Draw Venn Diagrams using matplotlib
gene_x 0 like s 840 view s
Tags: plot, python
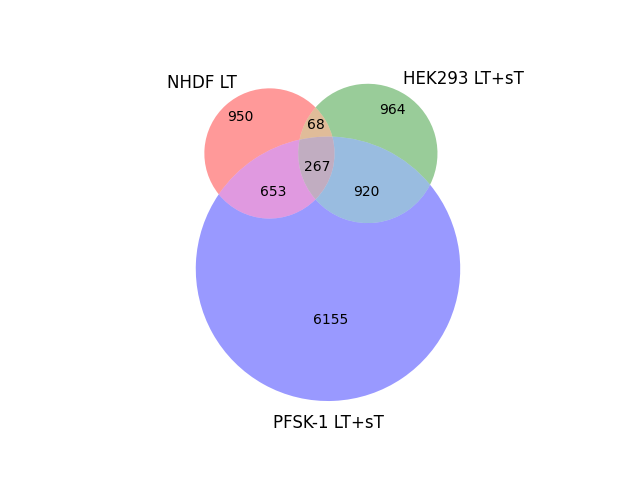
#peaks_PFSK-1.txt peaks_HEK293.txt peaks_NHDF.txt Total Name
# X 950 peaks_NHDF.txt
# X 964 peaks_HEK293.txt
# X X 68 peaks_HEK293.txt|peaks_NHDF.txt
#X 6155 peaks_PFSK-1.txt
#X X 653 peaks_PFSK-1.txt|peaks_NHDF.txt
#X X 920 peaks_PFSK-1.txt|peaks_HEK293.txt
#X X X 267 peaks_PFSK-1.txt|peaks_HEK293.txt|peaks_NHDF.txt
#-------
import matplotlib.pyplot as plt
from matplotlib_venn import venn3
# Define the sizes of the sets
set1_only = 950 # Size of NHDF
set2_only = 964 # Size of HEK293
set3_only = 6155 # Size of PFSK-1
# Define the sizes of the overlapping regions
shared_elements_12 = 68 # Size of the overlap between set 1 and set 2
shared_elements_13= 653 # Size of the overlap between set 1 and set 3
shared_elements_23 = 920 # Size of the overlap between set 2 and set 3
shared_elements_123 = 267 # Size of the overlap between all three sets
## Define the sizes of the datasets
#set1_size = 100
#set2_size = 80
#set3_size = 60
#shared_elements_12 = 30
#shared_elements_13 = 20
#shared_elements_23 = 15
#shared_elements_123 = 10
## Calculate the sizes of the individual and overlapping sets
#set1_only = set1_size - shared_elements_12 - shared_elements_13 - shared_elements_123
#set2_only = set2_size - shared_elements_12 - shared_elements_23 - shared_elements_123
#set3_only = set3_size - shared_elements_13 - shared_elements_23 - shared_elements_123
# Create the Venn diagram
venn3(subsets=(set1_only, set2_only, shared_elements_12, set3_only, shared_elements_13, shared_elements_23, shared_elements_123), set_labels=('NHDF LT', 'HEK293 LT+sT', 'PFSK-1 LT+sT'))
#venn3(subsets=(set1_size, set2_size, set1_set2_size, set3_size, set1_set3_size, set2_set3_size, set1_set2_set3_size), set_labels=('NHDF', 'HEK293', 'PFSK-1'))
# Set the title and labels
plt.title('', fontsize=16) #Venn Diagram of Three Datasets
plt.xlabel('Samples')
plt.ylabel('Count')
# Show the Venn diagram
#plt.show()
plt.savefig('Venn_Diagram_NHDF_vs_HEK293_vs_PFSK-1.png', dpi=300, bbox_inches='tight')
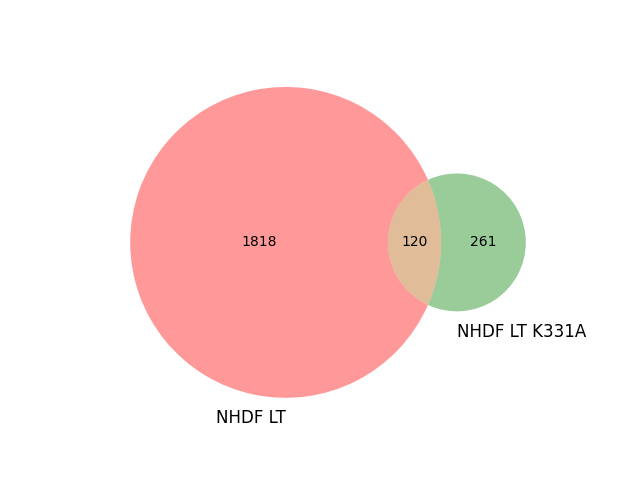
#peaks_K331A.txt peaks_NHDF.txt Total Name
# X 1818 peaks_NHDF.txt
#X 261 peaks_K331A.txt
#X X 120 peaks_K331A.txt|peaks_NHDF.txt
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
