K4me1/ -i Macrophage-IgG -region -size 150 -minDist 1000 > output.txt
#Recommend Parameters for variable length peaks (H3K4me1 at least).
findPeaks Macrophage-H3K4me1/ -i Macrophage-IgG -region -size 1000 -minDist 2500 > output.txt
#Effect on variable peaks if we increase minDist to 10000 (H3K4me1 at least).
findPeaks Macrophage-H3K4me1/ -i Macrophage-IgG -region -size 1000 -minDist 10000 > output.txt
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Calling peaks using findPeaks of HOMER
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Should the inputs for GSVA be normalized or raw?
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- Kraken2 Installation and Usage Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
- 基因组与表型特性研究:一株药物敏感的鲍曼不动杆菌显示出增强的毒力相关特性和应激耐受性
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
How to use H3K27me3 and H3K4me3 to identify transcription factors?