# Define the sizes of the datasets
set1_size = 1938
set2_size = 381
shared_elements = 120
# Calculate the sizes of the individual and overlapping sets
set1_only = set1_size - shared_elements
set2_only = set2_size - shared_elements
# Create the Venn diagram
venn2(subsets=(set1_only, set2_only, shared_elements), set_labels=('NHDF LT', 'NHDF LT K331A'))
# Set the title and labels
plt.title('') #NHDF LT vs NHDF K331A
plt.xlabel('Number of Elements')
plt.ylabel('')
# Display the Venn diagram
#plt.show()
plt.savefig('Venn_Diagram_NHDF-LT_vs_NHDF-LT-K331A.png', dpi=300, bbox_inches='tight')
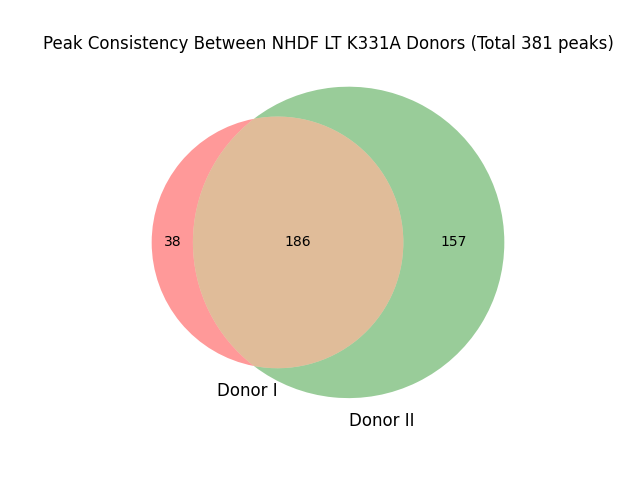
## under directory "results_ChIPseq_NHDF_LT_K331A_PFSK-1_HEK293_hg38/homer"
#grep "NHDF_K331A_DonorI/peaks.txt" peaks_K331A_LT.txt | wc -l # 224-186=38
#grep "NHDF_K331A_DonorII/peaks.txt" peaks_K331A_LT.txt | wc -l # 343-186=157
#grep "NHDF_K331A_DonorI/peaks.txt|NHDF_K331A_DonorII/peaks.txt" peaks_K331A_LT.txt | wc -l #186
##38+157+186=381
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
venn2(subsets=(38, 157, 186), set_labels=('Donor I', 'Donor II'))
plt.title('Peak Consistency Between NHDF LT K331A Donors (Total 381 peaks)')
plt.xlabel('Number of Elements')
plt.ylabel('')
plt.savefig('Venn_Diagram_Peak_Consistency_Between_NHDF-LT-K331A_Donors.png', dpi=300, bbox_inches='tight')
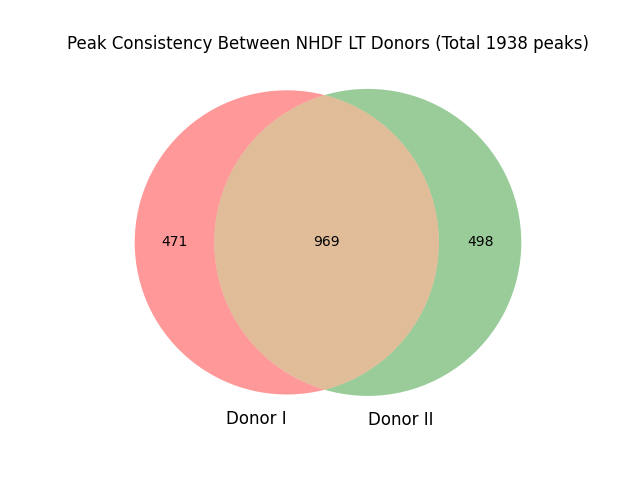
#grep "NHDF_LT_DonorI/peaks.txt" peaks_NHDF_LT.txt | wc -l # 1440-969=471
#grep "NHDF_LT_DonorII/peaks.txt" peaks_NHDF_LT.txt | wc -l # 1467-969=498
#grep "NHDF_LT_DonorI/peaks.txt|NHDF_LT_DonorII/peaks.txt" peaks_NHDF_LT.txt | wc -l #969
##471+498+969=1938
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
venn2(subsets=(471, 498, 969), set_labels=('Donor I', 'Donor II'))
plt.title('Peak Consistency Between NHDF LT Donors (Total 1938 peaks)')
plt.xlabel('Number of Elements')
plt.ylabel('')
plt.savefig('Venn_Diagram_Peak_Consistency_Between_NHDF-LT_Donors.png', dpi=300, bbox_inches='tight')
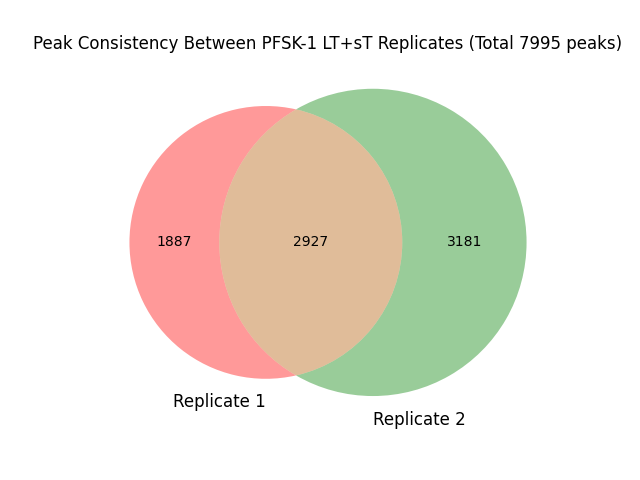
#grep "PFSK-1B_LT+sT_r1/peaks.txt" peaks_PFSK-1_LT+sT.txt | wc -l # 4814-2927=1887
#grep "PFSK-1B_LT+sT_r2/peaks.txt" peaks_PFSK-1_LT+sT.txt | wc -l # 6108-2927=3181
#grep "PFSK-1B_LT+sT_r1/peaks.txt|PFSK-1B_LT+sT_r2/peaks.txt" peaks_PFSK-1_LT+sT.txt | wc -l #2927
##1887+3181+2927=7995
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
venn2(subsets=(1887, 3181, 2927), set_labels=('Replicate 1', 'Replicate 2'))
plt.title('Peak Consistency Between PFSK-1 LT+sT Replicates (Total 7995 peaks)')
plt.xlabel('Number of Elements')
plt.ylabel('')
plt.savefig('Venn_Diagram_Peak_Consistency_Between_PFSK-1-LT+sT_Replicates.png', dpi=300, bbox_inches='tight')
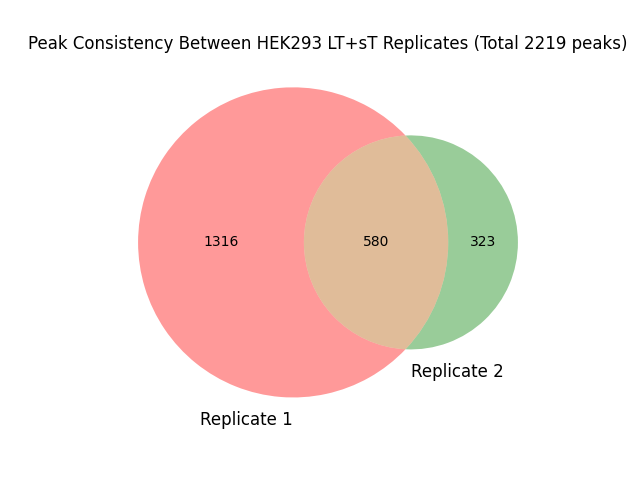
#grep "HEK293_LT+sT_r2/peaks.txt" peaks_HEK293_LT+sT.txt | wc -l # 1896-580=1316
#grep "HEK293_LT+sT_r3/peaks.txt" peaks_HEK293_LT+sT.txt | wc -l # 903-580=323
#grep "HEK293_LT+sT_r2/peaks.txt|HEK293_LT+sT_r3/peaks.txt" peaks_HEK293_LT+sT.txt | wc -l #580
##1316+323+580=2219
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
venn2(subsets=(1316, 323, 580), set_labels=('Replicate 1', 'Replicate 2'))
plt.title('Peak Consistency Between HEK293 LT+sT Replicates (Total 2219 peaks)')
plt.xlabel('Number of Elements')
plt.ylabel('')
plt.savefig('Venn_Diagram_Peak_Consistency_Between_HEK293-LT+sT_Replicates.png', dpi=300, bbox_inches='tight')
# Draw Venn Diagram using Set rather than Set Size
##38+186+157=381
#import matplotlib.pyplot as plt
#from matplotlib_venn import venn2
#
#set1 = set(range(1, 225))
#set2 = set(range(39, 382))
#
#venn2([set1, set2], set_labels=('Replicate 1', 'Replicate 2'), set_colors=('skyblue', 'lightgreen'))
#plt.title('Peak Consistency Between NHDF K331A Replicates (total 381 peaks)', fontsize=16)
#plt.xticks(fontsize=12)
#plt.yticks(fontsize=12)
#
##plt.show() # Display the Venn diagram
#plt.savefig('Venn_Diagram_Peak_Consistency_Between_NHDF_K331A_Replicates.png') # Save the Venn diagram as an image file
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Should the inputs for GSVA be normalized or raw?
- Setup conda environments
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Kraken2 Installation and Usage Guide
- File format for single channel analysis of Agilent microarray data with Limma?
最新文章
- Viral genome assembly and recombination analysis for Data_Sophie_HDV_Sequences
- 阳光房漏水怎么办?丁基胶带才是最佳密封选择
- DAMIAN Post-processing for Flavivirus and FSME
- All tools and services of BV-BRC
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
YopQ Secretion Boxplot and Fitting Function
RNA-seq skin organoids on GRCh38+chrHsv1 (final)
Genomic Organization of Herpes Simplex Virus type 1 (HSV-1 s17)